Abstract
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels increase in particulate fractions in association with cell death in HEK293 cells, S49 cells, primary thymocytes, PC12 cells, and primary cerebral cortical neuronal cultures. Subcellular fractionation and immunocytochemistry reveal that this increase primarily reflects nuclear translocation. Nuclear GAPDH is tightly bound, resisting extraction by DNase or salt treatment. Treating primary thymocytes, PC12 cells, and primary cortical neurons with antisense but not sense oligonucleotides to GAPDH prevents cell death. Because cell-death-associated nuclear translocation of GAPDH and antisense protection occur in multiple neuronal and nonneuronal systems, we propose that GAPDH is a general mediator of cell death and uses nuclear translocation as a signaling mechanism.
The glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), displays other activities (1), such as uracil DNA glycosylase activity that mediates DNA repair (2) as well as binding to transfer RNA (3) and DNA (4, 5). Because of its abundance, GAPDH is often regarded as a static marker protein contrasted to the dynamic alterations of other proteins. However, GAPDH expression is also dynamic and changes with hypoxia (6), calcium influx induced by an ionophore (7), in oncogene-transformed and growth-accelerated states (8) as well as in human tumors (9), and as a function of the cell cycle (10).
A role for GAPDH in neurotoxicity has been suggested by observations that nitric oxide (NO), which mediates neurotoxicity associated with vascular stroke (11), stimulates auto-ADP ribosylation of GAPDH (12–14) or direct attachment of NAD to the enzyme (15). Moreover, GAPDH can bind to Huntingtin, the protein whose glutamine repeats may mediate the symptoms of Huntington disease (HD) (16), as well as to the gene products with glutamine repeats involved in other neurodegenerative diseases such as spinocerebellar ataxia type-I (17), spinobulbar muscular atrophy (17), and dentatorubral pallidoluysian atrophy (16). Furthermore, GAPDH can bind to the cytoplasmic domain of amyloid precursor protein (18).
Ishitani and associates (19–22) have obtained evidence that overexpression of GAPDH participates in cell death of cerebellar granule cells and cerebral cortical neurons in culture, because antisense oligonucleotides to GAPDH prevent such cell death. We sought to elucidate mechanisms for the protective actions of GAPDH and explore its relevance to nonneuronal as well as neuronal cells. We report that GAPDH overexpression in particulate fractions occurs in multiple forms of cell death and is associated with translocation of the enzyme to the nucleus. Antisense oligonucleotides to GAPDH prevent cell death in several systems.
MATERIALS AND METHODS
Materials.
Unless noted, chemicals were obtained from Sigma. Primary antibodies (Abs) were obtained from the following companies or as generous gifts: anti-MAP2 monoclonal Ab (Boehringer Mannheim) (dilution 1:300), anti-MAP2 polyclonal Ab (Dr. Y. Ihara, University of Tokyo, Japan) (dilution 1:300), anti-glial fibrillary acidic protein (GFAP) Ab (Dako) (dilution 1:1,000), anti-GAPDH monoclonal Ab (Chemicon) (dilution 1:200–1,200), anti-GAPDH polyclonal Ab (Charles River Breeding Laboratories) (dilution 1:200–1,200). Peroxidase-conjugated anti-mouse, rabbit, or goat IgG Abs were used as secondary Abs for immunoblot (Boehringer Mannheim; 1:2,000 dilution). FITC-conjugated anti mouse IgG Ab (dilution 1:200) (Jackson ImmunoResearch) for GAPDH and MAP2, FITC-conjugated anti-goat IgG Ab (dilution 1:200) (Jackson ImmunoResearch) for lactate dehydrogenase (LDH), and Texas red-conjugated anti-rabbit IgG Ab (dilution 1:200) (Jackson ImmunoResearch) for MAP2 and GFAP were used as secondary Abs for immunocytochemistry. For nuclear staining, Hoechst 33258 (1:500 dilution) or propidium iodide (1:1,000 dilution) (Molecular Probes) was used.
Antisense phosphorothioate oligonucleotides to GAPDH were produced in a DNA analysis facility in the Johns Hopkins University. These rat sequences were 5′-GACCTTCACCATCTTGTCTA-3′ for antisense 1, 5′-GTGGATGCAGGGATGATGTT-3′ for antisense 2, and 5′-TAGACAAGATGGTGAAGGTC-3′ for sense oligo.
Cell Culture.
