Summary
Kinetochores are proteinaceous complexes containing dozens of components; they are assembled at centromeric DNA regions and provide the major microtubule attachment site on chromosomes during cell division. Recent studies have defined the kinetochore components comprising the direct interface with spindle microtubules, primarily through structural and functional analysis of the Ndc80 and Dam1 complexes. These studies have facilitated our understanding of how kinetochores remain attached to the end of dynamic microtubules and how proper orientation of a kinetochore-microtubule attachment is promoted on the mitotic spindle. In this article, we review these recent studies and summarize their key findings.
Introduction
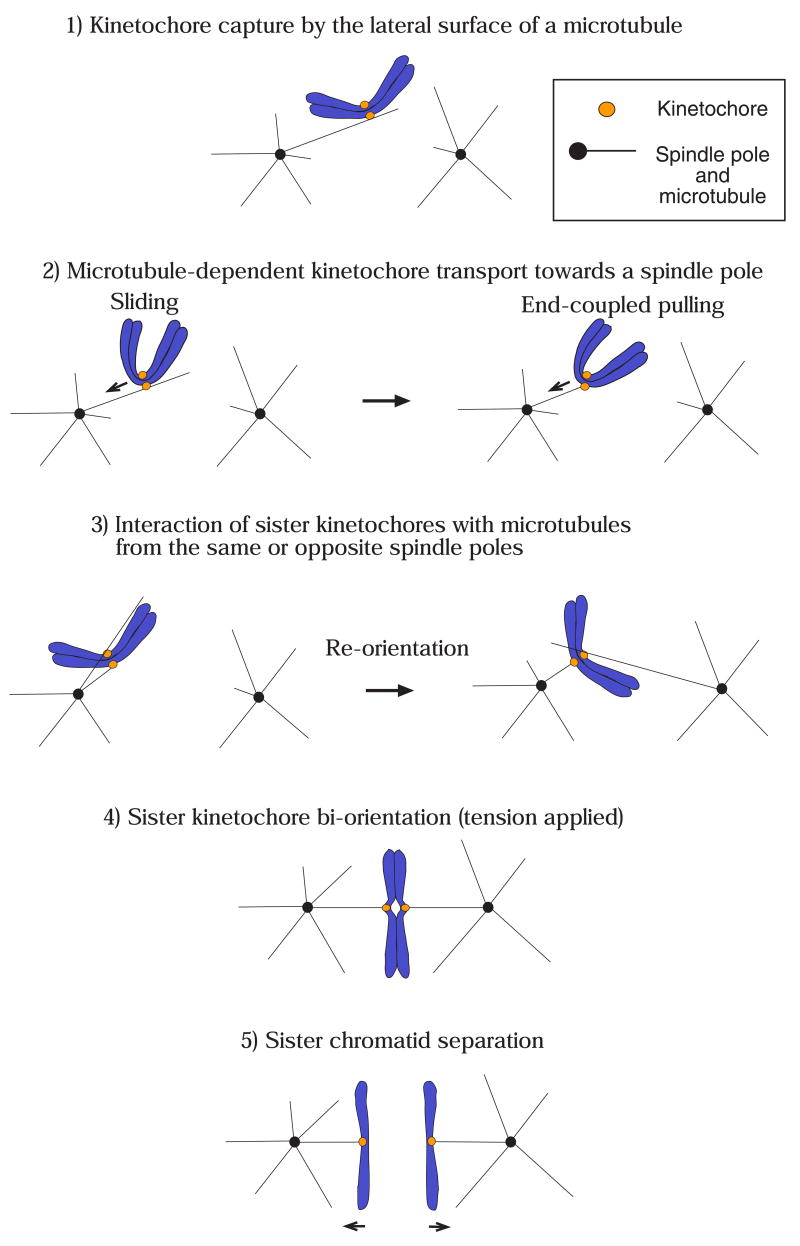
To maintain their genetic integrity, eukaryotic cells must segregate their replicated chromosomes properly during mitosis. Chromosome segregation is dependent on interactions between microtubules and the kinetochore, a large proteinaceous complex assembled on the centromere regions of chromosomes during mitotic entry (reviewed in [1–3]). For high-fidelity chromosome segregation, kinetochores must capture spindle microtubules and connect the sister chromatids of all chromosomes to opposite spindle poles prior to anaphase onset. Proper kinetochore capture of spindle microtubules is achieved in a stepwise manner (reviewed in [4,5]; Fig 1). Kinetochores initially attach to the surface of a single microtubule that extends from either spindle pole [6–8] (Fig 1, step 1). Once bound, kinetochores are transported poleward along microtubules (Fig 1, step 2). During or after this transport, both sister kinetochores interact with microtubules.
