Abstract
Background
Rieske non-heme iron aromatic ring-hydroxylating oxygenases (RHOs) are multi-component enzyme systems that are remarkably diverse in bacteria isolated from diverse habitats. Since the first classification in 1990, there has been a need to devise a new classification scheme for these enzymes because many RHOs have been discovered, which do not belong to any group in the previous classification. Here, we present a scheme for classification of RHOs reflecting new sequence information and interactions between RHO enzyme components.
Result
We have analyzed a total of 130 RHO enzymes in which 25 well-characterized RHO enzymes were used as standards to test our hypothesis for the proposed classification system. From the sequence analysis of electron transport chain (ETC) components of the standard RHOs, we extracted classification keys that reflect not only the phylogenetic affiliation within each component but also relationship among components. Oxygenase components of standard RHOs were phylogenetically classified into 10 groups with the classification keys derived from ETC components. This phylogenetic classification scheme was converted to a new systematic classification consisting of 5 distinct types. The new classification system was statistically examined to justify its stability. Type I represents two-component RHO systems that consist of an oxygenase and an FNRC-type reductase. Type II contains other two-component RHO systems that consist of an oxygenase and an FNRN-type reductase. Type III represents a group of three-component RHO systems that consist of an oxygenase, a [2Fe-2S]-type ferredoxin and an FNRN-type reductase. Type IV represents another three-component systems that consist of oxygenase, [2Fe-2S]-type ferredoxin and GR-type reductase. Type V represents another different three-component systems that consist of an oxygenase, a [3Fe-4S]-type ferredoxin and a GR-type reductase.
Conclusion
The new classification system provides the following features. First, the new classification system analyzes RHO enzymes as a whole. RwithSecond, the new classification system is not static but responds dynamically to the growing pool of RHO enzymes. Third, our classification can be applied reliably to the classification of incomplete RHOs. Fourth, the classification has direct applicability to experimental work. Fifth, the system provides new insights into the evolution of RHO systems based on enzyme interaction.
Background
Microorganisms play indispensable roles in the degradation and detoxification of polycyclic aromatic hydrocarbons (PAHs) in the environment [1,2]. The initiation of the aerobic microbial degradation of PAHs is an oxidative attack [3,4]. The enzymes that catalyze insertion of molecular oxygen into aromatic benzene rings are termed oxygenases [5]. They require transition metals, such as iron and heme, as catalytic centers. Oxygenases that utilize non-heme Fe(II) are called Rieske-type non-heme iron aromatic ring-hydroxylating oxygenase (RHO) whereas others that use heme are cytochrome P450s [6,7]. The term RHO is used herein to denote the Rieske-type non-heme iron ring-hydroxylating oxygenase.
Although RHOs mostly use NAD(P)H as an electron donor and catalyze the same oxygenation reaction, they are remarkably diverse with respect to their structure [3,4,8]. RHOs are multi-component enzymes of two or three protein components consisting of an electron transport chain (ETC) and an oxygenase. Oxygenase components are either homo- (αn) or hetero-oligomers (αnβn) and in each case, the α subunit, called large subunit, contains two conserved regions, a Rieske [2Fe-2S] center and non-heme mononuclear iron. The α subunits are known to be the catalytic components involved in the transfer of electrons to oxygen molecules. The ETC that transfers reducing equivalents from NAD(P)H to the oxygenase components consists of either a flavoprotein reductase or a flavoprotein reductase and a ferredoxin [3,4]. An interaction between oxygenase and ETC components is required for the enzyme system to transfer electrons from the electron donor to aromatic hydrocarbon electron acceptor. The RHO enzyme system has been extensively studied in many different microorganisms since the initial reaction mostly determines the aromatic substrate for degradation [9-15].
Classification of RHOs is essentially an effort to organize the information into a system that is useful for understanding the relationship between various aspects of sequence, structure, function and evolution. A three-class system (class I, II and III) was initially instituted by Batie et al. [16]. Based on the number of constituent components and the nature of the redox centers, this classification was able to give systematic information about RHOs. We will refer to this approach as "the traditional classification". With the recent tremendous accumulation of new sequence information on RHOs, there is a current need for a new classification strategy that can transform the multitude of complex data into useful organized information. In this regard, computational phylogenetic analysis of molecular sequence was imperative, which we term "the phylogenetic classification". Several challenges have been introduced using this method. Werlen et al. [17] grouped RHOs into four families based on substrate specificities and sequence alignments with associated distance calculations. This classification emphasized the structure-function relationship of the oxygenase component. However, some RHOs appear not to fit in this scheme probably because of the small RHO sample pool which resulted in the partial observation. Nam et al. [18] also proposed a clustering system based on the homology of the amino acid sequences of terminal oxygenase components. This classification system was more inclusive and well reflected the phylogenetic affiliation among RHOs.
In recent years, we characterized 3 oxygenases, NidAB, NidA3B3 and PhtAaAb, from M. vanbaalenii PYR-1 involved in the oxidation of aromatic hydrocarbons [9,10,15,19]. Phylogenetic analysis of these oxygenase components, NidA, NidA3 and PhtAa, showed that they clustered with a new group of α subunits found in Nocardioides, Rhodococcus, Terrabacter, Arthrobacter and other Mycobacterium spp. [20-25]. Interestingly, RHO genes found in this group of bacteria appear to have features in common; genes for ETC components are not always closely positioned with oxygenase genes (genetic discreteness) and limited numbers of ferredoxin and reductase components are shared by multiple oxygenases (numerical imbalance). Interestingly, ferredoxin components, that are compatible with the oxygenase enzymes from this bacterial group, were often identified to be a [3Fe-4S] cluster type [20,22,23]. The [3Fe-4S] type of ferredoxin has been recently introduced as an ETC component for RHO enzyme systems, the other ferredoxin components being the [2Fe-2S] type. In fact, NidAB and NidA3B3 from PYR-1 were also shown to be compatible with the [3Fe-4S]-type ferredoxin, PhtAc and ferredoxin reductase (PhtAd) [19]. However, the classification system proposed by Batie et al. and Nam et al. can not explain these new RHO features, the [3Fe-4S]-type ferredoxin, genetic discreteness of ETC components and the numerical imbalance between oxygenase and ETC component. In addition, considering that RHOs are multi-component enzyme systems, ETC components along with oxygenases components are also necessary for their functional understanding. Therefore, the new classification system was launched with the aim of analyzing the RHO components as a whole. The classification system not only systematically incorporates each component data but also basically reflects the previous "traditional" and "phylogenetic" classification.
Methods
Sequences retrieving
The amino acid sequences of RHO enzymes were retrieved by BLAST searches from public databases including NCBI nonredundant (NR) protein database [26]. From several hundreds of RHO enzymes that were recovered, we first ended up with the selection of 130 RHO enzymes. Among them, 25 "standard" RHO enzymes were chosen (Table 1) and used to construct the new classification system. These standard RHO enzymes are all well known with respect to genetics and enzyme functions. We also took sequence identities among RHO enzymes into consideration for the selection of standard RHO samples, in which highly identical RHOs were only once selected, so as for the samples to be a full representative of RHO enzymes. The size of the 25 standard RHOs as well as their quality as an RHO representative were statistically examined. To evaluate the new classification scheme, 38 RHO samples (Table 2) were selected as test enzyme systems. For some of these test enzymes, the genetic information for both the oxygenase and ETC components are available whereas for others only oxygenase information is available. To apply the new classification system, a total of 130 RHO enzymes, which include additional 67 RHO enzymes, were classified.
Table 1.
Standard RHO enzymes used in this study.
