Abstract
A method for site-specific, nitrobenzyl-induced photochemical proteolysis of diverse proteins expressed in living cells has been developed based on the chemistry of the unnatural amino acid (2-nitrophenyl)glycine (Npg). Using the in vivo nonsense codon suppression method for incorporating unnatural amino acids into proteins expressed in Xenopus oocytes, Npg has been incorporated into two ion channels: the Drosophila Shaker B K+ channel and the nicotinic acetylcholine receptor. Functional studies in vivo show that irradiation of proteins containing an Npg residue does lead to peptide backbone cleavage at the site of the novel residue. Using this method, evidence is obtained for an essential functional role of the “signature” Cys128–Cys142 disulfide loop of the nAChR α subunit.
Keywords: [nicotinic acetylcholine receptor; 2-nitrophenylglycine; K+ channel; signature disulfide loop; site-specific, nitrobenzyl-induced photochemical proteolysis]
A powerful tool for probing protein structure and function is the incorporation of unnatural amino acids by the method of nonsense suppression, a technique developed for in vitro translation systems by Schultz and coworkers (1–3). Recent work has expanded the scope of this methodology by establishing procedures for unnatural amino acid incorporation into proteins expressed in living cells (4–7). This allows the evaluation of neuroreceptors, ion channels, and related proteins that are studied most effectively in vivo using heterologous expression systems such as the Xenopus oocyte. The suppression methodology is especially promising for such integral membrane proteins, which are not yet generally amenable to the methods of high resolution structure determination (e.g., NMR, x-ray crystallography).
In this paper we (i) describe the unnatural amino acid 2-(nitrophenyl)glycine (Npg) and show that it can be efficiently incorporated into proteins expressed in vivo; (ii) establish that in vivo irradiation of an Npg-containing protein leads to site-specific, nitrobenzyl-induced photochemical proteolysis (SNIPP) (Fig. 1); and (iii) illustrate the usefulness of this residue by cleavage of the Cys128–Cys142 “signature” disulfide loop of the nicotinic acetylcholine receptor, establishing a crucial functional role for this structural feature.
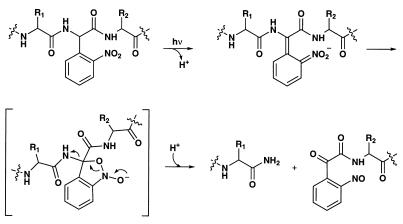
Figure 1.
Schematic of the photochemical cleavage of a protein backbone using the Npg residue.
The design of the unnatural amino acid Npg was based on the photochemistry of 2-nitrobenzyl derivatives. Compounds of this type, including Npg itself, have been used as protecting groups in organic synthesis and to produce “caged” neurotransmitters, ions, and second messengers that can then be liberated photochemically, but, to our knowledge, Npg has not been incorporated into peptides or proteins (8–17). As shown in Fig. 1, incorporation of Npg into a protein, followed by irradiation of the protein, was anticipated to produce peptide backbone cleavage (18) by analogy to other 2-nitrobenzyl systems (For an example of photochemically initiated protein self-splicing reaction, see ref. 18.)
METHODS
Synthesis of Npg–tRNA.
d,l-(2-Nitrophenylglycine)Hydrochloride. The unnatural amino acid d, l-(2-nitrophenylglycine) hydrochloride was prepared according to published procedures (19–20).
N-4PO-d,l-(2-Nitrophenyl)Glycine.
The amine of N-4PO-d,l-(2-nitrophenyl)glycine was protected as the N-pent-4-enoyl (4PO) derivative using standard conditions (21, 22). Na2CO3 (111 mg, 1.05 mmol) was added to a room temperature solution of (2-nitrophenyl)glycine hydrochloride (82 mg, 0.35 mmol) in H2O:dioxane (0.75 ml:0.5 ml), followed by a solution of pent-4-enoic anhydride (70.8 mg, 0.39 mmol) in dioxane (0.25 ml). After 3 h, the mixture was poured into saturated NaHSO4 and extracted with CH2Cl2. The organic phase was dried over anhydrous Na2SO4 and concentrated in vacuo. The residual oil was purified by flash silica gel column chromatography to yield the title compound (73.2 mg, 75.2%) as a white solid. 1H NMR (300 MHz, CD3OD) δ 8.06 (dd, J = 1.2, 8.1 Hz, 1H), 7.70 (ddd, J = 1.2, 7.5, 7.5 Hz, 1H), 7.62–7.53 (m, 2H), 6.21 (s, 1H), 5.80 (m, 1H), 5.04–4.97 (m, 2H), and 2.42–2.28 (m, 4H). High resolution mass spectometry calculated for C13H14N2O5 279.0981 found 279.0992.
N-4PO-d,l-(2-Nitrophenyl)Glycinate Cyanomethyl Ester.