Primary thymocytes taken from the thymus of 1- to 2-month-old male Sprague–Dawley rats (Charles River Breeding Laboratories) were cultured in RPMI 1640 medium (GIBCO/BRL) plus 10% fetal bovine serum (FBS). S49 cells were maintained in the same medium as for primary thymocytes. HEK293 cells were maintained in DMEM plus 10% FBS. After two washes with DMEM, the medium was replaced with serum-free DMEM to induce cell death.
PC12 cells were grown and differentiated as described previously (23). Briefly, PC12 cells were maintained in DMEM plus 5% FBS and 10% horse serum (HS). Differentiation was initiated by addition of nerve growth factor (NGF) and changing FBS and HS concentrations to 2% and 1%, respectively. Five days after differentiation, the medium was completely replaced with medium lacking NGF and serum. Primary cortical cultures were prepared from fetal Sprague–Dawley rats (19- to 20-day gestation). The dissection and plating procedures were described previously (24). The dissociated cells were grown in B27-supplemented serum-free media (25). More than 95% of cells were neurons, as estimated by double staining using anti-MAP2 Ab and anti-GFAP Ab.
Oligonucleotides were applied at 5-μM (thymocytes), 3-μM (PC12), and 2-μM (cortical neurons) concentrations.
Cell Viability.
The viability of primary thymocytes, S49, and PC12 cells was monitored by the trypan blue exclusion assay (24). The blue and non-blue cells was counted blindly by two independent observers. The viability of primary cortical neurons was monitored by counting the remaining MAP2 positive neurons under immunofluorescent microscopy after immunostaining. Statistical analysis was done by using a Student’s t test with at least five independent experiments. In each group, 500–1,000 cells were counted per experiment.
Subcellular Fractionation.
S49 cells were resuspended in buffer A (100 mM Hepes/1 mM phenylmethylsulfonyl fluoride (PMSF)/1 mM DTT) followed by sonication (strength 2, three times for 15 sec). After sonication, extracts were centrifuged at 10,000 × g for 10 min, providing a low-speed pellet (LP). Supernatants were centrifuged at 100,000 × g for 30 min, providing a supernatant (S) and high-speed pellet (HP).
Subfractionation using sucrose gradients was as described (26) with minor modifications. Briefly, cells (1 × 107) were resuspended in 700 μl of buffer B (0.25 M sucrose/10 mM MgCl2/10 mM KCl/2 mM DTT/1 mM PMSF) for 10 min on ice followed by homogenization with 30 strokes in a Dounce homogenizer, addition of 0.1% deoxycholic acid and 0.4% NP40, and gentle mixing on ice. Following centrifugation at 700 × g for 10 min, the pellet was resuspended in buffer B and washed twice. After resuspending this pellet in 100 μl of buffer B and layering it on 1.5 ml of a sucrose cushion (2.2 M sucose/10 mM MgCl2/10 mM KCl/2 mM DTT/1 mM PMSF) in polystyrene tubes, the mixture was centrifuged at 15,000 × g for 20 min, providing a nuclear pellet (N). The postnuclear supernatant was centrifuged at 14,000 × g for 30 min, providing a pellet (P2). The supernatant was then centrifuged at 100,000 × g for 3 hr, yielding a final supernatant (S) and pellet (P3).
The purity of the N fraction was monitored by the exclusion of the enzymatic activities of LDH and cytochrome c oxidase. LDH activity was measured by the LDH kit from Sigma. Cytochrome c oxidase activity was assayed as described (27). GAPDH activity was assayed as described (12). Protein concentration of each fraction was determined with BSA as a standard with the Coomassie protein assay reagent (Pierce).
Immunoblots and Immunochemistry.
SDS/PAGE was performed on 1.5-mm-thick 12% gels, and the separated proteins were transferred to nitrocellulose membranes. Blots were blocked with 5% skim milk in PBS with 0.01% Tween 20 before incubation at 25°C for 2 hr with primary antibodies. Primary antibodies were incubated in PBS with 3% BSA and 0.01% Tween 20. Blots were washed three times for 5 min each in 5% milk in PBS with 0.01% Tween 20 and then incubated with secondary antibodies. Blots were then washed twice in PBS and developed by Renaissance kit (DuPont/NEN). immunocytochemistry was done as described (28). Confocal microscopy, in which the immunofluorescent staining is superimposed on phase contrast images, employed a Noran OZ (Noran Instruments, Middleton, WI) confocal laser-scanning system, fitted to an Olympus IX-50 fluorescence microscope.
RESULTS
Increased GAPDH Expression in Particulate Fractions of Nonneuronal Cells Mediates Cell Death.