| Oxygenase | Ferredoxin | Reductase | ||||
| Standard RHO enzyme system | Gene | Prosthetic group | Prosthetic group | Prosthetic group | Structure | Accession number |
| Carbazole 1,9a-dioxygenase (P. resinovorans CA10) [44] | carAa | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | D89064 |
| Phenoxybenzoate dioxygenase (Alcaligenes sp. BR60) [45] | cbaA | [2Fe-2S]/Fe2+ | None | FMN/[2Fe-2S] | FNRC-type | U18133 |
| Phenoxybenzoate dioxygenase (P. pseudoalcaligenes POB310) [46] | pobA | [2Fe-2S]/Fe2+ | None | FMN/[2Fe-2S] | FNRC-type | X78823 |
| Phthalate dioxygenase (B. cepacoa DBO1) [13] | ophA2 | [2Fe-2S]/Fe2+ | None | FMN/[2Fe-2S] | FNRC-type | AF095748 |
| p-Toluenesulfonate monooxygenase (C. testosterone T-2) [47] | tsaM | [2Fe-2S]/Fe2+ | None | FMN/[2Fe-2S] | FNRC-type | U32622 |
| Aniline dioxygenase (Acinetobacter sp. YAA) [48] | atdA | [2Fe-2S]/Fe2+ | None | FAD/[2Fe-2S] | FNRC-type | D86080 |
| Aniline oxygenase (P. putida UCC22) [49] | tdnA1 | [2Fe-2S]/Fe2+ | None | FAD/[2Fe-2S] | FNRC-type | D85415 |
| 2-Halobenzoate 1,2-dioxygenase (P. cepacia 2CBS) [50] | cbdA | [2Fe-2S]/Fe2+ | None | FAD/[2Fe-2S] | FNRN-type | X79076 |
| Benzoate 1,2-dioxygenase (Acinetobacter. sp. ADP1) [51] | BenA | [2Fe-2S]/Fe2+ | None | FAD/[2Fe-2S] | FNRN-type | AF009224 |
| Anthranilate dioxygenase (Acinetobacter. sp. ADP1) [51] | antA | [2Fe-2S]/Fe2+ | None | FAD/[2Fe-2S] | FNRN-type | AF071556 |
| Phenanthrene dioxygenase (Nocardioides sp. KP7) [11] | phdA | [2Fe-2S]/Fe2+ | [3Fe-4S] | FAD | GR-type | AB017795 |
| Phthalate dioxygenase (Terrabacter sp. DBF63) [22] | phtA1 | [2Fe-2S]/Fe2+ | [3Fe-4S] | FAD | GR-type | AB084235 |
| Phthalate dioxygenase (A. keyseri 12B) [23] | phtAa | [2Fe-2S]/Fe2+ | [3Fe-4S] | FAD | GR-type | AF331043 |
| Phthalate dioxygenase (M. vanbaalenii PYR-1) [10] | phtAa | [2Fe-2S]/Fe2+ | [3Fe-4S] | FAD | GR-type | AY365117 |
| Phthalate dioxygenase (Rhodococcus sp. RHA1) [34] | padAa2 | [2Fe-2S]/Fe2+ | [3Fe-4S] | FAD | GR-type | AB154537 |
| 3,4-Dihydroxyphenanthrene dioxygenase (A. faecalis AFK2) [52] | phnAc | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | AB024945 |
| Naphthalene dioxygenase (Pseudomonas sp. 9816-4) [12] | nahAc | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | U49496 |
| PAH dioxygenase (P. putida OUS82) [53] | pahAc | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | AB004059 |
| Carbazole dioxygenase (Sphingomonas sp. CB3) [54] | carAa | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD | GR-type | AF060489 |
| Dioxin dioxygenase (Sphingomonas sp. RW1) [14] | dxnA1 | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD | GR-type | X72850 |
| Biphenyl dioxygenase (Rhodococcus sp. RHA1) [55] | bphA1 | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD | GR-type | D32142 |
| Toluene dioxygenase (P. putida F1) [56] | todC1 | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD | GR-type | J04996 |
| Biphenyl dioxygenase (Pseudomonas sp. KKS102) [57] | bphA1 | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD | GR-type | D17319 |
| Biphenyl 2,3-dioxygenase (B. xenovorans LB400) [58] | bphA | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD | GR-type | M86348 |
| Biphenyl dioxygenase (P. pseudoalcaligenes KF707) [59] | bphA1 | [2Fe-2S]/Fe2+ | [2Fe-2S] | FAD | GR-type | M83673 |
Table 2.
Test RHO enzymes used in this study.
| Oxygenase | Ferredoxin | Reductase | ||||
| Test RHO enzyme system | Gene | Prosthetic group | Prosthetic group | Prosthetic group | Structure | Accession number |
| PAH dioxygenase (M. vanbaalenii PYR-1) [9] | orf25 | [2Fe-2S]/Fe2 | unknown | unknown | unknown | AY365117 |
| nidA | [2Fe-2S]/Fe2 | unknown | unknown | unknown | AF249301 | |
| nidA3 | [2Fe-2S]/Fe2 | unknown | unknown | unknown | DQ028634 | |
| PAH dioxygenase (Mycobacterium sp. 6PY1) [25] | pdoA1 | [2Fe-2S]/Fe2 | unknown | unknown | unknown | AJ494745 |
| PAH dioxygenase (Mycobacterium sp. S65) [24] | pdoA | [2Fe-2S]/Fe2 | unknown | unknown | unknown | AF546905 |
| PAH dioxygenase (Rhodococcus strain I24) [21] | nidA | [2Fe-2S]/Fe2 | unknown | unknown | unknown | AF121905 |
| PAH dioxygenase (Pseudomonas abietaniphila BKME-9) [60] | ditA1 | [2Fe-2S]/Fe2 | [3Fe-4S] | unknown | unknown | AF119621 |
| PAH dioxygenase (N. aromaticivorans F199) [37] | xylX | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | AF079317 |
| bphA1a | [2Fe-2S]/Fe2 | |||||
| bphA1b | [2Fe-2S]/Fe2 | |||||
| bphA1c | [2Fe-2S]/Fe2 | |||||
| bphA1d | [2Fe-2S]/Fe2 | |||||
| bphA1e | [2Fe-2S]/Fe2 | |||||
| bphA1f | [2Fe-2S]/Fe2 | |||||
| PAH dioxygenase (Sphingomonas sp. P2) [36] | ahdA1a | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | AB091693 |
| ahdA1b | [2Fe-2S]/Fe2 | |||||
| ahdA1c | [2Fe-2S]/Fe2 | |||||
| ahdA1d | [2Fe-2S]/Fe2 | |||||
| ahdA1e | [2Fe-2S]/Fe2 | |||||
| xylX | [2Fe-2S]/Fe2 | |||||
| PAH dioxygenase (Sphingomonas sp. CHY-1) [42] | phnA1a | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | AJ633551 |
| phnA1b | [2Fe-2S]/Fe2 | |||||
| PAH dioxygenase (Cycloclasticus sp. A5) [43] | phnA1 | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | FNRN-type | AB102786 |
| Aniline dioxygenase (D. acidovorans 7N) [61] | orf7NC | [2Fe-2S]/Fe2 | None | FAD/[2Fe-2S] | FNRC-type | AB177545 |
| Phthalate dioxygenase (P. putida) [62] | pht3 | [2Fe-2S]/Fe2 | None | FMN/[2Fe-2S] | FNRC-type | D13229 |
| Vanillate O-demethylase oxygenase (Pseudomonas sp. HR199) [63] | vanA | [2Fe-2S]/Fe2 | None | FMN/[2Fe-2S] | FNRC-type | Y11521 |
| Anthranilate 1,2-dioxygenase (P. resinovorans CA10) [64] | antA | [2Fe-2S]/Fe2 | None | FAD/[2Fe-2S] | FNRN-type | AB047548 |
| Naphthalene dioxygenase (Ralstonia sp. U2) [38] | nagAc | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | AF036940 |
| Salicylate 5-hydroxylase (Ralstonia sp. U2) [65] | nagG | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | AF036940 |
| Salicylate 5-hydroxylase (P. aeruginosa JB2) [66] | hybB | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | AF087482 |
| Naphthalene dioxygenase (P. aeruginosa Pak1) [67] | pahA3 | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | D84146 |
| Carbazole 1,9a-dioxygenase (Pseudomonas sp. XLDN4-9) | carAa | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD/[2Fe-2S] | FNRN-type | DQ060076 |
| PAH dioxygenase (R. erythropolis TA421) [68] | bphA1 | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | D88021 |
| Chllorobenzene dioxygenase (Pseudomonas sp. P51) [17] | tcbAa | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | U15298 |
| Cumene dioxygenase (P. fluorescens IP01) [69] | cumA1 | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | D37828 |
| p-Cumate dioxygenase (R. palustris No.7) [70] | psbAc | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | AB022919 |
| p-Cumate dioxygenase (P. putida F1) [71] | cmtAb | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | U24215 |
| Anthranilate 1,2-dioxygenase (B. cepacoa DBO1) [72] | andAc | [2Fe-2S]/Fe2 | [2Fe-2S] | FAD | GR-type | AY223539 |
Sequence analysis
ClustalX [27] was used to obtain multiple alignments followed by distance calculations for each RHO component. Pairwise and multiple alignments were carried out with the default parameters. The weight matrix, Gonnet 250, was used to obtain the pairwise distance (PD) matrix. Alignments were visualized and examined using GENEDOC [28]. Phylogenetic trees were constructed by NJ method [29] from distance data and visualized and manipulated with TREEVIEW 1.6.6 [30]. Tree was also generated using iTOL: Interactive Tree Of Life, an online phylogenetic tree viewer and Tree Of Life resource [31]. The reliability of the trees obtained was evaluated by 1000 bootstrap replications. The domain structures for reductase components were analyzed using the Conserved Domain (CD) Database of NCBI [32], which compares primary sequences with all of the known domain structures in the databases.
Statistical analysis
Matlab™ (The Mathworks, Inc.) was used to implement the proposed algorithm and to perform the simulation. The program under Matlab generates random subsets among 43 RHO samples, performs cluster analysis using the pairwise distance (PD) matrices and obtains the PD value which maximizes the classification accuracy. In the program, ClustalX [27] was used to obtain multiple alignments and PD matrices. The total running time spent for the simulation used in this paper is less than an hour and the matlab source code (.m) of the program is available upon request.