N-4PO-d,l-(2-nitrophenyl)glycine was activated as the cyanomethyl ester using standard conditions (5, 23). NEt3 (95 μl, 0.68 mmol) was added to a room temperature solution of the acid (63.2 mg, 0.23 mmol) in anhydrous DMF (1 ml), followed by addition of ClCH2CN (1 ml). After 16 h, the mixture was diluted with Et2O and extracted against H2O. The organic phase was washed with saturated NaCl, dried over anhydrous Na2SO4, and concentrated in vacuo. The residual oil was purified by flash silica gel column chromatography to yield the title compound (62.6 mg, 85.8%) as a yellow solid. 1H NMR (300 MHz, CDCl3) δ 8.18 (dd, J = 1.2, 8.1 Hz, 1H), 7.74–7.65 (m, 2H), 7.58 (ddd, J = 1.8, 7.2, 8.4 Hz, 1H), 6.84 (d, J = 7.8 Hz, 1H), 6.17 (d, J = 6.2 Hz, 1H), 5.76 (m, 1H), 5.00 (dd, J = 1.5, 15.6 Hz, 1H), 4.96 (dd, J = 1.5, 9.9 Hz, 1H), 4.79 (d, J = 15.6 Hz, 1H), 4.72 (d, J = 15.6 Hz, 1H), and 2.45–2.25 (m, 4H). High resolution mass spectometry calculated for C16H17N3O5 317.1012 found 317.1004.
N-4PO-(2-Nitrophenyl)Glycine-dCA.
The dinucleotide dCA N-4PO-(2-nitrophenyl)glycine-dCA was prepared as reported by Schultz (24) with the modifications described by Kearney (5). The cyanomethyl ester was coupled to dCA according to a standard protocol (5, 23). N-4PO-d,l-(2-nitrophenyl)glycinate cyanomethyl ester (16.3 mg, 51.4 μmol) was added to a room temperature solution of dCA (tetrabutylammonium salt, 20 mg, 16.6 μmol) in anhydrous DMF (400 μl) under argon. The resulting deep red solution was stirred for 1 h and then quenched with 25 mM NH4OAc (20 μl, pH 4.5). The crude product was purified by reverse-phase semi-preparative HPLC (Whatman Partisil 10 ODS-3 column, 9.4 mm × 50 cm) using a gradient from 25 mM NH4OAc (pH 4.5) to CH3CN. The appropriate fractions were combined and lyophilized. The resulting solid was redissolved in 10 mM HOAc/CH3CN and lyophilized to afford 4PO–Npg–dCA (3.9 mg, 8.8%) as a pale yellow solid: electrospray ionization-MS: M− 896 (31) [M-H]− 895 (100); calculated for C32H36N10O17P2 896. The material was quantified by UV absorption (ɛ260 ≈ 37,000).
N-4PO-(2-Nitrophenyl)Glycine-tRNA.
The tRNA gene used was Tetrahymena thermophila tRNAGlnCUA having a G at position 73 in the plasmid pTHG73 (6). This gene contains an upstream T7 RNA polymerase promotor and a downstream Fok I restriction site. Digestion of pTHG73 with Fok I provided the linearized template DNA lacking the 3′-terminal CA at positions 75 and 76. In vitro transcription of the linearized transcript and purification of the truncated THG73 tRNA product was performed as described (6). Ligation of 4PO–Npg-dCA to the THG73 runoff transcript was accomplished using T4 RNA ligase (New England Biolabs) as described (6).
(2-Nitrophenyl)Glycine-tRNA.
Deprotection of 4PO–Npg–tRNA was performed just before injection according to the method described by Fraser–Reid (21). A saturated solution of iodine (0.50 μl, 1.2 mM) was added to a room temperature solution of 4PO–Npg–tRNA (0.5 μl). After 10 min, the resulting Npg–tRNA was immediately mixed with the desired mRNA (1.0 μl).
Mutagenesis and mRNA Synthesis.
PCR mutagenesis was used to generate cassettes containing the Shaker B (ShB)-Leu47TAG, ShB-Pro64TAG, and α-Val132TAG amber mutations. Cassettes were trimmed with appropriate restriction enzymes, purified, and ligated into the parent construct (ShB/pAMV-PA or α/pAMV-PA) that had been previously digested with the same restriction enzymes and dephosphorylated. Mutations were verified by automated sequencing over the entire amplified region and the ligation sites. The βLeu9′TAG amber mutation was prepared using the CLONTECH Transformer kit and was transferred into pAMV-PA. The pAMV-PA vector is a modified pBS (S/K+) vector containing an alfalfa mosaic virus region directly upstream from the coding region of the insert and an A50 sequence downstream from the insert. Plasmid DNAs were linearized with NotI, and mRNA was transcribed using the Ambion (Austin, TX) T7 mMESSAGE mMACHINE machine Kit. mRNAs were dissolved in sterile water and quantitated by UV absorption (A260). Restriction enzymes were purchased from Boehringer, and Pfu polymerase was purchased from Stratagene.