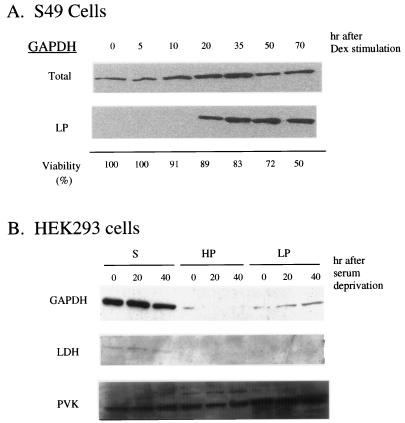
S49 T cells undergo apoptotic death after dexamethasone (Dex) stimulation (29). GAPDH levels in a LP fraction, absent initially, became apparent at 20 hr and increased substantially at 30 and 50 hr, whereas only modest increases were evident in the total cell extract (Fig. 1A). We also explored relationships between these changes in GAPDH expression and cell viability following Dex. Cell death occurred more slowly than GAPDH changes, with 11 and 50% decreases in viability at 20 and 70 hr, respectively (Fig. 1A).
Figure 1.
Translocation of GAPDH into particulate fractions in nonneuronal cells during cell death. (A) S49 cells undergoing apoptotic cell death after Dex stimulation. GAPDH in a low-speed particulate fraction LP increases with Dex (1 μM) stimulation as early as 20 hr, when 90% of cells are still viable by trypan blue exclusion assay. Ten micrograms of proteins were loaded on each lane. (B) After serum deprivation, HEK293 cells undergo apoptotic cell death. GAPDH in LP increases 20 and 40 hr after serum deprivation with no changes in lactate dehydrogenase (LDH) and pyruvate kinase (PVK). Ten micrograms of proteins were loaded on each lane.
HEK293 cells also undergo apoptotic death following serum deprivation. In these cells we demonstrated DNA ladder formation on agarose gels stained with ethidium bromide as well as nuclear fragmentation observed by immunofluorescent microscopy with Hoechst 33258 and propidium iodide (data not shown). GAPDH levels in LP were increased at 20 hr and more so at 40 hr, whereas no increase was evident in supernatant or HP fractions (Fig. 1B). The augmentation of GAPDH was selective, because LDH and pyruvate kinase (PVK) protein levels were unchanged in all fractions (Fig. 1B).
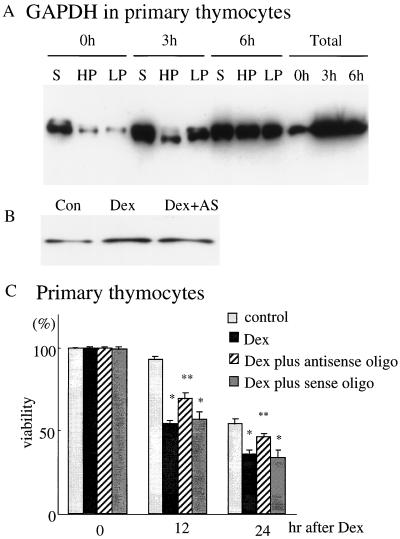
To ascertain whether increased expression of GAPDH was causally associated with cell death, we utilized primary thymocyte cultures treated with Dex to elicit cell death. In these cultures, GAPDH protein levels increased as early as 3 hr following Dex treatment, with somewhat greater increases at 6 hr (Fig. 2A). Augmentations were pronounced in HPs and LPs following a substantial overexpression in supernatant fraction (Fig. 2A). To lower GAPDH expression, we employed antisense oligonucleotide treatment, which reduced GAPDH protein levels by about 30% (Fig. 2B). After 24 hr of incubation with Dex, cell viability was reduced by about 40%. GAPDH antisense treatment provided protection with levels restored to about half-way to control values. By contrast, GAPDH sense oligonucleotide treatment had no effect (Fig. 2C).
Figure 2.
Overexpression of GAPDH followed by its translocation into particulate fractions and the protective effect of antisense oligonucleotides to GAPDH against cell death in primary thymocytes. (A) After Dex stimulation (3 μM), GAPDH levels in total extracts are greatly elevated as early as 3 hr after stimulation. Modest cytosolic overexpression of GAPDH at 3 hr is followed at 6 hr by particulate augmentation, especially in LP. Twenty micrograms of proteins were loaded on each lane. (B) Antisense oligonucleotides (5 μM) to GAPDH reduce the augmented GAPDH protein level after Dex. The antisense oligonucleotide was added 6 hr before Dex stimulation, and cell extracts were taken 12 hr after treatment. Five micrograms of proteins were loaded on each lane. Con, without treatment; Dex, only Dex addition; Dex+AS, antisense treatment prior to Dex addition. (C) Antisense oligonucleotides to GAPDH protect, with levels increased half of the way to control levels. By contrast, GAPDH sense oligonucleotides have no effect.