Results
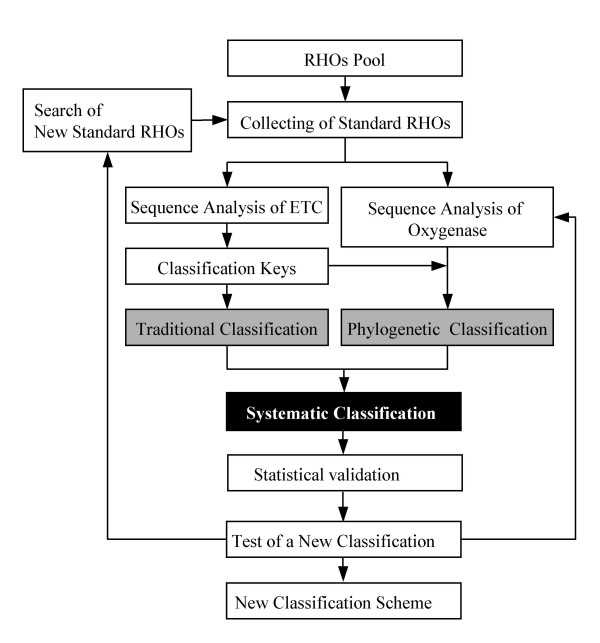
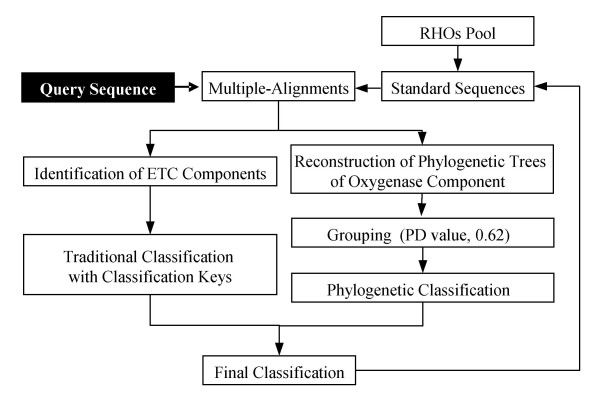
Construction of the new classification scheme was accomplished as shown in the flowchart (Figure 1). Briefly, we first analyzed a total of 130 RHO enzymes in which 25 well-characterized RHO enzymes were selected as standards to test our hypothesis for the proposed classification system (Table 1). From the sequence analysis of ETC components of the standard RHO samples, we extracted the classification keys that reflect the phylogenetic relationship among ETC components, which turned into the grouping criteria in the following classifications. Next, oxygenases were phylogenetically classified with the classification keys obtained from ETC analyses. Finally we constructed the new systematic classification through statistical justification and tested its applicability with 38 RHO samples.
Figure 1.
Flowchart for a new classification scheme.
Step 1: Sampling standard RHOs
Table 1 shows the list of the 25 standard RHO samples whose genetic and functional information were completely available. It includes the members of Batie's three classes and Nam's four groups [16,18] and others that are not mentioned in those classifications. They are phenanthrene dioxygenase of Nocardioides sp. KP7 [11] and phthalate dioxygenases of Terrabacter sp. DBF63 [33], Arthrobacter keyseri 12B [23], M. vanbaalenii PYR-1 [10] and Rhodococcus sp. RHA1 [34]. The ferredoxin components of these RHO enzymes are the [3Fe-4S] type ferredoxin; only [2Fe-2S] type ferredoxin components were analyzed in the previous classification.
Step 2: Sequence analysis of ETC components from standard RHO samples
In step 2, phylogenetic information was obtained from the amino acid sequences of RHO ETC components, which in turn were converted into classification keys for grouping of oxygenase components for step 3 analyses.
Reductase components
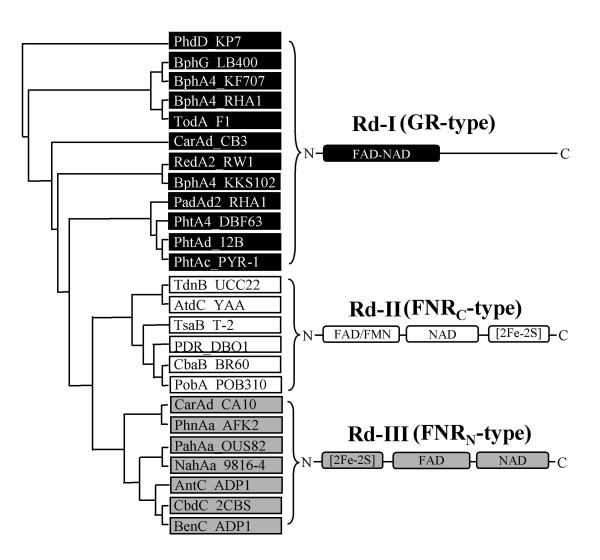
The phenogram with the domain arrangements of reductases is presented in Figure 2. The 25 reductases were divided into three groups, Rd-I, Rd-II and Rd-III, based on the conserved domain arrangements. When grouped with PD value of 0.85, the 25 reductases can also be grouped into the same three groups. Therefore, the PD values obtained by using Gonnet weight matrix were less than 0.85 within each group which has the same arrangement of conserved domains. Group Rd-I consists of glutathione reductase (GR) type reductases that show over 28% amino acid identity to one another, while groups Rd-II and Rd-III include the ferredoxin-NADP+ reductase (FNR) type reductases that show over 15 and 23% amino acid identity within each group, respectively. Groups Rd-II and Rd-III share the same three domains for flavin, NAD and [2Fe-2S] binding, but show different domain arrangements. In group Rd-II, the [2Fe-2S] ferredoxin domains are connected to the C-terminus of NAD domains, but to the N-terminus of flavin-binding domains in group Rd-III. The overall degree of sequence identity between the Group Rd-II and Rd-III is generally no more than 14%. Accordingly, group Rd-I, Rd-II and Rd-III are designated as GR-type, FNRC-type and FNRN-type reductases, respectively, and were selected as classification keys for the reductase components of RHO enzymes.
Figure 2.
Grouping of reductase components from 25 standard RHO enzymes with schematic representation of the conserved domain structures. The names of bacterial strains are indicated after the enzyme names. GR-type, FNRC-type, and FNRN-type reductases are shown in the boxes with black, gray, and white background, respectively. Designations: FAD-Flavin adenine dinucleotide; NAD-Nicotinamide adenine dinucleotide.
Ferredoxin Components
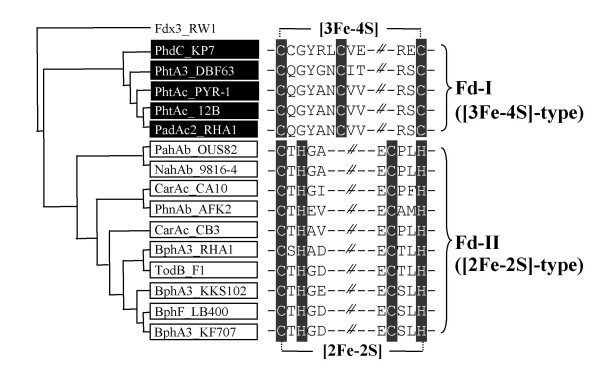
Figure 3 shows the result of phylogenetic analysis for the ferredoxin components of standard RHO systems. Conserved amino acid residues were also shown to reveal the classification keys for ferredoxins. For the sequence alignment of ferredoxins, we initially aligned [2Fe-2S]-type and [3Fe-4S]-type ferredoxins separately and identified conserved amino acids. Multiple sequence alignment for the all ferredoxin sequences was then performed, from which we evaluated the validity of alignment. The tree shows that the 16 ferredoxins are divided into two distinct groups based on the type of iron-sulfur clusters, designated group Fd-I and Fd-II. The PD values within each group were less than 0.716. While the overall degree of amino acid sequence identity between groups is no more than 20%, members in each group show over 29% sequence identity to one another. In this analysis, Fdx3, a putidaredoxin-type ferredoxin isolated from Sphingomonas wittichii RW1, was used as an outgroup because Fdx3 is phylogenetically unrelated to those of RHO enzymes [35]. The group Fd-I comprises the [3Fe-4S] cluster-containing ferredoxins. Sequence alignments of this group revealed three conserved cysteine residues which serve as ligands for the [3Fe-4S] cluster. Group Fd-II consists of the [2Fe-2S] cluster-containing ferredoxins containing a highly conserved [2Fe-2S]-binding motif, CXHXnCX2H. Accordingly, group Fd-I and Fd-II are referred as [3Fe-4S]- and [2Fe-2S]-type ferredoxins, respectively, which in turn were selected as classification keys for ferredoxin components of RHOs.
Figure 3.
Grouping of ferredoxin components from standard RHO enzymes with the conserved sequence alignments. The amino acid residues involved in binding to [2Fe-2S] (CXHXnCXXH) and [3Fe-4S] (CX5CXnC) cluster are indicated by highlighted characters. Fdx3 was used as an outgroup. The names of bacterial strains are indicated after the enzyme names.
Step 3: Sequence analysis and grouping of oxygenase components from standard RHO samples
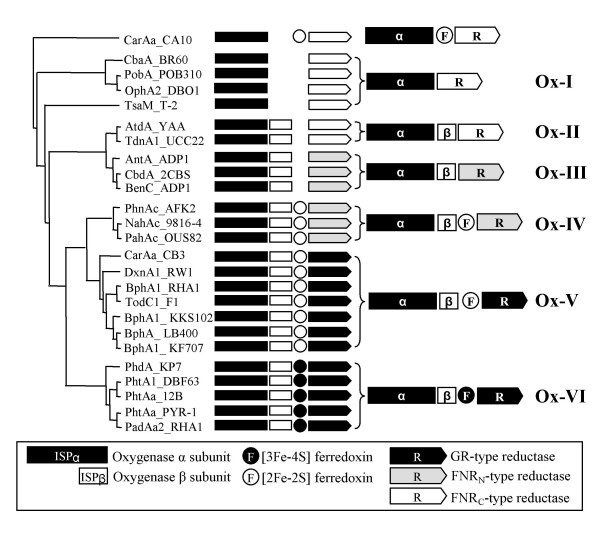
This step involves integration of the phylogenetic data of oxygenase components with respect to the classification keys obtained from ETC components. Figure 4 shows the dendrogram of 25 oxygenase components of standard RHO samples by the neighbor-joining (NJ) approach with CarAa from Pseudomonas resinovorans CA10 as an outgroup. The 25 oxygenase components can be clustered into two groups of homo (αn) and hetero (αnβn) oligomers (Figure 4). The homo-oligomer oxygenases include CbaA from Alcaligenes sp. BR60, PobA from Pseudomonas pseudoalcaligenes POB310, OphA2 from Burkholderia cepacia DBO1, TsaM from Comamonas testosteroni T-2 and CarAa from the strain CA10. However, when grouped with the classification keys from the phylogenetic analysis of ETC components, the 25 oxygenases are clustered into six distinct groups, designated Ox-I (an FNRC-type reductase and a homo-oligomer oxygenase (αn)), Ox-II (an FNRC-type reductase and a hetero-oligomer type oxygenase (αnβn)), Ox-III (an FNRN-type reductase and an oxygenase (αnβn)), Ox-IV (a hetero-oligomer oxygenase (αnβn), a [2Fe-2S]-type ferredoxin and an FNRN-type reductase), Ox-V (a hetero-oligomer oxygenase (αnβn), a [2Fe-2S]-type ferredoxin and a GR-type reductase) and Ox-VI (a hetero-oligomer oxygenase (αnβn), a [3Fe-4S]-type ferredoxin and a GR-type reductase) (Figure 4). Groups Ox-I, Ox-II and Ox-III consist of two-component RHO systems, while groups Ox-IV, Ox-V and Ox-VI include three-component RHO systems.