Oocyte Injections.
Oocytes were removed from Xenopus laevis as described (25) and maintained at 18°C in ND96 solution (96 mM NaCl/2 mM KCl/1.8 mM CaCl2/1 mM MgCl2/5 mM Hepes/2.5 mM sodium pyruvate/0.5 mM theophylline/50 μg/ml gentamycin, pH 7.5, with NaOH). Just before microinjection, the 4PO–Npg–tRNA was deprotected (as described above) and combined with the appropriate mRNA. Oocytes were microinjected (50 nl) with the following mRNA and tRNA concentrations: ShB-Pro64TAG (0.4 ng/nl) or ShB-Leu47TAG (0.4 ng/nl) and Npg–tRNA (1.0 ng/nl); ShB-wild type [1 pg/nl; Fig. 2B (all concentrations are divided by two for Fig. 2C)]; nAChR-β9′TAG (0.017 ng/nl total mRNA; α:β:γ:δ ratio, 2:1:1:1); Npg–tRNA (1.0 ng/nl); wild-type nAChR (1.6 pg/nl); nAChR-αVal132TAG (0.094 ng/nl total mRNA; α:β:γ:δ ratio, 12:1:1:1); and Npg–tRNA (1.0 ng/nl). For the nAChR-αVal132TAG experiments, oocytes were injected three times (at 0, 24, and 48 h) before recording and [125I]-α-bungarotoxin binding (at 72 h). Oocytes were assayed ≈24 h after injection (ShB-Pro64TAG, ShB-Leu47TAG, ShB-wild type, nAChR-β9′TAG, wild-type nAChR) or ≈72 h after injection (nAChR-αVal132TAG).
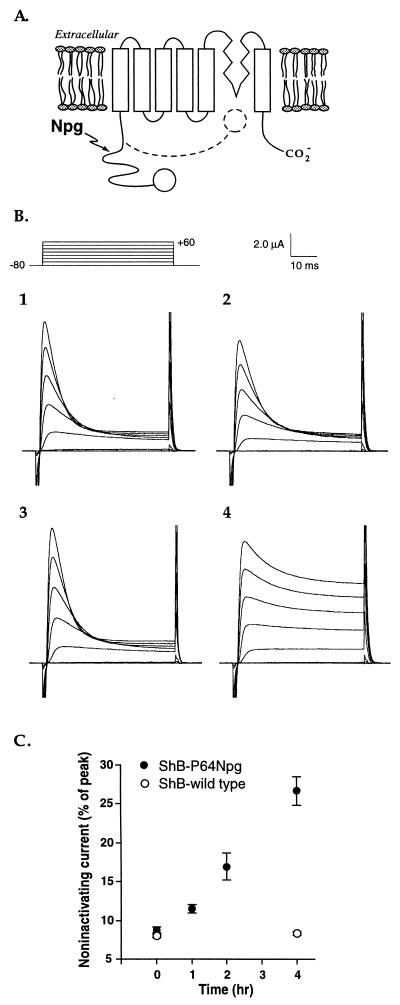
Figure 2.
(A) Schematic of the ShB channel, with the ball indicated as an open circle. The inactivation process involves movement of the ball and chain as suggested by the dashed line. The general region of Npg incorporation is also noted. (B) Effect of irradiation on ShB inactivation. Voltage–clamp currents for oocytes expressing ShB wild-type and mutant channels during depolarizing jumps from a holding potential of −80 mV to various test potentials between −80 and +60 mV. Leakage currents have been subtracted with a P/4 procedure. (1) Wild-type channels before irradiation; (2) Wild-type channels after irradiation (4 h); (3) Pro64Npg mutant channel before irradiation; (4) Pro64Npg mutant channels after irradiation (4 h). Note that each panel corresponds to a different oocyte, so absolute current values cannot be directly compared. (C) Photochemically induced increase in noninactivating current for Pro64Npg mutant and wild-type channels vs. irradiation time. Fractional noninactivating current values were measured at +60 mV by dividing the steady-state current (45 msec) by the peak current. More oocytes were irradiated for the experiment of C than for B. Therefore, the UV lamp was defocused to provide a uniform light intensity over a larger area. As such, light intensity for C was severalfold reduced relative to B.
Oocyte Irradiation and Electrophysiology.