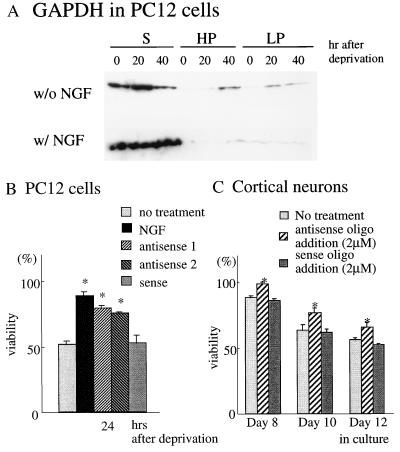
We examined the role of GAPDH in cell death of two neuronal systems. In PC12 cells, NGF deprivation elicits cell death (30). NGF deprivation led to GAPDH increases in HP and LP fractions but not in the supernatant fraction, and NGF treatment prevented these changes (Fig. 3A). We utilized two distinct antisense oligonucleotides to GAPDH to evaluate effects upon viability. Both of the antisense preparations restored viability. By contrast, sense oligonucleotide treatment was without effect (Fig. 3B). The application of antisense oligonucleotide to cells without Dex treatment did not influence viability (data not shown).
Figure 3.
Prevention of cell death by antisense oligonucleotides to GAPDH in neuronal systems. (A) PC12 cells after nerve growth factor (NGF) deprivation show translocation of GAPDH into particulate fractions, which is rescued by NGF addition. Ten micrograms of proteins were loaded on each lane. (B) Two distinct antisense oligonucleotides (3 μM) to GAPDH partially restore the viability of PC12 after NGF deprivation, whereas sense oligonucleotides have no effect. (C) Cortical neurons gradually die after 5 days in culture. Application of antisense oligonucleotides (2 μM) to GAPDH every 2 days from day 5 attenuates cell death, whereas sense oligonucleotides have no effect.
Cerebral cortical neurons in primary culture die following prolonged culture. In this system, GAPDH antisense treatment prevents cell death (20). We utilized serum-free media to establish purer neuronal cultures (25). GAPDH antisense treatment attenuated cell death, whereas sense oligonucleotides were not effective (Fig. 3C).
Translocation of GAPDH to the Nucleus Associated with Cell Death.
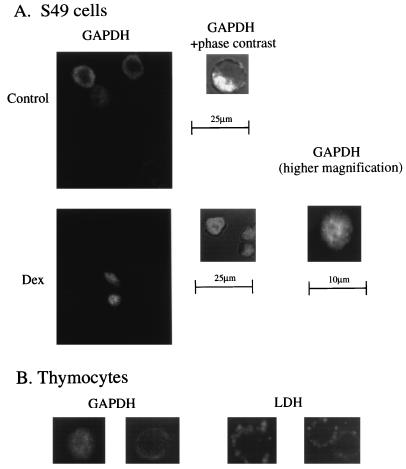
To ascertain the intracellular compartment responsible for the particulate translocation of GAPDH during cell death, we utilized confocal microscopy associated with GAPDH immunocytochemistry in S49 cells. Prior to Dex treatment GAPDH was localized in a donut-like configuration in the periphery of cells, reflecting a cytosolic localization, with no staining in the large central nucleus, findings confirmed by superimposition on a phase contrast image (Fig. 4A). Under basal conditions, staining was heterogeneous with 5–10% of the cells staining much more then the rest, but no nuclear staining occurred in any cells (data not shown). Dex treatment led to uniform cellular staining including the nuclear region (Fig. 4A). Nuclear translocation of GAPDH preceded cell death. When more than 70% cells were still intact by trypan blue exclusion assay, almost all cells already displayed nuclear staining. Some dying cells revealed nuclear fragmentations when stained by Hoechst 33258 or propidium iodide (data not shown). In primary thymocytes treated with Dex, GAPDH staining was also uniformly distributed over the cell including the nucleus. By contrast, GAPDH staining in cells treated with antisense oligonucleotides retained a donut-shaped cytosolic disposition even after Dex stimulation (Fig. 4B). Nuclear translocation appeared to be GAPDH-specific, because LDH staining was nonnuclear both before and after Dex treatment (Fig. 4B).