Figure 4.
Phylogenetic clustering of oxygenase components from standard RHO enzymes with regard to the type of their ETC components. The tree was constructed by the NJ method with CarAa from P. resinovorans CA10 as an outgroup.
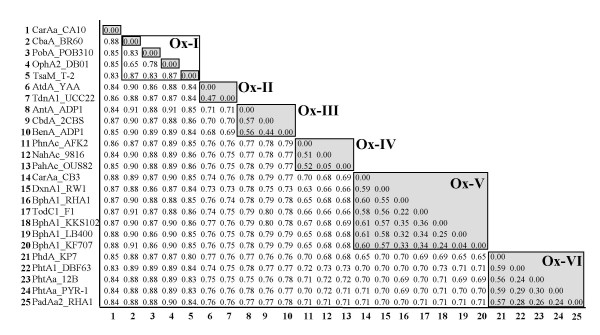
In Figure 5, the PD values within each group that shares the same classification keys were less than 0.61 with the exception of group Ox-I. This group can be further divided into four subgroups if using 0.61 as a PD value, where 25 oxygenases can be clustered into 10 groups including the outgroup of CarAa from CA10. Therefore, the PD value, 0.61, is a suitable criterion for grouping oxygenases based on our classification keys.
Figure 5.
PD matrix of oxygenase components from standard RHO enzymes by Gonnet 250 weight matrix. The PD value for grouping in Figure 4 is 0.61. Each group is shown in shadow box with the exception of Ox-I. The group Ox-I can be further divided into four subgroups at the PD value of 0.61.
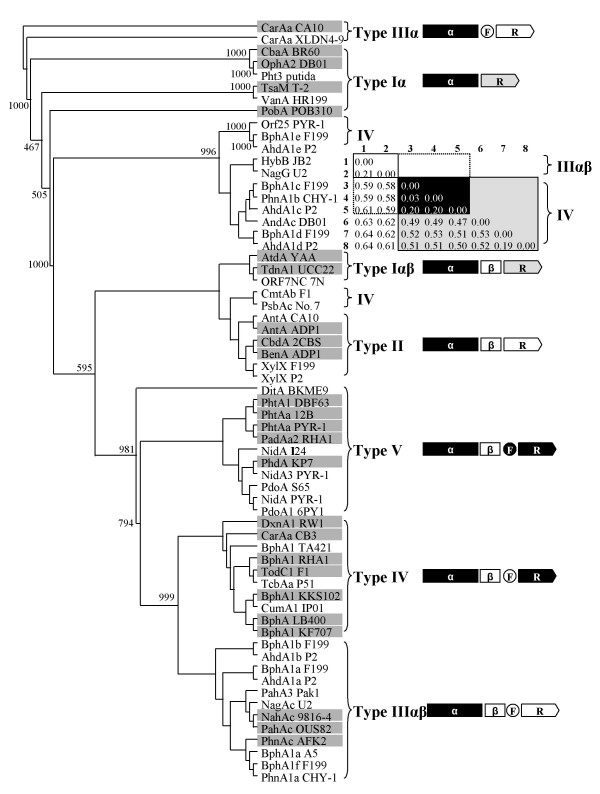
Step 4: Construction of the new systematic classification
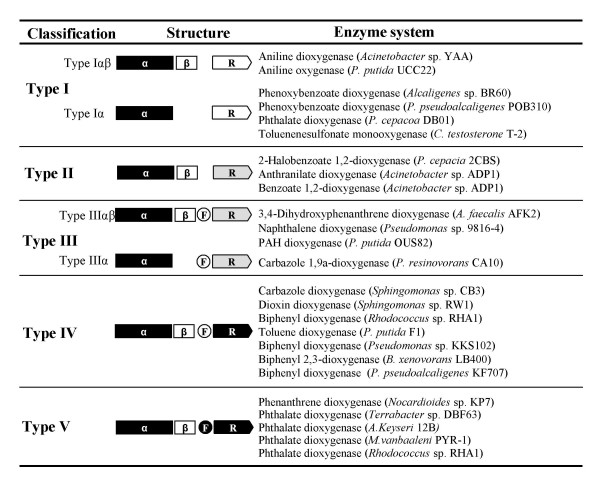
In this step, the integrated classification data were converted to a systematic classification. As shown in Figure 6, the standard 25 RHO samples can be organized into 5 distinct types from 10 groups which clustered at the PD value of 0.61, designated type I, II, III, IV and V. All the members within each type share the same classification keys of ETC components. Type I and III can be further divided into two subtypes, being designated αβ and α according to the type of oxygenase components in the RHO system; oxygenase components in the subtype αβ and α are hetero-oligomers (αnβn) and homo-oligomers (αn), respectively.
Figure 6.
A systematic classification scheme for standard RHO enzymes. Schematic diagram of the structures of RHO systems are as in Figure 4.
The type I system represents two-component RHOs that consist of an FNRC-type reductase and an oxygenase, which is either hetero-oligomer (type Iαβ) or homo-oligomer (type Iα). The RHO systems of group Ox-I and Ox-II belong to the type Iαβ and type Iα, respectively. Type II represents the other two-component systems that consist of an oxygenase and a FNRN-type reductase. The RHO enzymes in group Ox-III fall into type II. The well-characterized system for type II is benzoate 1,2-dioxygenase (BenABC) from Acinetobacter sp. ADP1. Type III represents three-component systems that consist of an oxygenase, a [2Fe-2S]-type ferredoxin and an FNRN-type reductase. All of group Ox-IV belong to type IIIαβ. The carbazole 1,9a-dioxygenase (CarAaAcAd) from P. resinovorans CA10 used as an outgroup for reconstructing the dendrogram of oxygenase components was determined to be type IIIα. The type IV systems represent another three-component systems that consist of an oxygenase, a [2Fe-2S]-type ferredoxin and a GR-type reductase. Type IV was shown to be the biggest group for the known RHO enzyme systems. Type V represents another different three-component systems that consist of an oxygenase, a [3Fe-4S]-type ferredoxin and a GR-type reductase. The RHO samples of the group Ox-VI were belonged to type V. The well-known example of the type V enzyme is phenanthrene dioxygenase (PhdABCD) from Nocardioides sp. KP7, in which the PhdC was the first [3Fe-4s]-type ferredoxin to be found in RHO enzyme systems.
Step 5: Statistical justification of the new systematic classification
In this step, we statistically examined the classification scheme; the proposed PD value and the size and quality of the standard RHOs were evaluated using in-house program. Here, the learning accuracy is defined as the ratio of the number of correctly classified observations and the total number of observations.
First, we evaluated which PD value maximizes the classification accuracy for the 25 standard RHO set by the following algorithm:
INPUT: N oxygenases
(1) Select randomly four key oxygenases a2, a3, a4 and a5 from each categories II, III, IV, V respectively.
(2) At each of the N-1 steps p0, p1, ..., pn, the closest two clusters are merged into a single cluster.
(3) Define cut-off distances as qi = (pi-1 + pi)/2 for each i = 1,..., n. For each qi, a cluster is classified as category I, if there is no a2~a5 in the cluster. If one or more of ai's (i = 2, ..., 5) are present in the cluster, the cluster is classified randomly as one of their category. Calculate the learning accuracy and find the cutoff which maximizes the learning accuracy.
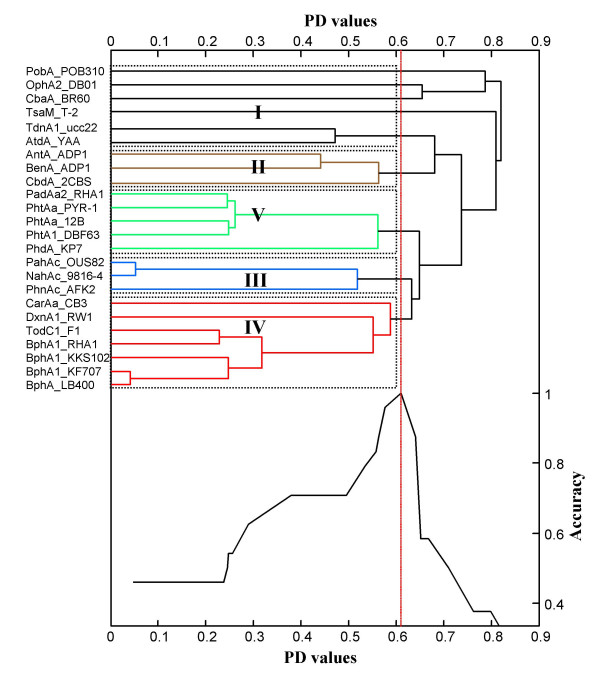
Figure 7 is an output data showing the relations among learning accuracy, PD value and RHO classification result deduced from the 25 standard RHOs. This data indicates clearly that the PD value 0.611 satisfies demand for the classification of 25 standard RHO set with a maximum learning accuracy 1.0.