Figure 1. Overview of kinetochore-microtubule interactions.
The figure depicts kinetochore-microtubule interactions during prometaphase (steps 1–3), metaphase (step 4) and anaphase A (step 5).
1) Kinetochores are initially captured by the lateral surface of single microtubules that extend from one of the spindle poles [6–8]. The initial kinetochore encounter with microtubules happens quickly following nuclear envelope breakdown (metazoan cells) [6,7] or once kinetochore assembly is complete (budding yeast: note that spindle poles of this organism have not yet separated in step 1–2) [53].
2) Once captured, kinetochores are transported along the lateral surface of single microtubules toward the spindle pole (sliding) [6–8]. Subsequently, at least in budding yeast, kinetochores are tethered at the end of the single microtubules and transported further as the microtubules shrink (end-coupled pulling) [40,53].
3) As kinetochores approach spindle poles, both sister kinetochores attach to microtubules. If both kinetochores attach to microtubules from the same spindle pole, kinetochore-spindle pole connections by microtubules are re-oriented until proper bi-orientation is established [5,9].
4) Cessation of re-orientation is dependent on the tension that is generated by microtubules upon establishment of bi-orientation [5,9]. The number of microtubules whose plus ends attach to a single kinetochore increases when tension is applied in metazoan cells [107], while only a single microtubule is thought to attach to a each kinetochore in budding yeast [62] (the latter case is shown here for simplicity).
5) Once all kinetochores bi-orient on the spindle, cohesion between sister chromatids is removed, causing sister chromatid segregation to opposite spindle poles during anaphase A [11]. Kinetochores are end-coupled and pulled poleward as the microtubules depolymerize [12,13].
If kinetochores are wrongly attached, as occurs during syntelic attachment (where both sister kinetochores connect to microtubules from the same spindle pole), the kinetochore-spindle pole connections must be re-oriented (Fig 1, step 3) to convert to proper bi-orientation (i.e. attachments of sister kinetochores to microtubules extending from the opposite spindle poles; step 4), before anaphase onset is triggered (reviewed in [5,9]).
During the course of achieving bi-orientation, kinetochores exhibit two distinct types of associations with spindle microtubules. Initially kinetochores interact laterally with the microtubules lattice. Subsequently they are tethered at the microtubule plus end and exhibit motility directly coupled to microtubule polymerization and depolymerization (end-coupled attachment). A major question in the field has been to define the molecular mechanisms operating during these two types of attachments.
After bi-orientation, kinetochores and chromosome arms are aligned on the equatorial plate of the mitotic spindle (metaphase plate); this process is called congression [10]. Once all kinetochores bi-orient and congress on the spindle, cohesion between sister chromatids is removed (reviewed in [11]). Then each sister kinetochore, attached to the plus end of microtubules, is pulled towards the opposite spindle poles during anaphase (Fig 1, step 5). Sister separation during anaphase A, when chromosome-pole distance decreases, is coupled to microtubule depolymerization that occurs at the kinetochore (microtubule plus end) and, in the case of metazoan cells, also near centrosomes (microtubule minus end; reviewed in [12,13]).