Oocytes were irradiated at 4°C in 1/2-dram Pyrex vials with a 288-W Hg lamp (BLAK-RAY Longwave Ultraviolet Lamp, Ultraviolet Products, San Gabriel, CA) equipped with a 360-nm band pass filter at a distance of 15–30 cm for 4 h unless otherwise indicated. Nonirradiated oocytes were maintained at 4°C for the corresponding period of time. Whole-cell currents from oocytes were measured using a Geneclamp 500 amplifier (Axon Instruments, Foster City, CA) in the 2-electrode voltage–clamp configuration. Current and voltage electrodes were filled with 3 M KCl to yield resistances ranging from 0.5 to 1.5 mΩ. Holding potential was −80 mV. Oocytes were continuously perfused with a nominally calcium-free bath solution consisting of 96 mM NaCl, 2 mM KCl, 1 mM MgCl2 and 5 mM Hepes (pH 7.5). Atropine (1 μM) was added to the bath solution for the nAChR recordings. ShB peak and steady-state current values were obtained using clampfit from pclamp 6.0 (Axon Instruments). Macroscopic acetylcholine-induced currents were recorded in response to bath application of the indicated agonist concentration at −80 mV. All numerical and plotted data are from measurements obtained from several oocytes (three to eight for K+ channel studies and at least eight for nAChR studies) and are reported as mean ± SEM.
[125I]-α-Bungarotoxin Binding.
[125I]-α-bungarotoxin (10 nM) binding was carried out in PBS (1 mM CaCl2) for 1.5 h at room temperature.
RESULTS AND DISCUSSION
To establish the viability of SNIPP, Npg was incorporated into the Drosophila ShB K+ ion channel (26–31). The ShB channel is formed from four identical subunits, each of which contains six transmembrane domains and the reentrant “P” region (Fig. 2). A hallmark of this voltage-gated channel is rapid (N-type) inactivation, which terminates openings induced by changes in the transmembrane potential. Aldrich and coworkers established the molecular transitions that underlie N-type inactivation as involving a “ball-and-chain” mechanism (Fig. 2) (32–35). The first 20 amino acids in the NH2 terminus form a structural domain that interacts with part of the open channel to cause inactivation on a millisecond time scale. This structural domain, or “ball” region, is connected to the rest of the protein by a “chain” sequence of 60 or more amino acids that tethers the inactivation ball near the channel pore. Deletion of a section of this region (Δ6–46) produces a well characterized channel termed “Shaker-IR” (IR = inactivation removed) that does not inactivate on a millisecond time scale.
With the intent of transforming ShB subunits to Shaker-IR subunits by SNIPP in an intact cell, we introduced Npg into either of two sites (Leu47 and Pro64) in the chain region of ShB (Fig. 2). Xenopus oocytes thus expressed mutant ShB K+ channels comprised of either ShB-Leu47Npg (i.e., ShB with a TAG codon at position 47, normally a Leu residue, suppressed by Npg-tRNA, which has the CUA anticodon) or ShB-Pro64Npg subunits. These mutant channels displayed activation and inactivation kinetics equivalent to those of wild-type ShB channels. Irradiation of oocytes expressing either mutant channel resulted in K+ currents with reduced levels and rates of inactivation, and both of these effects increased in the expected manner (35) with increased irradiation time (Fig. 2). Irradiation of wild-type ShB K+ channels expressed in Xenopus oocytes produced no change in either level or rate of inactivation.
Given the established photochemistry of the 2-nitrobenzyl group (8–16) and the well characterized Shaker-IR phenotype (26–35), these results provide convincing evidence that irradiation of Npg leads to peptide backbone cleavage of functional ion channels in vivo. Photolysis of the Npg-containing channels resulted in the liberation of the inactivation ball from the remainder of the channel, thereby slowing inactivation. Because each channel contains four independent balls, with only a single ball necessary to produce inactivation, the rate and extent of inactivation depend on the number of balls (35). The photolyzed channels are indistinguishable from channels produced in systems expressing an appropriate mixture of ShB and Shaker-IR, and, based on those studies, we estimated that we have removed on average two of the four balls from ShB channels after irradiation for 4 h.
Having established that Npg functions as designed using ShB, we next turned our attention toward using Npg to probe functional domains of the nicotinic acetylcholine receptor (nAChR). The nAChR is the prototypical ligand-gated ion channel, being the best studied member of a class that also includes receptors for γ-aminobutyric acid, glycine, and serotonin (36–39). Structurally, the nAChR is formed by five subunits arranged in a roughly pentagonal array. The muscle form of the receptor has four homologous subunits with α2βγδ stoichiometry. Each subunit is thought to have a long extracellular N-terminal region, followed by four transmembrane segments and an extracellular C terminus (Fig. 3). Having established that Npg can function when placed in a cytoplasmic region of an ion channel protein, we wished to determine whether it would also function in a transmembrane region. Indeed, we found that incorporation of Npg at the crucial 9′ site (40–44) (Leu262) in the second transmembrane region (M2) of the β subunit (βLeu9′Npg) gives a functional receptor.§ Irradiation reduces the ACh-induced whole-cell current by 50% (Fig. 3). As a control, oocytes expressing wild-type nAChR were subjected to the same irradiation conditions, and they exhibited no reduction in whole-cell current or change in EC50.