Figure 7.
Accuracy plot and dendrogram of phylogenetic clustering of oxygenase components from standard RHO enzymes using the proposed classification algorithm. The right vertical axis represents the accuracy of classification based on clustering with the corresponding PD values on the horizontal axis. The labels along the left vertical axis display 25 standard RHO enzymes. At PD value of 0.61 (dashed vertical line), the standard enzymes have the maximum accuracy of 1.0.
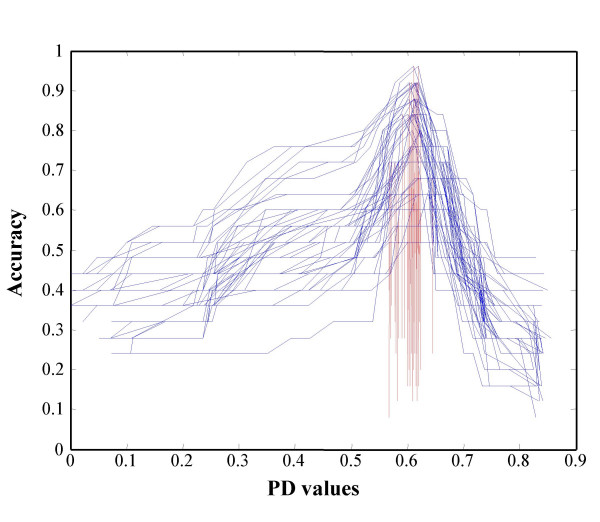
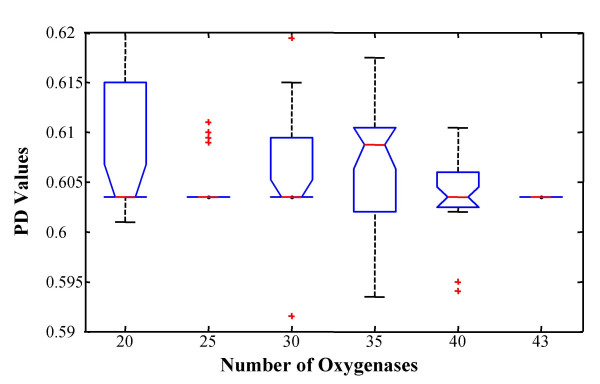
Based on the estimation of PD value, we devised a simulation to determine the suitable set of oxygenases which may minimize the classification error during the prediction. For the purpose of simulation, we used a total of 43 oxygenases which includes 25 standard and 18 test RHO samples. The training sets of 20, 25, 30, 35 and 40 RHOs were randomly selected from a total of 43 oxygenases for 50 times for each training set. Then, we evaluated the PD values which attain the maximum learning accuracy using the above algorithm for each set. Table 3 indicates that the mean deviation of such PD values is close to 0.61 and the values are not much affected by the number of observations, although mean PD values diminishes as the number of oxygenases increases. Figure 8 shows the simulation result based on 50 repetitions for 25 randomly selected RHOs, which indicates that the maximum accuracy is obtained at the mean PD value of 0.619. In Figure 9, it was further shown that the PD values of 25 oxygenases are most stable except for the set of entire 43 oxygenases, although there are some outliers which increase the variance. This simulation test can give us reliability of the 25 standard RHO set and PD value 0.611 for our classification.
Table 3.
Mean and standard deviation of PD values for randomly selected 20, 25, 30, 35, 40, and 43 oxygenases based on 50 repetitions.
| Number of oxygenases | Mean PD value | Standard deviation |
| 20 | 0.6096 | 0.0191 |
| 25 | 0.6119 | 0.0146 |
| 30 | 0.6088 | 0.0117 |
| 35 | 0.6036 | 0.0137 |
| 40 | 0.6024 | 0.0096 |
| 43 | 0.5980 | 0.0105 |
Figure 8.
Accuracy plot using the proposed classification algorithm based on 50 repetitions of 25 randomly selected RHOs. The vertical axis represents the accuracy and the horizontal axis represents the PD values. Blue solid lines represent the accuracy of classification based on clustering with the corresponding PD values on the horizontal axis. Dashed vertical lines indicate the PD values at which the maximum accuracy is obtained for each repetition.
Figure 9.
Box and whisker plot of PD values for randomly selected 20, 25, 30, 35, 40, and 43 oxygenases based on 50 repetitions. The box has lines at the lower quartile, median, and upper quartile values. Outliers are displayed with a red + sign.
Step 6: Test of the new classification systems
The reliability of the new classification system was evaluated by examining its applicability and usefulness in the classification of 38 RHO enzymes that ranged broadly over the various type of RHO enzymes (Table 2). The selection criteria are as follows. At first, 14 complete RHO enzymes for which both genetic and functional information were well-known were selected. They were chosen for the purpose of verification of the classification systems, which are the same case as those standard RHO samples. Next, the 24 incomplete RHO enzymes were further selected. These incomplete samples include some of the RHO enzymes which have been functionally characterized by gene complementation with compatible source of ETC components and others that have other known equivalent homologs from which ETC information could be deduced.
Figure 10 shows the strategy used for the classification of test RHO enzymes. Basically, the strategy depends on whether RHOs sequence information is limited. In the case of complete RHO samples, query sequences were first subjected to multiple alignments followed by systematic classification; oxygenase components of RHOs undergo a phylogenetic classification whereas ETC sequences go through traditional classification route which are lastly consolidated into the final classification. In contrast, when analyzing incomplete RHOs, due to the lack of sequence information of the ETC component, only the oxygenase part can be classified according to the phylogenetic analysis. One of the merits of this classification strategy is that it responds dynamically to the growing pool of RHO enzymes. As shown in Figure 10, the output (final classification) of the classification system is returned to increase the standard sequence pool. As the standard RHO pool is growing, the classification gets more stable and objective and could possibly expand its coverage for many other RHO enzymes.
Figure 10.
Strategy for query sequence classification used in this study.
Figure 11 shows the result of systematic/phylogenetic analysis for the classification of a total of 63 RHOs which comprised 25 standard and 38 test enzyme samples. This tree has a similar branching structure to that of the standard RHOs samples (Figure 3). It also mirrors well our classification scheme. When analyzing complete test RHO enzymes, the phylogenetic classification results of oxygenase components were in agreement with those of the traditional classification of ETC components. That is to say, ETC sequences of these complete RHO samples were identified by the classification keys to be the same types to which the respective oxygenase components were assigned. For example, both oxygenase (PahA3) and ETC components (PahA1A2) from P. aeruginosa Pak1 naphthalene dioxygenase were shown to be typed IIIA by phylogenetic and traditional classification, respectively. It indicates that our systematic classification reflects exactly the relationship/partnership between oxygenase and ETC, which strongly demonstrates that our classification scheme has a potential to classify incomplete RHOs.
Figure 11.
The resultant tree of the systematic/phylogenetic analysis for 63 RHO enzymes and PD matrix of the type IIIαβ group (HybB and NagG) and type IV group (BphA1c, PhnA1b, AhdA1c, BphA1d, and AhdA1d). The standard RHO enzymes are shown in shadow box. Schematic diagram of the structures of RHO systems are as in Figure 4. The tree was constructed by the NJ method with CarAa from P. resinovorans CA10 as an outgroup.
The 24 incomplete test RHO samples were all classified and assigned to each type by phylogenetic analysis. For instance, NidA and NidA3 from M. vanbaalenii PYR-1 were clustered together with phenanthrene dioxygenase (PhdABCD) from Nocardioides sp. KP7 being allocated to type V with a high bootstrap score and stable PD support. Oxygenases from Novosphingobium aromaticivorans F199 (BphA1a, BphA1b and BphA1f), Sphingomonas sp. P2 (AhdA1a and AhdA1b), Cycloclasticus sp. A5 (PhnA1) and Sphingomonas sp. CHY-1 (PhnA1a) were clustered into type IIIαβ with high bootstrap and stable PD values. In case of NagG from Ralstonia sp. U2, it was classified as type III with the complete RHO, HybB, from P. aeruginosa JB2. Another incomplete RHO samples, BphA1c and BphA1d from the strain F199, AhdA1c and AhdA1d from P2 and PhnA1b from CHY-1 were clustered as type IV together with the complete RHO, AndAc, from DBO1. Interestingly, BphA1c from the strain F199, AhdA1c from P2 and PhnA1b from CHY-1 of type IV can also be grouped into type IIIαβ with HybB from JB2 (2-hydroxybenzoate 5-salicylate hydroxylase) based on the PD value of 0.61 (Figure 8). This means that those three type IV RHOs are in the intersection with type IIIαβ. Both XylX oxygenase components from the strain F199 and P2 were grouped into the type II. Notably, ORF25, BphA1e and AhdA1e from the strain PYR-1, F199 and P2, respectively, formed a single distinctive separate cluster with a stable PD value; no standard or complete RHO is clustered together. In this case, although the genetic information of ETC for these three oxygenases are lacking, they were tentatively classified as type IV since BphA1e from the strain P2 was shown to be functionally active when complemented with the ETC component from type IV [36]. In fact, if all the classification keys are combined, we would be able to have 9 possible types for both two- and three-component systems. However, only 5 types were actually applied to classify RHO enzymes and we would need type X (alphabet) for the temporary classification of incomplete RHOs which do not have any standard RHO for comparison.