Figure 4.
Translocation of GAPDH to the nucleus; immunocytochemical studies in S49 cells and primary thymocytes. (A) Prior to Dex treatment, GAPDH staining is excluded from the nucleus in S49 cells. (Left) A confocal image of GAPDH staining. (Right) A confocal section of the phase contrast image superimposed on GAPDH staining. Nuclear exclusion is detected in almost all of more than 500 cells observed. After Dex treatment, GADPH staining is uniform over S49 cells. These pictures were taken 48 hr after stimulation, when more than 70% of cells are still intact by trypan blue exclusion assay. Left shows a confocal image of GAPDH staining. Center shows a confocal section of the phase contrast image superimposed on GAPDH staining. Right shows GAPDH staining at higher magnification. No nuclear exclusion of GAPDH is detected in more than 500 cells examined with independent confocal sections at 0.5-μm intervals. (B) In primary thymocytes treated with Dex, GAPDH staining is uniformly distributed in nuclei and cytosol, whereas antisense oligonucleotide treatment (5 μM) restores nuclear exclusion (Left). LDH staining is nonnuclear both before and after Dex treatment. After stimulation, cell size is reduced slightly with no change in the staining pattern of LDH (Right).
To assess the role of nuclear translocation in neuronal cells, we utilized cerebral cortical neurons in culture. After 5 days in culture, when neurons are maximally viable, GAPDH staining was predominantly cytoplasmic, though some was evident in nuclei, whereas MAP2 staining was exclusively nonnuclear and extended robustly into neurites (data not shown). At 12 days in culture, when substantial cell death was taking place, nuclear staining was augmented, compared with 5 days in culture, and some neuritic staining was evident. GAPDH antisense treatment abolished nuclear staining (Fig. 5).
Figure 5.
Participation of nuclear translocation of GADPH in cell death of primary cortical neurons. After day 12 in culture, about 50% of neurons are dead. In untreated viable cells GAPDH is nuclear as well as cytosolic, whereas antisense treatment (2 μM) abolishes nuclear staining.
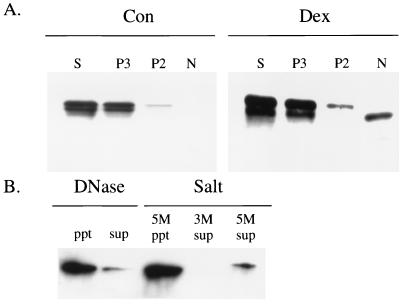
We also utilized subcellular fractionation techniques to examine a nuclear association of GAPDH. In S49 cells, nuclei were purified from a crude nuclear pellet by subfractionation on a sucrose gradient. Microscopic examination confirmed that nuclei were the only major organelle present in the purified nuclei fraction (data not shown). Before Dex treatment GAPDH occurred in the supernatant and the P3, high-speed pellet, with a modest amount in the P2, low-speed pellet, and none in N, the purified nuclei (Fig. 6A). Treatment with Dex greatly enhanced the GAPDH content of N with lesser increases in P2, P3, and cytosol (Fig. 6A). In most fractions GAPDH staining comprised two bands, with the upper more prominent. By contrast, the emergent nuclear GAPDH after Dex treatment corresponded only to the lower band. The same results were obtained with distinct antibodies to GAPDH (polyclonal and monoclonal with independent epitopes). Cytochrome c oxidase activity was equally distributed in P2 and P3 fractions, indicating that mitochondria are most concentrated in P2 and P3. LDH, a soluble enzyme, was confined to the supernatant fraction with almost complete exclusion from N (data not shown).
Figure 6.
Biochemical characterization of nuclear GAPDH in dexamethasone (Dex)-treated S49 cells. (A) Before stimulation, GAPDH is mainly in cytosolic and P3 fractions with less in P2 and no staining in the nuclear fraction (N). In supernatant (S) and P3 fractions, two distinct bands are detected, with the upper more abundant. In the P2 fraction, only the upper band is seen. After Dex stimulation, only the lower band of GAPDH emerges in N with some augmentation of GAPDH also in S, P3, and P2. Twenty micrograms of proteins were loaded on each lane. Con, before stimulation (control); Dex, 48 hr after dexathasone treatment. (B) Nuclear GAPDH resists extraction; DNase I treatment or high salt treatment (up to 5 M NaCl) fail to elute GAPDH from the nuclear fraction.