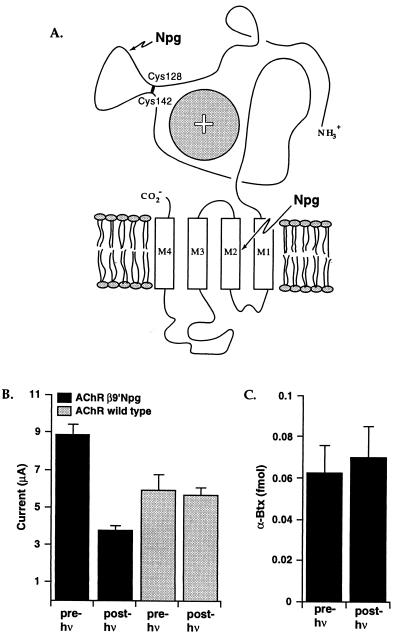
Figure 3.
(A) Schematic of a nAChR subunit, showing the extracellular N-terminal region and the four proposed transmembrane regions. The sphere with a + indicates the agonist binding site region (α subunits only) as suggested by affinity labeling and mutagenesis studies. [Adapted from a figure by Changeux (38).] Also indicated are the two sites where Npg was incorporated: V132 in the Cys128–Cys142 disulfide loop (α subunits) or the 9′ position within the second transmembrane (M2) region (β subunit). (B) Effect of irradiation on the βLeu9′Npg mutant. Oocytes were irradiated for 7.5 h. Whole-cell currents are shown for mutant and wild-type receptors. ACh concentrations were 2.5 and 50 μM for mutant and wild type, respectively, roughly equal to the EC50 values for the two (see text). When the mutant receptor is activated with 25 μM ACh, comparable diminutions of current are seen (data not shown). (C) [125I]-α-bungarotoxin (α-Btx) binding to βLeu9′Npg mutant receptor before and after irradiation, using the same oocytes from B.
Loss of ACh-induced current could result from a destruction of the agonist binding site, a disruption of the gating pathway, or some combination of the two. Alternatively, photolysis could lead to a complete disruption of the protein, causing the disassembly of the multisubunit structure. To probe this issue, we studied the binding of α-bungarotoxin and found that receptors containing the β9′ Npg mutation bind bungarotoxin well. That the β9′ mutation in M2 does not disrupt α-bungarotoxin binding agrees with present ideas that this site is buried in a transmembrane region, and the α-bungarotoxin binding site substantially overlaps the agonist binding site, which is thought to be well removed from the 9′ site (36–39). In addition, photolysis does not destroy toxin binding (Fig. 3). Surface α-bungarotoxin binding requires a properly folded, assembled receptor (45–47), so this result establishes that Npg-induced photochemical proteolysis does not lead to total destruction of the target protein but instead represents a local distortion: cleavage of the peptide backbone.
We next turned to an intriguing feature of the nAChR that seemed especially well suited to interrogation with Npg. A highly conserved feature of the ligand-gated ion channel family is the N-terminal disulfide loop, which in the muscle α subunit runs from Cys128 to Cys142 (48–52). The function of this structure has not been established. Several groups have attempted to address this issue by conventional mutagenesis, replacing one or more cysteines with serines, thereby preventing loop formation. However, this mutation leads to little or no expression of functional protein, and it is now well understood that disulfide loop formation is critical for proper subunit folding and assembly (45–52). Similarly, selective reduction of the α disulfide loop in the presence of analogous loops in the other subunits and the highly reactive and functionally crucial α C192-C193 disulfide is not feasible.
The key question is whether the signature loop has any functional role in the receptor or whether its sole function is to assist folding and assembly. The SNIPP approach allows a natural way to address this issue. As shown in Fig. 3, introducing Npg within the disulfide loop would, after photolysis, lead to a relatively local perturbation—destruction of a relatively small loop while maintaining the disulfide bond. The overall layout of the subunit would be intact because the disulfide bond still exists. In contrast, the introduction of Cys-to-Ser mutations could grossly perturb the structure of the extracellular domain, and so it is not surprising that little or no functional expression was seen in previous experiments involving such mutations.
Oocytes expressing nAChR channels containing an αVal132Npg mutation showed ACh-induced currents with an EC50 equal to that of wild-type nAChR channels. Thus, channels containing the Npg mutation and an intact disulfide loop assemble, fold, and function properly. Irradiation of these oocytes led to an almost complete loss (≈90%) of ACh-induced, whole-cell current (Fig. 4). As noted above, oocytes expressing wild-type nAChR also were subjected to UV irradiation, and they exhibited no reduction in whole-cell current or change in EC50. These results establish that receptor function requires the loop structure between the Cys128–Cys142 disulfide bond in the α subunit. That is, in addition to playing a key role in folding and assembly of the receptor, this conserved structure is also essential for receptor function.