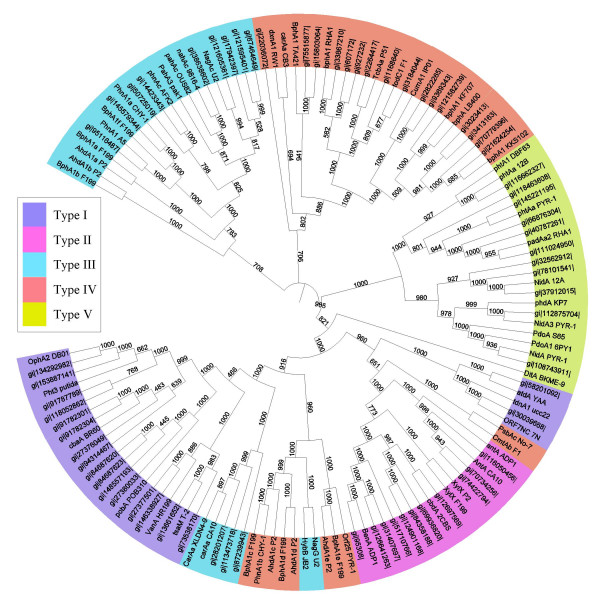
Step 7: classification of a total of 130 RHO enzymes
As a final, a total of 130 RHO enzymes, which include 67 new RHO sequences, were classified using our classification system. Figure 12 shows an output based on the new classification system, which has a similar branching structure to that of the test set shown in step 6. Although RHO enzymes seem to have a grouping tendency according to their substrate specificities, the classification clearly indicates that the phylogenetic affiliations of RHO enzymes are determined mainly by their relationship with ETC components. For example, as shown in Type V, PhtAa (phthalate dioxygenase) and NidA3 (fluoranthene dioxygenase) from PYR-1 are grouped together. Although their substrates have been reported to be different, their ETC components have been experimentally shown to be the same as [3Fe-4S]-type ferredoxin and GR-type reductase [9,10,15,19]. In this respect, the results presented in Figure 12 well reflects the phylogenetic affiliations which are integrated with the interactions between RHO enzyme components.
Figure 12.
Classification of a total of 130 RHO enzymes. Some RHO sequences are provided with gi numbers whose information can be found in NCBI database.
Discussion
In many cases, RHO oxygenases have not been identified together with ETC components, thus it has been a challenging task to find a cognate ETC partner that can function in cooperation with these "incomplete" oxygenase components [9,35-39]. Since some ETCs are interchangeable with each other and can be often replaced by outside ETC sources and it is the terminal oxygenase component that has the catalytic active site, less emphasis was given to ETC components in Nam's classification. For example, DitA from P. abietaniphila BKME-9 and NidA from Rhodococcus sp. strain I24 were clustered in group III together with NahAc from Pseudomonas sp. NCIB9816-4 and PahAc from P. putida OUS82. However, these two oxygenases, DitA and NidA, which lacked ETC information (only ferredoxin for DitA is known), initially formed a group different from that of NahAc and PahAc in the phylogenetic classification route, being classified as type V that represents the three-component systems consisting of an oxygenase, a [3Fe-4S]-type ferredoxin and a GR-type reductase. Whereas the NahAc and PahAc were classified as type III which consists of an oxygenase, a [2Fe-2S]-type ferredoxin and an FNRN-type reductase. The discrepancy between these two classification's results comes from ETC information that reflects the relationship among oxygenases. The classification in Nam's scheme was based on pairwise sequence homology of oxygenase components, in which four percentage sets, 0–14, 15–24, 25–34 and 35–99%, were used. In the new classification, however, the grouping criterion for oxygenase components, which is a PD value of 0.62, was objectively determined based on ETC information. This PD value finally determines the two oxygenases, DitA and NidA, differently from the NahAc and PahAc. The results clearly show that information on oxygenases whose ETC components are not available can be used to draw ETC data in our classification system.
This new classification system provides insights into the evolutionary changes and relationships between oxygenase and ETC components. Seven oxygenase components, BphA1 [a-f] and XylX from N. aromaticivorans F199 [37] are of particular interest in this context. Although other ETC components are maybe involved, those oxygenases from F199, which were found to be scattered throughout several gene clusters, seem to function with a limited number of ETC components. Two ferredoxins (Rieske- [2Fe-2S]-type BphA3 and plant- [2Fe-2S]-type XylT) and two reductases (GR-type BphA4 and FNR-type XylM) have been identified on the aromatic catabolic plasmid (pNL1) of F199. These oxygenases were classified as type II (XylX), IIIA (BphA1 [a,b,f]) and IV (BphA1 [c-e]) (Figure 8). Although there is no functional data available directly from the strain F199, genetic information with functional evidence have been reported from other sphingomonads which matches with those of F199. For example, sets of genes for an oxygenase from Sphingobium sp. P2 (AhdA1 [a-e]) [36], S. yanoikuyae B1 (BphA1c and XylX) [40,41] and Sphingobium sp. CHY-1 (PhnA1a and PhnA1b) [42] were reported to have the same gene arrangement with 63–97% sequence identity when compared to F199. Consequently, they were classified as the same types with those from F199; type II for XylX (B1), type IIIαβ for AhdA1 [a,b] (P2), PhnA1a (CHY-1) and type IV for BphA1c (B1), AhdA1 [c-e] (P2) and PhnA1b (CHY-1). We found that, without an exception, they were experimentally shown to use type IV ETC (all named as BphA3A4) that consists of [2Fe-2S]-type ferredoxin and GR-type reductase (Figure 6). These ETCs are highly homologous (> 79%) to the BphA3A4 from F199. Therefore, it is reasonable to predict that the 7 oxygenase components (BphA1 [a-f] and XylX) from F199 might be sharing the same type of ETC, BphA3A4. This implies that the limited ETC (BphA3A4) probably makes for strong selective pressure, favoring oxygenase components that can rapidly adapt by increasing their tolerance toward alternative ETC. This adaptation should be accompanied by the change of genetic information, which suggesting that RHO enzymes are probably evolving under the direction of selective pressure derived from ETCs. In fact, although BphA1c from F199 was clustered in type IV, it intersected with type III, the group of 2-hydroxybenzoate 5-salicylate hydroxylase (HybABCD) from P. aeruginosa JB2 (Figure 8). It suggests that BphA1c from F199 is probably evolving under the selective pressure derived from limited ETC. The XylX from the strain B1 is also worth noting, XylX belongs to type II, the two-component RHO enzymes, which normally functions with an FNR-type reductase. However, it was unexpectedly shown to be compatible with BphA3A4 [40,41]. Although both ferredoxins from type III and IV are [2Fe-2S]-type, the ferredoxin component (BphA3) from B1 seems phylogenetically inclined to be branched with ferredoxins which can complement oxygenases belong to type IIIαβ. The inclination of the ferredoxin component (BphA3) toward type IIIαβ also can be seen in the RHO enzyme from Cycloclasticus sp. A5, of which PhnA1 and PhnA3 components showed high identities over 62% with those (BphA1f and BphA3, respectively) from the strain F199 [43]. In this case, the oxygenase (PhnA1) belongs to type IIIαβ with the ETC components consist of a [2Fe-2S]-type ferredoxin (PhnA3) and an FNRN-type reductase (PhnA4). Taken together, the 7 oxygenases from F199, belonging to type II, IIIA and IV, were classified by our system to work with the three different ETC groups, FNRN-type reductase, [2Fe-2S]-type ferredoxins/FNRN-type reductase and [2Fe-2S]-type ferredoxins/GR-type reductase, respectively. However, all those oxygenases are assumed to function with [2Fe-2S]-type ferredoxin and GR-type reductase. From these observations question may arise about the degree of specificity in recognition between redox partners.
RHO pool is believed to respond dynamically to the environmental transitions such as substrate changes. In addition, relative tolerance between oxygenase and ETC components is also thought to be one of the important selective forces in evolution. In this context, even though little is known as to the role of ferredoxin component under selective pressure, it seems likely to be deeply involved in evolutionary changes. A possible benefit for using ferredoxin comes from its potential to promptly enhance the tolerance toward new redox partners. Cognizant of "the shorter, the faster and more effective", because of the relatively short sequence and simple structure of ferredoxin than that of reductase component, it has an evolutionary merit to adapt rapidly to dynamic environments. Hence, it might be an attractive hypothesis that ferredoxin has been evolutionarily chosen as a buffer between reductase and oxygenase component for rapid adaptation toward selective force. It might also be a strong driving force to affect the direction of evolution from two-component RHO to/toward three-component. Considering this two-way communication flow in RHO system, the adoption of ferredoxin as a new redox partner must have led to the changes in other partners, reductase and oxygenase, in RHO system. In this context, type III RHO seems likely to be a living fossil that has recorded evolutionary changes because they show transitional properties from two-component to three-component system. Two-component systems for type I and II share FNR-type reductase, FNRC- and FNRN-type, respectively, which consist of three domains (flavin, NAD and [2Fe-2S] binding). On the other hand, three-component systems for type IV and V, except for type III, share a GR-type reductase, which have no [2Fe-2S] cluster (Figure 2 and Figure 6). A [2Fe-2S] cluster of reductases separates the two-component system (type I and II) from the three-component system (type IV and V). It strongly demonstrates that the joining of ferredoxin to the two-component RHO system was probably the start of the type III three-component structure. During evolution, the type III FNRN-type reductases might have been gradually changed toward type IV and V GR-type reductases, although it is not sure whether it was recruited from other systems. In some cases, it might also be possible under extreme conditions that type I and II RHO systems evolved directly to type IV (or V) as seen in the XylX from S. yanoikuyae B1. Taken together, it may be postulated that RHOs are evolving under selective pressures from type I and II (two-component system) toward type IV and V (three-component), in which this change moves toward efficiency and keeps going continuously under dynamic environments.