In this article, we review recent papers (over the last 2 years), focusing on the following aspects of kinetochore-microtubule interactions: 1) The kinetochore is a large complex composed of dozens of proteins (reviewed in [3,14,15]); which of these proteins form the direct interface with spindle microtubules during lateral and end-coupled attachments? 2) How do kinetochores remain attached to the ends of depolymerizing microtubules? 3) While microtubule depolymerization is an important driving force for kinetochore movement, how do microtubule motor proteins contribute to kinetochore motility, especially in the initial steps of kinetochore-microtubule interactions? 4) How is sister kinetochore bi-orientation promoted on the mitotic spindle by re-orientation of kinetochore-spindle pole connections?
When addressing these questions, we will emphasize work on two protein complexes that have been the subject of intense recent study: the Ndc80 complex and the Dam1 complex. We will not discuss kinetochore composition and assembly [3,14,15] the spindle-assembly checkpoint [16], dynamics of spindle and kinetochore microtubules [17–19], or kinetochore-microtubule interactions in meiosis [20,21]. These topics have been recently reviewed in the indicated references.
The Ndc80 complex: A key component of the kinetochore-microtubule interface
Recent studies have revealed that the Ndc80 complex, an outer kinetochore component conserved from yeast to humans (reviewed in [3,22]), comprises a centrally important constituent of the kinetochore that directly interacts with microtubules. Depletion or inactivation of the Ndc80 complex causes the most severe chromosome segregation defect observed following inhibition of an outer kinetochore component. The complex is composed of four proteins; Ndc80 (also called Hec1 in mammals), Nuf2, Spc24 and Spc25 (Fig 2A, top). Ndc80-Nuf2 and Spc24-Spc25 form heterodimers with a globular domain at one side and a coiled-coil shaft at the other [23–25]. The two heterodimers are held together by interaction of their coiled-coil shafts, making a heterotetrameric ~55 nm long rod structure with globular domains at both ends. The Spc24/25 globular domain is