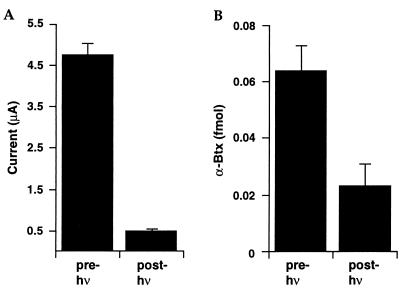
Figure 4.
Effects of irradiation on the αVal132Npg mutant receptor. Oocytes were irradiated for 7.5 h. (A) Change in whole-cell current in response to 200 μM ACh; (B) [125I]-α-bungarotoxin (α-Btx) binding before and after irradiation using the same oocytes from A.
As before, loss of function could result from a destruction of the agonist binding site, a disruption of the gating pathway, or some combination of the two. This mutation is in the N-terminal extracellular domain of the α subunit, a region that is thought to contain most (or all) of the agonist binding site. However, extensive affinity labeling and mutagenesis studies have failed to establish any role in agonist or antagonist binding for the signature disulfide loop. Nevertheless, although the intact nAChR with the αVal132Npg mutation binds α-bungarotoxin well, irradiation of the Npg-containing receptor (but not the wild type) substantially reduces toxin binding (Fig. 4). This suggests that there may be some overlap between the toxin binding site and the signature disulfide loop or that cleavage of the loop leads to conformational changes elsewhere in the α subunit that impact α-bungarotoxin binding.
The present results clearly establish that irradiation of the Npg residue leads to cleavage of the peptide backbone with significant functional consequences that can be evaluated electrophysiologically, but several quantitative issues remain. The long irradiation times required here indicate that Npg photochemistry may be less efficient than in other related systems (8–16). Additional work from our labs establishes that more conventional nitrobenzyl caging groups can undergo very efficient photolysis when incorporated into ion channel proteins expressed in Xenopus oocytes (J. Miller, S. K. Silverman, and P.M.E., unpublished work). Further work will be required to characterize the source of these differences. Another issue is the extent of photolysis, as indicated by loss of current. In the nAChR, one site (αVal132Npg) produces a ≈90% reduction in current, and another (βLeu9′Npg) gives only a ≈50% reduction under similar conditions. In both cases, controls involving injection of full length, but uncharged, tRNA (4–6) gave signals <5% as large as those from oocytes injected with Npg–tRNA, suggesting that essentially all receptors do contain Npg. Future studies will address these issues.
CONCLUSIONS
The unnatural amino acid Npg can be efficiently incorporated into ion channel proteins expressed in Xenopus oocytes, using the in vivo nonsense suppression methodology. Irradiation of Npg-containing proteins lead to site-specific cleavage of the peptide backbone, regardless of whether the residue was located in an intracellular (ShB positions 47 and 64), extracellular (nAChR position α132), or transmembrane (nAChR position βM2 9′) region. Using SNIPP, we have established that the signature disulfide loop of the α subunit of the nAChR has a role beyond subunit folding and receptor assembly. The loop must be intact for the receptor to function.
A notable feature of many ion channels and neuroreceptors is their modular nature, often containing distinct domains that can be assigned specific functional roles such as gating vs. ligand binding. Conventional efforts to evaluate the separate roles of such domains, typically by expressing two separately synthesized peptides, often, with a few notable exceptions (53–55), lead to failure of expression and/or function of the receptor. The SNIPP strategy overcomes this limitation by first producing intact, functional, properly localized protein and subsequently introducing the backbone lesion photochemically. Future challenges include modification of the Npg residue to improve the efficiency and rate of photolysis, but already Npg provides a valuable structural probe for proteins expressed in intact cells. We anticipate application of SNIPP in further studies of ion channels and also in evaluating the roles of specific proteins in signal transduction pathways.
Acknowledgments
We thank P. Reinhart, P. Kearney, M. Nowak, and S. Silverman for suggestions and helpful discussions and B. Henkle and H. Li for assistance with oocytes. This work was supported by the National Institutes of Health (NS-34407, NS-11756, and a National Research Service Award to P.M.E.) and the University of California Tobacco-Related Disease Research Program.