Conclusion
In this study, we have developed a new classification system to classify bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. The new classification system presented in this study not only reflects sequence information but also interactions between RHO enzyme components. The system is characterized by the features that include the following. First, the new classification system analyzes RHO enzymes as a whole, in which information on each RHO components is organized into a system that is useful for the understanding of various aspects with respect to sequence, structure and biochemical function. Second, our new classification system is not static but responds dynamically to the growing pool of RHO enzymes. As standard RHO samples increase, the classification system evolves, which gets more stable and objective and could increase its coverage of many other RHO enzymes. Third, our classification can be applied reliably to the classification of incomplete RHOs. Fourth, the classification has direct applicability to experimental work. Our classification can provide the information about cognate ETC partners for oxygenase catalytic activity. Fifth, the system provides new insights into the evolution of RHO systems based on enzyme interaction.
Abbreviations
PAH, polycyclic aromatic hydrocarbons; RHO, Rieske-type non-heme iron aromatic ring-hydroxylating oxygenase; ETC, electron transport chain; PD, pairwise distance; NJ, neighbor-joing; CD, conserved domain; NR, nonredundant; GR, glutathione reductase; FNR, ferredoxin-NADP+ reductase.
Authors' contributions
All authors participated in the design of the study and writing of the manuscript. CEC directed the whole research and critically revised the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank H. Chen, CA Elkins and F. Rafii for critical review of on the manuscript. This work was supported in part by an appointment to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U. S. Department of Energy and the U. S. Food and Drug Administration. The views presented in this article do not necessarily reflect those of the Food and Drug Administration
Contributor Information
Ohgew Kweon, Email: oh-gew.kweon@fda.hhs.gov.
Seong-Jae Kim, Email: seong-jae.kim@fda.hhs.gov.
Songjoon Baek, Email: songjoon.baek@fda.hhs.gov.
Jong-Chan Chae, Email: chae@aesop.rutgers.edu.
Michael D Adjei, Email: michael.adjei@vdh.virginia.gov.
Dong-Heon Baek, Email: micro94@dku.edu.
Young-Chang Kim, Email: youngkim@chungbuk.ac.kr.
Carl E Cerniglia, Email: carl.cerniglia@fda.hhs.gov.
References
- Sutherland JB, Rafii F, Khan AA, Cerniglia CE. Microbial transformation and degradation of toxic organic chemicals. In: Young LY and Cerniglia CE, editor. Mechanisms of polycyclic aromatic hydrocarbon degradation. New York, Wiley-Liss Publication; 1995. pp. 269–306. [Google Scholar]
- Cerniglia CE. Biodegradation of polycyclic aromatic hydrocarbons. Curr Opin Biotechnol. 1993;4:331–338. doi: 10.1016/0958-1669(93)90104-5. [DOI] [Google Scholar]
- Mason JR, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- Butler CS, Mason JR. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol. 1997;38:47–84. doi: 10.1016/s0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- Hayaishi O. History and scope. In: Hayaishi O, editor. Oxygenases. New York, Academic Press; 1962. p. 1–29. [Google Scholar]
- Feig AL, Lippard SJ. Reactions of non-heme iron (II) centers with dioxygen in biology and chemistry. Chem Rev. 1994;94:124–133. doi: 10.1021/cr00027a011. [DOI] [Google Scholar]
- Raag R, Poulos TL. Crystal structure of the carbon monoxide-substrate-cytochrome P-450CAM ternary complex. Biochemistry. 1989;28:7586–7592. doi: 10.1021/bi00445a013. [DOI] [PubMed] [Google Scholar]
- Harayama S, Kok M, Neidle EL. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- Stingley RL, Khan AA, Cerniglia CE. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaalenii PYR-1. Biochem Biophys Res Commun. 2004;322:133–146. doi: 10.1016/j.bbrc.2004.07.089. [DOI] [PubMed] [Google Scholar]
- Stingley RL, Brezna B, Khan AA, Cerniglia CE. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaalenii PYR-1. Microbiology. 2004;150:3749–3761. doi: 10.1099/mic.0.27263-0. [DOI] [PubMed] [Google Scholar]
- Saito A, Iwabuchi T, Harayama S. Characterization of genes for enzymes involved in the phenanthrene degradation in Nocardioides sp. KP7. Chemosphere. 1999;38:1331–1337. doi: 10.1016/S0045-6535(98)00534-7. [DOI] [PubMed] [Google Scholar]
- Gibson DT, Resnick SM, Lee K, Brand JM, Torok DS, Wackett LP, Schocken MJ, Haigler BE. Desaturation, dioxygenation, and monooxygenation reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain 9816-4. J Bacteriol. 1995;177:2615–2621. doi: 10.1128/jb.177.10.2615-2621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HK, Zylstra GJ. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J Bacteriol. 1998;180:6529–6537. doi: 10.1128/jb.180.24.6529-6537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud J, Happe B, Timmis KN. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J Bacteriol. 1998;180:3954–3966. doi: 10.1128/jb.180.15.3954-3966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Wang RF, Cao WW, Doerge DR, Wennerstrom D, Cerniglia CE. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl Environ Microbiol. 2001;67:3577–3585. doi: 10.1128/AEM.67.8.3577-3585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batie CJ, Ballou DP, Correll CC. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. III. Boca Raton, FL, CRC Press; 1992. pp. 543–556. [Google Scholar]
- Werlen C, Kohler HP, van der Meer JR. The broad substrate chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase of Pseudomonas sp. strain P51 are linked evolutionarily to the enzymes for benzene and toluene degradation. J Biol Chem. 1996;271:4009–4016. doi: 10.1074/jbc.271.8.4009. [DOI] [PubMed] [Google Scholar]
- Nam JW, Nojiri H, Yoshida T, Habe H, Yamane H, Omori T. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci Biotechnol Biochem. 2001;65:254–263. doi: 10.1271/bbb.65.254. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kweon OG, Freeman JP, Jones RC, Adjei MD, Jhoo JW, Edmondson RD, Cerniglia CE. Molecular cloning and expression of genes encoding a novel dioxygenase Involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol. 2006;72:1045–1054. doi: 10.1128/AEM.72.2.1045-1054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Iwabuchi T, Harayama S. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli. J Bacteriol. 2000;182:2134–2141. doi: 10.1128/JB.182.8.2134-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treadway SL, Yanagimachi KS, Lankenau E, Lessard PA, Stephanopoulos G, Sinskey AJ. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl Microbiol Biotechnol. 1999;51:786–793. doi: 10.1007/s002530051463. [DOI] [PubMed] [Google Scholar]
- Nojiri H, Kamakura M, Urata M, Tanaka T, Chung JS, Takemura T, Yoshida T, Habe H, Omori T. Dioxin catabolic genes are dispersed on the Terrabacter sp. DBF63 genome. Biochem Biophys Res Commun. 2002;296:233–240. doi: 10.1016/S0006-291X(02)00873-2. [DOI] [PubMed] [Google Scholar]
- Eaton RW. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J Bacteriol. 2001;183:3689–3703. doi: 10.1128/JB.183.12.3689-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sho M, Hamel C, Greer CW. Two distinct gene clusters encode pyrene degradation in Mycobacterium sp. strain S65. FEMS Microbiol Ecol. 2004;48:209–220. doi: 10.1016/j.femsec.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J Bacteriol. 2003;185:3828–3841. doi: 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/S0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HBJ, Deerfield DW. GeneDoc: Analysis and visualization of genetic variation. EMBNEW NEWS. 1997;4:14. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, Liebert CA, Liu C, Madej T, Marchler GH, Mazumder R, Nikolskaya AN, Panchenko AR, Rao BS, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Vasudevan S, Wang Y, Yamashita RA, Yin JJ, Bryant SH. CDD: a curated Entrez database of conserved domain alignments. Nucl Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe H, Miyakoshi M, Chung J, Kasuga K, Yoshida T, Nojiri H, Omori T. Phthalate catabolic gene cluster is linked to the angular dioxygenase gene in Terrabacter sp. strain DBF63. Appl Microbiol Biotechnol. 2003;61:44–54. doi: 10.1007/s00253-002-1166-6. [DOI] [PubMed] [Google Scholar]
- Kitagawa W, Suzuki A, Hoaki T, Masai E, Fukuda M. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci Biotechnol Biochem. 2001;65:1907–1911. doi: 10.1271/bbb.65.1907. [DOI] [PubMed] [Google Scholar]
- Armengaud J, Timmis KN. Molecular characterization of Fdx1, a putidaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase of Sphingomonas sp. RW1. Eur J Biochem. 1997;247:833–842. doi: 10.1111/j.1432-1033.1997.00833.x. [DOI] [PubMed] [Google Scholar]