GAPDH in the nuclei appears to be tightly bound. Treatment with DNase I failed to liberate GAPDH from the nuclear fraction (Fig. 6B). Extensive washing with 3 M and 5 M NaCl also failed to remove GAPDH from the nuclear pellet (Fig. 6B). No GAPDH catalytic activity was demonstrable in purified S49 nuclei before or after Dex treatment (data not shown). Thus translocation of GAPDH to the nucleus somehow inactivates its catalytic capacity.
DISCUSSION
Ishitani and colleagues (19–22) reported increased GAPDH expression associated with cell death in cerebellar granule cells and cerebral cortical cultures. We have shown that this phenomenon is not restricted to neuronal systems but can be demonstrated in HEK293 cells, in S49 T cells, and in thymocytes in primary culture. Ishitani et al. (19–22) found that GAPDH antisense oligonucleotides protect against cell death in cerebellar and cortical cells in culture. We confirm the protection in cortical cultures and also observe protection in thymocytes and PC12 cells.
The increased levels of GAPDH are most notable in particulate fractions, as observed independently by Saunders et al. (31) in a preliminary report. Utilizing confocal microscopy and subcellular fractionation, we demonstrate a selective augmentation in the nucleus. In Dex-treated S49 cells, mobility of this nuclear GAPDH on SDS/PAGE differs from other fractions, suggesting that posttranslational modifications accompany nuclear translocation. Within the nucleus the GAPDH is bound tightly, resistant to extraction with high salt concentrations and DNase treatment. Whether the GAPDH in the nucleus is covalently linked to another protein or tightly bound by some other means to the nuclear scaffold is unclear.
The ability of antisense oligonucleotides to prevent cell death suggests that GAPDH mediates decreases in cell viability. Though protection is only partial, the antisense treatment reduces GAPDH protein levels only about 30%, similar to the levels of protection.
The striking augmentation of GAPDH in the nucleus suggests that a nuclear pool of GAPDH is principally responsible for decreases in cell viability. Nuclear functions that might be influenced by GAPDH include its binding DNA (4, 5) and its uracil glycosylase activity (2).
GAPDH is overexpressed during tumorogenesis and phases of the cell cycle that require more energy, (8–10). In 5–10% of healthy-growing S49 cells, GAPDH is overexpressed exclusively outside nuclei. Conceivably, these changes reflect cell-cycle alterations and indicate that cytosolic overexpression of GAPDH is not related to cell death.
The selective nuclear translocation of GAPDH argues for at least two discrete pools of the enzyme: one associated with glycolysis and another with nuclear and/or other functions. Epner and Coffey (32) recently described multiple forms of GAPDH in prostatic cancer cells. Conceivably, the unique mobility of nuclear GAPDH in our experiments reflects a novel form of the enzyme. The notion that enzymes of carbohydrate metabolism may display other, sometimes nuclear, functions, has precedence. With reduced tissue iron levels (33) or glutamate neurotransmission (34), cytosolic aconitase loses its catalytic activity and binds to mRNA of various proteins. This is reminiscent of the loss of GAPDH catalytic activity after nuclear translocation. Aldolase also has been reported to display nuclear localizations (35).
GAPDH may associate with other cellular compartments, such as lysozymes (36), synaptic vesicles (37), the cytoskeleton (38), and the plasma membrane (39). As a small soluble protein, GAPDH might serve as a chaperone, translocating proteins between intracellular sites. A similar chaperone function has been reported for the abundant, small molecular immunophilin protein FKBP12, which serves as a scaffold, linking large proteins such as the inositol-1,4,5-trisphophate receptor and calcineurin (40). AKAP (A-kinase anchoring protein) similarly links cAMP-dependent protein kinase and calcineurin (41).
Which proteins might bind to GAPDH in association with such a postulated chaperone function? GAPDH binds to glutamine repeats in Huntingtin and other polyglutamine-containing proteins associated with neurodegenerative diseases (16, 17). GAPDH also binds to amyloid precursor protein (18). Interestingly, polyglutamine repeats provide a β-pleated structure, and amyloid precursor protein also contains a β-pleated sheet. Overexpression of the polyglutamine repeat itself can elicit neurotoxicity (42). Conceivably, GAPDH binds to such β-pleated structures and translocates them. Neuronal intranuclear inclusions containing Huntingtin occur in transgenic mice with the HD mutation and in juvenile-onset HD (43). GAPDH might serve as a chaperone for such a nuclear translocation.