ABBREVIATIONS
- Npg
2-(nitrophenyl)glycine
- SNIPP
site-specific, nitrobenzyl-induced photochemical proteolysis
- 4PO
N-pent-4-enoyl
- ShB
Shaker B
- nAChR
nicotinic acetylcholine receptor
Footnotes
References
- 1.Noren C J, Anthony-Cahill S J, Griffith M C, Schultz P G. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 2.Cornish V W, Mendel D, Schultz P G. Angew Chem Int Ed Engl. 1995;34:621–633. [Google Scholar]
- 3.Steward L E, Collins C S, Gilmore M A, Carlson J E, Ross J B A, Chamberlin A R. J Am Chem Soc. 1997;119:6–11. [Google Scholar]
- 4.Nowak M W, Kearney P C, Sampson J R, Saks M E, Labarca C G, Silverman S K, Zhong W, Thorson J, Abelson J N, Davidson N, Schultz P G, Dougherty D A, Lester H A. Science. 1995;268:439–442. doi: 10.1126/science.7716551. [DOI] [PubMed] [Google Scholar]
- 5.Kearney P C, Nowak N W, Zhong W, Silverman S, K, Lester H A, Dougherty D A. Mol Pharmacol. 1996;50:1401–1412. [PubMed] [Google Scholar]
- 6.Saks M E, Sampson J R, Nowak M W, Kearney P C, Du F, Abelson J N, Lester H A, Dougherty D A. J Biol Chem. 1996;271:23169–23175. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- 7.Turcatti G, Nemeth K, Edgerton M D, Meseth U, Talabot F, Peitsch M, Knowles J, Vogel H, Chollet A. J Biol Chem. 1996;271:19991–19998. doi: 10.1074/jbc.271.33.19991. [DOI] [PubMed] [Google Scholar]
- 8.Milburn T, Matsubara N, Billington A P, Udgaonkar J B, Walker J W, Carpenter B K, Webb W W, Marque J, Denk W, McCray J A, Hess G P. Biochemistry. 1989;28:49–55. doi: 10.1021/bi00427a008. [DOI] [PubMed] [Google Scholar]
- 9.Wieboldt R, Gee K R, Niu L, Ramesh D, Carpenter B K, Hess G P. Proc Natl Acad Sci USA. 1994;91:8752–8756. doi: 10.1073/pnas.91.19.8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wieboldt R, Ramesh D, Carpenter B K, Hess G P. Biochemistry. 1994;33:1526–1533. doi: 10.1021/bi00172a032. [DOI] [PubMed] [Google Scholar]
- 11.Ramesh D, Wieboldt R, Billington A P, Carpenter B K, Hess G P. J Org Chem. 1993;58:4599–4605. [Google Scholar]
- 12.McCray J A, Trentham D R. Annu Rev Biophys Chem. 1989;18:239–270. doi: 10.1146/annurev.bb.18.060189.001323. [DOI] [PubMed] [Google Scholar]
- 13.Gurney A M, Lester H A. Physiol Rev. 1987;67:583–617. doi: 10.1152/physrev.1987.67.2.583. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan J H, Somlyo A P. Trends Neurosci. 1989;12:54–59. doi: 10.1016/0166-2236(89)90136-7. [DOI] [PubMed] [Google Scholar]
- 15.Adams S R, Tsien R Y. Annu Rev Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 16.Padwa A, editor. Organic Photochemistry. New York: Dekker; 1987. [Google Scholar]
- 17.Mendel D, Ellman J A, Schultz P G. J Am Chem Soc. 1991;113:2758–2760. [Google Scholar]
- 18.Cook S N, Jack W E, Xiong X, Danley L E, Ellman J A, Schultz P G, Noren C J. Angew Chem Int Ed Engl. 1995;34:1629–1630. [Google Scholar]
- 19.Davis A L, Smith D R, McCord T J. J Med Chem. 1973;16:1043–1045. doi: 10.1021/jm00267a020. [DOI] [PubMed] [Google Scholar]
- 20.Muralidharan S, Nerbonne J M. J Photochem Photobiol B. 1995;27:123–137. doi: 10.1016/1011-1344(94)07063-t. [DOI] [PubMed] [Google Scholar]
- 21.Madsen R, Roberts C, Fraser-Reid B. J Org Chem. 1995;60:7920–7926. [Google Scholar]
- 22.Lodder M, Golvine S, Hecht S M. J Org Chem. 1997;62:778–779. [Google Scholar]
- 23.Robertson S A, Ellman J A, Schultz P G. J Am Chem Soc. 1991;113:2722–2729. [Google Scholar]
- 24.Robertson S A, Noren C J, Anthony-Cahill S J, Griffin M C, Schultz P G. Nucleic Acids Res. 1989;17:9649–9660. doi: 10.1093/nar/17.23.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quick M W, Lester H A. In: Ion Channels of Excitable Cells. Narahashi T, editor. London: Academic; 1994. pp. 261–279. [Google Scholar]
- 26.Iverson L E, Tanouye M A, Lester H A, Davidson N, Rudy B. Proc Natl Acad Sci USA. 1988;85:5723–5727. doi: 10.1073/pnas.85.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamb A, Iverson L E, Tanouye M A. Cell. 1987;50:405–413. doi: 10.1016/0092-8674(87)90494-6. [DOI] [PubMed] [Google Scholar]