- Pinyakong O, Habe H, Yoshida T, Nojiri H, Omori T. Identification of three novel salicylate 1-hydroxylases involved in the phenanthrene degradation of Sphingobium sp. strain P2. Biochem Biophys Res Commun. 2003;301:350–357. doi: 10.1016/S0006-291X(02)03036-X. [DOI] [PubMed] [Google Scholar]
- Romine MF, Stillwell LC, Wong KK, Thurston SJ, Sisk EC, Sensen C, Gaasterland T, Fredrickson JK, Saffer JD. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Britt-Compton B, Williams PA. The naphthalene catabolic (nag) genes of Ralstonia sp. strain U2 are an operon that is regulated by NagR, a LysR-type transcriptional regulator. J Bacteriol. 2003;185:5847–5853. doi: 10.1128/JB.185.19.5847-5853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud J, Timmis KN. The reductase RedA2 of the multi-component dioxin dioxygenase system of Sphingomonas sp. RW1 is related to class-I cytochrome P450-type reductases. Eur J Biochem. 1998;253:437–444. doi: 10.1046/j.1432-1327.1998.2530437.x. [DOI] [PubMed] [Google Scholar]
- Kim E, Zylstra GJ. Functional analysis of genes involved in biphenyl, naphthalene, phenanthrene, and m-xylene degradation by Sphingomonas yanoikuyae B1. J Ind Microbiol Biotechnol. 1999;23:294–302. doi: 10.1038/sj.jim.2900724. [DOI] [PubMed] [Google Scholar]
- Cho O, Choi KY, Zylstra GJ, Kim YS, Kim SK, Lee JH, Sohn HY, Kwon GS, Kim YM, Kim E. Catabolic role of a three-component salicylate oxygenase from Sphingomonas yanoikuyae B1 in polycyclic aromatic hydrocarbon degradation. Biochem Biophys Res Commun. 2005;327:656–662. doi: 10.1016/j.bbrc.2004.12.060. [DOI] [PubMed] [Google Scholar]
- Demaneche S, Meyer C, Micoud J, Louwagie M, Willison JC, Jouanneau Y. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2004;70:6714–6725. doi: 10.1128/AEM.70.11.6714-6725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y, Shindo K, Harayama S, Misawa N. Molecular characterization and substrate preference of a polycyclic aromatic hydrocarbon dioxygenase from Cycloclasticus sp. strain A5. Appl Environ Microbiol. 2003;69:6688–6697. doi: 10.1128/AEM.69.11.6688-6697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato SI, Nam JW, Kasuga K, Nojiri H, Yamane H, Omori T. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J Bacteriol. 1997;179:4850–4858. doi: 10.1128/jb.179.15.4850-4858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu CH, Wyndham RC. Cloning and expression of the transposable chlorobenzoate-3,4-dioxygenase genes of Alcaligenes sp. strain BR60. Appl Environ Microbiol. 1993;59:3625–3633. doi: 10.1128/aem.59.11.3625-3633.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmel U, Engesser KH, Timmis KN, Dwyer DF. Cloning, nucleotide sequence, and expression of the gene encoding a novel dioxygenase involved in metabolism of carboxydiphenyl ethers in Pseudomonas pseudoalcaligenes POB310. Arch Microbiol. 1995;163:35–41. doi: 10.1007/BF00262201. [DOI] [PubMed] [Google Scholar]
- Locher HH, Leisinger T, Cook AM. 4-Toluene sulfonate methyl-monooxygenase from Comamonas testosteroni T-2: purification and some properties of the oxygenase component. J Bacteriol. 1991;173:3741–3378. doi: 10.1128/jb.173.12.3741-3748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Takeo M, Maeda Y. Plasmid-encoded genes specifying aniline oxidation from Acinetobacter sp. strain YAA. Microbiology. 1997;143:93–99. doi: 10.1099/00221287-143-1-93. [DOI] [PubMed] [Google Scholar]
- Fukumori F, Saint CP. Nucleotide sequences and regulational analysis of genes involved in conversion of aniline to catechol in Pseudomonas putida UCC22(pTDN1) J Bacteriol. 1997;179:399–408. doi: 10.1128/jb.179.2.399-408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak B, Fetzner S, Lingens F. Cloning, nucleotide sequence, and expression of the plasmid-encoded genes for the two-component 2-halobenzoate 1,2-dioxygenase from Pseudomonas cepacia 2CBS. J Bacteriol. 1995;177:667–675. doi: 10.1128/jb.177.3.667-675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy BM, Campbell AL, Neidle EL. Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobacter sp. strain ADP1. J Bacteriol. 1998;180:4466–4474. doi: 10.1128/jb.180.17.4466-4474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara H, Nagao K, Kouno K, Yano K. Phenanthrene-degrading phenotype of Alcaligenes faecalis AFK2. Appl Environ Microbiol. 1982;43:458–461. doi: 10.1128/aem.43.2.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa N, Kaida N, Torigoe S, Moritani T, Sawada T, Satoh S, Kiyohara H. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J Bacteriol. 1994;176:2444–2449. doi: 10.1128/jb.176.8.2444-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JM, Lloyd-Jones G. Novel carbazole degradation genes of Sphingomonas CB3: sequence analysis, transcription, and molecular ecology. Biochem Biophys Res Commun. 1998;247:129–135. doi: 10.1006/bbrc.1998.8750. [DOI] [PubMed] [Google Scholar]
- Masai E, Yamada A, Healy JM, Hatta T, Kimbara K, Fukuda M, Yano K. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 1995;61:2079–2085. doi: 10.1128/aem.61.6.2079-2085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett LP. Toluene dioxygenase from Pseudomonas putida F1. Methods Enzymol. 1990;188:39–45. doi: 10.1016/0076-6879(90)88010-8. [DOI] [PubMed] [Google Scholar]
- Ohtsubo Y, Nagata Y, Kimbara K, Takagi M, Ohta A. Expression of the bph genes involved in biphenyl/PCB degradation in Pseudomonas sp. KKS102 induced by the biphenyl degradation intermediate, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid. Gene. 2000;256:223–228. doi: 10.1016/S0378-1119(00)00349-8. [DOI] [PubMed] [Google Scholar]
- Erickson BD, Mondello FJ. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- Martin VJJ, Mohn WW. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J Bacteriol. 1999;181:2675–2682. doi: 10.1128/jb.181.9.2675-2682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata M, Uchida E, Nojiri H, Omori T, Obo R, Miyaura N, Ouchiyama N. Genes involved in aniline degradation by Delftia acidovorans strain 7N and its distribution in the natural environment. Biosci Biotechnol Biochem. 2004;68:2457–2465. doi: 10.1271/bbb.68.2457. [DOI] [PubMed] [Google Scholar]
- Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng. 1992;74:333–344. doi: 10.1016/0922-338X(92)90028-S. [DOI] [Google Scholar]
- Priefert H, Rabenhorst J, Steinbuchel A. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J Bacteriol. 1997;179:2595–2607. doi: 10.1128/jb.179.8.2595-2607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata M, Miyakoshi M, Kai S, Maeda K, Habe H, Omori T, Yamane H, Nojiri H. Transcriptional regulation of the ant operon, encoding two-component anthranilate 1,2-dioxygenase, on the carbazole-degradative plasmid pCAR1 of Pseudomonas resinovorans strain CA10. J Bacteriol. 2004;186:6815–6823. doi: 10.1128/JB.186.20.6815-6823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou NY, Al-Dulayymi J, Baird MS, Williams PA. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J Bacteriol. 2002;184:1547–1555. doi: 10.1128/JB.184.6.1547-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey WJ, Sabat G, Yuroff AS, Arment AR, Perez-Lesher J. Cloning, nucleotide sequencing, and functional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa strain JB2. Appl Environ Microbiol. 2001;67:4603–4609. doi: 10.1128/AEM.67.10.4603-4609.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa N, Iida T, Sawada T, Yamauchi K, Wang YW, Fukuda M, Kiyohara H. Nucleotide sequences and characterization of genes encoding naphthalene upper pathway of pseudomonas aeruginosa PaK1 and Pseudomonas putida OUS82. J Biosci Bioeng. 1999;87:721–731. doi: 10.1016/S1389-1723(99)80144-3. [DOI] [PubMed] [Google Scholar]
- Kosono S, Maeda M, Fuji F, Arai H, Kudo T. Three of the seven bphC genes of Rhodococcus erythropolis TA421, isolated from a termite ecosystem, are located on an indigenous plasmid associated with biphenyl degradation. Appl Environ Microbiol. 1997;63:3282–3285. doi: 10.1128/aem.63.8.3282-3285.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe H, Kasuga K, Nojiri H, Yamane H, Omori T. Analysis of cumene (isopropylbenzene) degradation genes from Pseudomonas fluorescens IP01. Appl Environ Microbiol. 1996;62:4471–4477. doi: 10.1128/aem.62.12.4471-4477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskas LG, Inui M, Kele Z, Yukawa H. Cloning of genes participating in aerobic biodegradation of p-cumate from Rhodopseudomonas palustris. DNA Seq. 2000;11:9–20. doi: 10.3109/10425170009033989. [DOI] [PubMed] [Google Scholar]
- Eaton RW. p-Cumate catabolic pathway in Pseudomonas putida Fl: cloning and characterization of DNA carrying the cmt operon. J Bacteriol. 1996;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HK, Mohseni P, Zylstra GJ. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J Bacteriol. 2003;185:5871–5881. doi: 10.1128/JB.185.19.5871-5881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]