oriented towards the inner kinetochore whereas the Ndc80/Nuf2 globular domain projects outwards and is positioned to interact with spindle microtubules. In metazoans, the Ndc80 complex associates with kinetochores prior to nuclear envelope breakdown [26,27]. Photobleaching studies indicate that this association is very stable [28,29]. In budding yeast, direct measurements have shown that 5–8 Ndc80 complexes are present per bound single kinetochore microtubule [29]; these observations suggest that multivalent interactions between kinetochore-associated Ndc80 complexes and spindle microtubules is critical for chromosome segregation.
Figure 2. The Ndc80 csomplex and the KMN network.
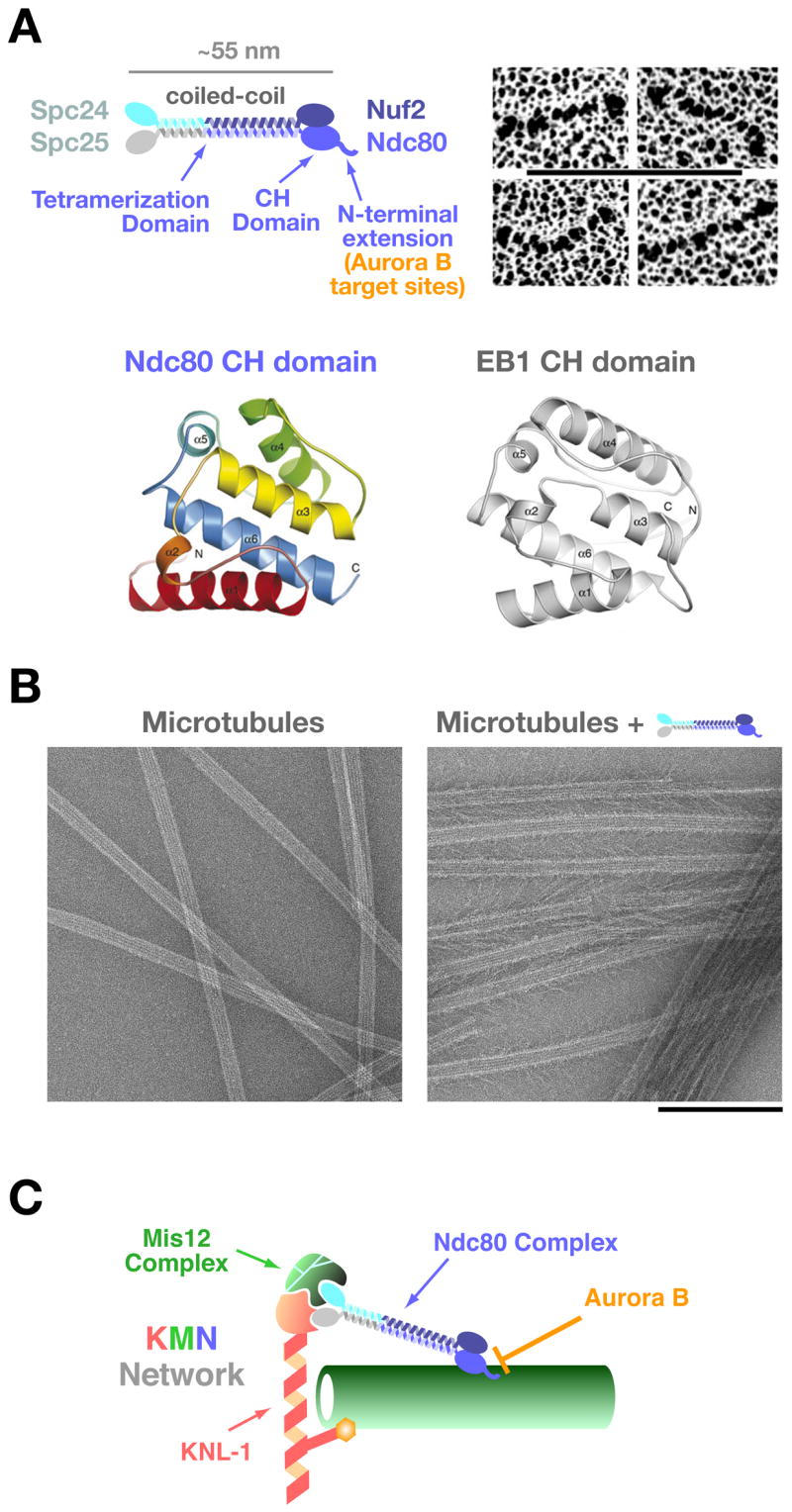
A) Schematic of the 4-subunit Ndc80 complex indicating the constituent parts and defined domains. Panels on the right show the rod-like structure of the complex visualized by electron microscopy of individual rotary-shadowed recombinant complexes (scale bar 100 nm; reprinted from ref. [23]). The ribbon diagrams below show the calponin-homology domain of the Ndc80 subunit (residues 81–196 of human Ndc80, also known as Hec1) and comparison with the EB1 CH domain (reprinted from [31]).
B) Microtubule-binding of the Ndc80 complex. The Ndc80 complex binds to the microtubule lattice with a fixed orientation forming “barbs” that extend away from the lattice. The microtubule-binding activity is located in the globular region of the Ndc80/Nuf2 dimer and is severely reduced by removal of the N-terminal extension on Ndc80. The “barbs” have a uniform polarity and binding angle indicating a specific binding site on the lattice (scale bar 200 nm; reprinted from [30]).
C) Schematic of the KNL-1/Mis12 complex/Ndc80 complex (KMN) complex network. Direct association of the Ndc80 complex with these other two kinetochore constituents is conserved in fungi, nematodes, insects, and vertebrates [3]. The Spc24/25 dimer is required for association with KNL-1 and the Mis12 complex. KNL-1 also directly binds to microtubules. Aurora B negatively regulates the microtubule-binding activity of the Ndc80 complex by phosphorylating the basic N-terminal extension of the Ndc80 subunit [30].