- 28.Papazian D M, Schwarz T L, Tempel B L, Jan Y N, Jan L Y. Science. 1987;237:749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- 29.Pongs O, Kecskemethy N, Muller R, Krah-Jentgens I, Baumann A, Koltz H H, Canal I, Llamazares S, Ferrus A. EMBO J. 1988;7:1087–1096. doi: 10.1002/j.1460-2075.1988.tb02917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timpe L C, Schwarz T L, Tempel B L, Papazian D M, Jan Y N, Jan L Y. Nature (London) 1988;331:143–145. doi: 10.1038/331143a0. [DOI] [PubMed] [Google Scholar]
- 31.Miller C. Science. 1991;252:1092–1096. doi: 10.1126/science.252.5009.1092. [DOI] [PubMed] [Google Scholar]
- 32.Hoshi T, Zagotta W N, Aldrich R W. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 33.Zagotta W N, Hoshi T, Aldrich R W. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 34.Demo S D, Yellen G. Neuron. 1991;7:743–753. doi: 10.1016/0896-6273(91)90277-7. [DOI] [PubMed] [Google Scholar]
- 35.MacKinnon R, Aldrich R W, Lee A W. Science. 1993;262:757–759. doi: 10.1126/science.7694359. [DOI] [PubMed] [Google Scholar]
- 36.Lester H A. Annu Rev Biophys Biomol Struct. 1992;21:267–292. doi: 10.1146/annurev.bb.21.060192.001411. [DOI] [PubMed] [Google Scholar]
- 37.Karlin A. Curr Opin Neurobiol. 1993;3:299–309. doi: 10.1016/0959-4388(93)90121-e. [DOI] [PubMed] [Google Scholar]
- 38.Devillers-Thiéry A, Galzi J L, Eiselé J L, Bertrand S, Bertrand D, Changeux J P. J Membrane Biol. 1993;136:97–112. doi: 10.1007/BF02505755. [DOI] [PubMed] [Google Scholar]
- 39.Stroud R M, McCarthy M P, Shuster M. Biochemistry. 1990;29:11009–11023. doi: 10.1021/bi00502a001. [DOI] [PubMed] [Google Scholar]
- 40.Revah F, Bertrand D, Galzi J-L, Devillers-Thiéry A, Mulle C, Hussy N, Bertrand S, Ballivet M, Changeux J-P. Nature (London) 1991;353:846–849. doi: 10.1038/353846a0. [DOI] [PubMed] [Google Scholar]
- 41.Labarca C, Nowak M W, Zhang H, Tang L, Deshpande P, Lester H A. Nature (London) 1995;376:514–516. doi: 10.1038/376514a0. [DOI] [PubMed] [Google Scholar]
- 42.Filatov G N, White M M. Mol Pharmacol. 1995;48:379–384. [PubMed] [Google Scholar]
- 43.Akabas M H, Stauffer D A, Xu M, Karlin A. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 44.Kearney P C, Zhang H, Zhong W, Dougherty D A, Lester H A. Neuron. 1996;17:1221–1229. doi: 10.1016/s0896-6273(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 45.Shtrom S S, Hall Z W. J Biol Chem. 1996;271:25506–25514. doi: 10.1074/jbc.271.41.25506. [DOI] [PubMed] [Google Scholar]
- 46.Green W N, Millar N S. Trends Neurosci. 1995;18:280–287. [PubMed] [Google Scholar]
- 47.Blount P, Smith M M, Merlie J P. J Cell Biol. 1990;111:2601–2611. doi: 10.1083/jcb.111.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kao P N, Dwork A J, Kaldany R R J, Silver M L, Wideman J, Stein S, Karlin A. J Biol Chem. 1984;259:11662–11665. [PubMed] [Google Scholar]
- 49.Mishina M, Tobimatsu T, Imoto K, Tanaka K, Fujita Y, Fukuda K, Kurasaki M, Takahashi H, Morimoto Y, Hirose T, Inayama S, Takahashi T, Kuno M, Numa S. Nature (London) 1985;313:364–369. doi: 10.1038/313364a0. [DOI] [PubMed] [Google Scholar]
- 50.Sumikawa K, Gehle V M. J Biol Chem. 1992;267:6286–6290. [PubMed] [Google Scholar]
- 51.Walcott E C, Sumikawa K. Mol Brain Res. 1996;41:289–300. doi: 10.1016/0169-328x(96)00122-2. [DOI] [PubMed] [Google Scholar]
- 52.Fu D-X, Sine S M. J Biol Chem. 1996;271:31479–31484. doi: 10.1074/jbc.271.49.31479. [DOI] [PubMed] [Google Scholar]
- 53.Sahin-Toth M, Kaback H R, Friedlander M. Biochemistry. 1996;35:2016–2021. doi: 10.1021/bi952496g. [DOI] [PubMed] [Google Scholar]
- 54.Wei A, Solaro C, Lingel C, Salkoff L. Neuron. 1994;13:671–681. doi: 10.1016/0896-6273(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 55.Stuhmer W, Conti F, Suzuki H, Wang W D, Noda M, Yahagi N, Kubo H, Numa S. Nature (London) 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]