In summary, in multiple neuronal and nonneuronal systems, GAPDH translocates to the nucleus in association with cell death. Antisense oligonucleotide treatment prevents cell death in several systems. Accordingly, we suggest that GAPDH, or novel posttranslationally altered isoform, serves as a mediator of cell death by translocating to the nucleus.
Acknowledgments
We thank M. Dellanoy for his technical assistance in taking confocal pictures. We thank C. A. Ross, T. M. Dawson, V. L. Dawson, C. V. Dang, H. Shim, J. Zhang, S. Blackshaw, J. K. Hurt, and M. Takahashi for useful comments. We also thank Y. Ihara for providing polyclonal Abs to MAP2. This paper was supported by U.S. Public Health Service Grant MH 18501 and Research Scientist Award DA 00074 to S.H.S., a Uehara Memorial Foundation postdoctoral fellowship (Japan), a Univers Foundation research grant (Japan), and a research grant from the Foundation for Total Health Promotion (Japan) to A.S.
ABBREVIATIONS
- Ab
antibody
- Dex
dexamethasone
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HD
Huntington disease
- LDH
lactate dehydrogenase
- NGF
nerve growth factor
- PMSF
phenylmethylsulfonyl fluoride
- HP
high-speed pellet
- LP
low-speed pellet
References
- 1.Sirover M A. Life Sci. 1996;58:2271–2277. doi: 10.1016/0024-3205(96)00123-3. [DOI] [PubMed] [Google Scholar]
- 2.Meyer-Siegler K, Mauro D J, Seal G, Wurzer J, DeRiel J K, Sirover M A. Proc Natl Acad Sci USA. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Green M R. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 4.Grosse F, Nasheuer H-P, Scholtissek S, Schomburg U. Eur J Biochem. 1986;160:459–467. doi: 10.1111/j.1432-1033.1986.tb10062.x. [DOI] [PubMed] [Google Scholar]
- 5.Morgenegg G, Winkler G C, Hubscher U, Heizmann C W, Mous J, Kuenzle C C. J Neurochem. 1986;47:54–62. doi: 10.1111/j.1471-4159.1986.tb02830.x. [DOI] [PubMed] [Google Scholar]
- 6.Graven K K, Troxler R F, Kornfeld H, Panchenko M V, Farber H W. J Biol Chem. 1994;269:24446–24453. [PubMed] [Google Scholar]
- 7.Chao C C-K, Yam W-C, Lin-Chao S. Biochem Biophys Res Commun. 1990;171:431–438. doi: 10.1016/0006-291x(90)91411-k. [DOI] [PubMed] [Google Scholar]
- 8.Persons D A, Schek N, Hall B L, Finn O J. Mol Carcinog. 1989;2:88–94. doi: 10.1002/mc.2940020207. [DOI] [PubMed] [Google Scholar]
- 9.Tokunaga K, Nakamura Y, Sakata K, Fujimori K, Ohkubo M, Sawada K, Sakiyama S. Cancer Res. 1987;47:5616–5619. [PubMed] [Google Scholar]
- 10.Mansur N R, Meyer-Siegler K, Wurzer J C, Sirover M A. Nucleic Acids Res. 1993;21:993–998. doi: 10.1093/nar/21.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Snyder S H. Annu Rev Pharmacol Toxicol. 1995;35:213–233. doi: 10.1146/annurev.pa.35.040195.001241. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Snyder S H. Proc Natl Acad Sci USA. 1992;89:9382–9385. doi: 10.1073/pnas.89.20.9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kots A Y, Skurat A V, Sergienko E A, Bulargina T V, Severin E S. FEBS Lett. 1992;300:9–12. doi: 10.1016/0014-5793(92)80153-8. [DOI] [PubMed] [Google Scholar]
- 14.Dimmeler S, Lottspeich F, Brune B. J Biol Chem. 1992;267:16771–16774. [PubMed] [Google Scholar]
- 15.McDonald L J, Moss J. Proc Natl Acad Sci USA. 1993;90:6238–6241. doi: 10.1073/pnas.90.13.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke J R, Enghild J J, Martin M E, Jou Y-S, Myers R M, Roses A D, Vance J M, Strittmatter W J. Nat Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 17.Koshy B, Matilla T, Burright E N, Merry D E, Fischbeck K H, Orr H T, Zoghbi H Y. Hum Mol Genet. 1996;5:1311–1318. doi: 10.1093/hmg/5.9.1311. [DOI] [PubMed] [Google Scholar]