Most significantly, biochemical analysis and electron microscopy of the reconstituted Ndc80 complex has shown that the Ndc80-Nuf2 subunits (in particular, their globular domains) directly interact with the microtubule lattice, albeit with a low affinity [30,31] (Fig 2B). The crystal structure of the globular domain of the Ndc80 subunit has revealed that it is folded into a calponin-homology (CH) domain (Fig 2A, bottom) [31]. Intriguingly, the microtubule-binding region of the plus-end-associated protein EB1 also forms a CH domain [32]; this similarity was not anticipated from the primary sequence and was revealed by the structure. CH domains were first discovered and characterized in actin-binding proteins such as α-actinin [33]. The presence of CH domains in EB1 and in the globular domain of Ndc80 indicates that this structural motif is also utilized in microtubule-binding proteins and suggests an ancient evolutionary origin for this fold; whether a similar domain protein interacts with prokaryotic actin and tubulin-like polymers will be interesting to investigate in future work.
For the Ndc80/Nuf2 dimer, an unstructured basic region of approximately 80–100 amino acid length that extends out of the CH domain of Ndc80 is critical for microtubule-association. Following deletion of this region, the affinity of the Ndc80/Nuf2 dimer for microtubules is precipitously decreased [31]. The association of the Ndc80 complex with microtubules also exhibits cooperativity [30], which may be important in the context of forming multivalent associations with single microtubules. The CH domain of EB1 interacts with microtubules preferentially along the microtubule seam [34] (where the two-dimensional protofilament sheet of a microtubule is finally closed [35]) whereas the Ndc80 complex appear to bind along the entire microtubule lattice [30,31]; the reason for this difference remains to be characterized.
Consistent with its lattice binding activity, the Ndc80 complex is required for kinetochore association with the microtubule lateral surface in vivo in budding yeast [8]. There is also evidence that the Ndc80 complex plays a critical role in end-coupled kinetochore attachments that predominate during bi-orientation, congression and segregation. Of particular note is a recent study in mammalian cells where injection of an antibody to an epitope in the globular domain of the Ndc80 subunit resulted in the opposite phenotype to Ndc80-complex inhibition [36]; instead of loss of kinetochore-microtubule interactions, the attachment between the kinetochore and the spindle was hyper-stabilized, resulting in reduced turnover of bound microtubules and increased stretching of centromeric chromatin between sister kinetochores.
The KMN Network: A Conserved Core Protein Group of the Outer Kinetochore
The Ndc80 complex is directly associated with KNL1 (Spc105/Spc7 in budding and fission yeasts, respectively) and the 4-subunit Mis12 complex, forming a larger interacting protein set termed the KMN network from the names of its constituent parts (reviewed in [3]; Fig 2C). The Mis12 complex does not bind directly to microtubules in vitro whereas KNL1 does, albeit weakly and with a high degree of cooperativity [30]; KNL1 family proteins may also bind to microtubules in vivo, as suggested by analysis in fission yeast [37]. Reconstituting the interaction of these three pieces by mixing purified complexes increases the net microtubule-binding affinity, either via generation of a locally high density of microtubule-binding sites or via allosteric regulation [30]. Thus, in vivo, the Ndc80 complex is acting in the context of this protein set, which not only directs its localization but may also influence its microtubule-binding activity. Based on these findings, it was proposed that microtubule ends are bound by a network composed of the KMN protein group [30]. Consistent with this proposal, electron tomography has recently shown that the microtubule ends are embedded in a fibrous network within the kinetochore outer plate [38].
Dam1 Complex Rings: Force Transducers for Pulling on Kinetochores
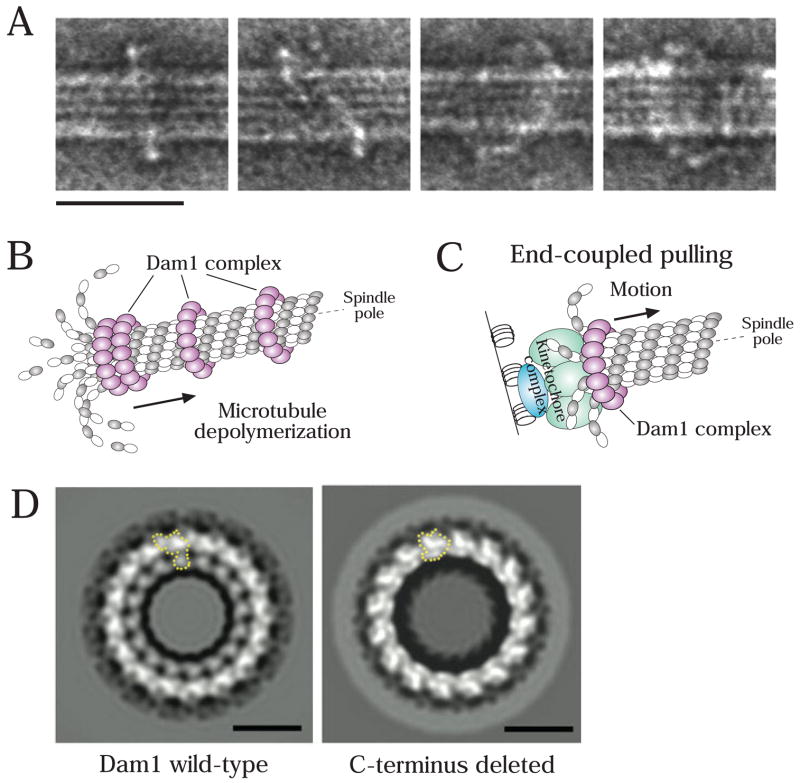
During end-coupled attachments, a special device is necessary to maintain kinetochore association with dynamic microtubule ends. Recent results suggest that the Dam1 complex has the properties to construct such a device. The Dam1 complex, also called DASH or DDD, is composed of at least 10 proteins, and has been identified in yeast (reviewed in [15]). This complex is not at kinetochores during their initial association with the microtubule lateral surface; however it subsequently is present at end-coupled attachments [39,40], possibly because of a direct Dam1 complex—Ndc80 complex interaction [41–43]. The Dam1 complex plays a crucial role in tethering kinetochores at the ends of depolymerizing microtubules in budding yeast [40]. Biochemical reconstitution has revealed that several (~16) Dam1 complexes associate together on the microtubule surface to form a ring that encircles the polymer [44,45] (Fig 3A). If this view is correct, Dam1 complex rings formed along a microtubule should accumulate at the microtubule plus end during the outward curling of protofilaments that accompanies microtubule depolymerization (Fig 3B); this was indeed the case not only in an in vitro reconstituted system [46], but also in in vivo observations [40].
Figure 3. Dam1 complexes oligomerize to form a ring encircling a microtubule.
A) Electron micrographs of negatively stained microtubules in the presence of the Dam1 complexes. Scale bar, 50 nm. Reprinted from [44].
B) If the Dam1 complexes formed rings around microtubules, they would be accumulated at the microtubule plus end during the outward curling of protofilaments that accompanies depolymerization; this accumulation was observed in vitro [46] and in vivo [40].
C) The Dam1 complex has a crucial role in tethering kinetochores at microtubule ends and in converting microtubule depolymerization into kinetochore pulling force [40,46,49].
D) Electron micrographs of rings (negative stain and 16-fold rotational average), formed by the Dam1 complexes containing either a wild-type Dam1 protein (left) or a C-terminus (140 residues)-deleted Dam1 protein. A single Dam1 complex is shown by the dotted line. Scale bars, 25 nm. Reprinted from [73].