- 18.Shulze H, Schuler A, Stuber D, Dobeli H, Langen H, Huber G. J Neurochem. 1992;60:1915–1922. doi: 10.1111/j.1471-4159.1993.tb13420.x. [DOI] [PubMed] [Google Scholar]
- 19.Ishitani R, Chuang D M. Proc Natl Acad Sci USA. 1996;93:9937–9941. doi: 10.1073/pnas.93.18.9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishitani R, Kimura M, Sunaga K, Katsube N, Tanaka M, Chuang D-M. J Pharmacol Exp Ther. 1996;278:447–454. [PubMed] [Google Scholar]
- 21.Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chuang D-M. J Neurochem. 1996;66:928–935. doi: 10.1046/j.1471-4159.1996.66030928.x. [DOI] [PubMed] [Google Scholar]
- 22.Ishitani R, Sunaga K, Tanaka M, Aishita H, Chuang D-M. Mol Pharmacol. 1997;51:542–550. doi: 10.1124/mol.51.4.542. [DOI] [PubMed] [Google Scholar]
- 23.Lyons W E, George E B, Dawson T M, Steiner J P, Snyder S H. Proc Natl Acad Sci USA. 1994;91:3191–3195. doi: 10.1073/pnas.91.8.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson V L, Dawson T M, London E D, Bredt D S, Snyder S H. Proc Natl Acad Sci USA. 1991;88:6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dore S, Kar S, Quirion R. Neuroscience. 1997;78:373–383. doi: 10.1016/s0306-4522(96)00594-5. [DOI] [PubMed] [Google Scholar]
- 26.Chauveau J, Moule Y, Rouiller C. Exp Cell Res. 1956;11:317–321. doi: 10.1016/0014-4827(56)90107-0. [DOI] [PubMed] [Google Scholar]
- 27.Madden E A, Storrie B. Anal Biochem. 1987;163:350–357. doi: 10.1016/0003-2697(87)90235-1. [DOI] [PubMed] [Google Scholar]
- 28.Khan A A, Soloski M J, Sharp A H, Schilling G, Sabatini D M, Li S, Ross C A, Snyder S H. Science. 1996;273:503–507. doi: 10.1126/science.273.5274.503. [DOI] [PubMed] [Google Scholar]
- 29.Caron-Leslie L-A M, Evans R B, Cidlowski J A. FASEB J. 1994;8:639–645. doi: 10.1096/fasebj.8.9.8005391. [DOI] [PubMed] [Google Scholar]
- 30.Batistatou A, Greene L A. J Cell Biol. 1991;115:461–471. doi: 10.1083/jcb.115.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders P A, Ishitani R, Chalecka-Franaszek E, Chuang D-M. Soc Neurosci Abstr. 1996;22:1484. [Google Scholar]
- 32.Epner D E, Coffey D S. Prostate. 1996;28:372–378. doi: 10.1002/(SICI)1097-0045(199606)28:6<372::AID-PROS6>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Klausner R D, Rouault T A, Harford J B. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 34.Jaffrey S R, Cohen N A, Roualt T A, Klausner R D, Snyder S H. Proc Natl Acad Sci USA. 1994;91:12994–12998. doi: 10.1073/pnas.91.26.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronai Z. Int J Biochem. 1993;25:1073–1076. doi: 10.1016/0020-711x(93)90123-v. [DOI] [PubMed] [Google Scholar]
- 36.Cuervo A M, Dice J F. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 37.Rogalski A, Steck T L, Waseem A. J Biol Chem. 1989;264:6438–6446. [PubMed] [Google Scholar]
- 38.Huitorel P, Pantaloni D. Eur J Biochem. 1985;150:265–269. doi: 10.1111/j.1432-1033.1985.tb09016.x. [DOI] [PubMed] [Google Scholar]
- 39.Schlafer M, Volknandt W, Zimmermann H. J Neurochem. 1994;63:1924–1931. doi: 10.1046/j.1471-4159.1994.63051924.x. [DOI] [PubMed] [Google Scholar]
- 40.Cameron A M, Steiner J P, Roskams J A, Ali S M, Ronnett G V, Snyder S H. Cell. 1995;83:463–472. doi: 10.1016/0092-8674(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 41.Klauck T M, Faux M C, Labudda K, Langeberg L K, Jaken S, Scott J D. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 42.Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A. Nat Genet. 1996;13:196–202. doi: 10.1038/ng0696-196. [DOI] [PubMed] [Google Scholar]
- 43.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Wanker E E, Lehrach H, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]