In vitro experiments with purified tubulin have indicated that protofilament curling at the microtubule plus end can produce forces that are more than sufficient to move chromosomes towards a spindle pole [47]. A mathematical model has predicted that a microtubule-encircling ring, if present, would facilitate this process [48]. Thus, the Dam1-complex ring is an ideal machine to link chromosome movement to polymerization dynamics at end-coupled kinetochore attachments. In vitro biophysical studies have revealed that the recombinant Dam1 complex can effectively harness the intrinsic microtubule depolymerization-generated force and is also able to promote polymerization when placed under tension, both attributes of end-coupled kinetochore attachments [49,50]. The process of kinetochore poleward movement coupled to depolymerization was recently visualized in vivo in budding and fission yeast cells [40,51–53] (end-coupled pulling; Fig 1, step 2, right; Fig 3C). End-coupled poleward movement of kinetochores was dependent on the Dam1 complex in vivo [40,52], as suggested by the in vitro studies. Presumably, this Dam1 function is also important during anaphase A, when end-coupled kinetochores are pulled polewards concomitant with depolymerization of plus ends at the kinetcohore [12,13,54] (Fig 1, step 5). Thus, the Dam1 complex is the perfect molecular device that, together with the Ndc80 complex, would help generate the special properties of end-coupled attachments that are central to chromosome segregation.
Do metazoans have functional counterparts of the Dam1 complex?
The principles of Dam1-complex function are likely to be important for kinetochore-microtubule interactions in all eukaryotic cells. However, convincing orthologues of the Dam1 complex have not been identified in metazoans either by sequence searches or using genome-wide functional analysis [55,56]. This discrepancy might be explained by either of the following two scenarios: first, Dam1-complex orthologues, which form a ring encircling a microtubule, might be present in metazoans but their peptide sequence might be too divergent to be identified. This scenario may be rebutted by a lack of a detectable ring structure in electron tomography [38]; however, such rings may not generate sufficient contrast for visualization by electron microscopy of cells and no rings have been observed at budding-yeast kinetochores [57,58], where the Dam1 complex is clearly present in numbers sufficient to form a ring and important for end-coupled attachments [29,40]. Second, the ring structure might be dispensable for kinetochore-microtubule interaction and therefore absent in metazoa; meanwhile other molecules may compensate for this function. Relevant to this scenario is the finding that Dam1-complex components are not essential for cell viability in fission yeast [59,60], in contrast to budding yeast; this difference may reflect the fact that a single versus multiple microtubules attach to a single kinetochore during metaphase of budding and fission yeasts, respectively [61,62].
Four possible functional counterparts (orthologs or compensatory factors) of the Dam1 complex have been suggested at vertebrate kinetochores (all are outer kinetochore components, except for kinesin-13). Kinesin-13 (MCAK etc), which facilitates microtubule depolymerization [63], is a substrate of Aurora B kinase [64–69], similar to Dam1 (see below), and can form rings/spirals encircling a microtubule at least in vitro [70–72]. However, the ring/spiral structures formed by MCAK are distinct from those formed by the Dam1 complex and structural comparisons suggest that they would not be as effective as the Dam1 complex in coupling to polymerization dynamics [73]. Ska1/2 are proteins identified in vertebrates that require kinetochore-microtubule attachment for their recruitment to kinetochores and, once recruited, somehow modulate this attachment [74], similarly to the Dam1 complex [39]. Cep57 also contributes to kinetochore-microtubule attachment and exhibits weak sequence similarity to Dam1 [69]. Bod1 is necessary for ensuring sister kinetochore bi-orientation [75], similarly to the Dam1 complex [41]. Further studies on these interesting proteins are needed to understand their mechanistic contributions to chromosome segregation as well as to establish their relationship to the fungal Dam1 complex.
Motor proteins associated with kinetochores
Several motor proteins are associated with kinetochores and play important roles in microtubule-dependent kinetochore motion (reviewed in [2,76]). In the initial stages of kinetochore-microtubule interactions, kinetochores associate with the microtubule lateral surface and are transported towards a spindle pole [6–8] (sliding; Fig 1, step 2, left). Kinetochore sliding is often converted to end-coupled attachment that exerts a poleward pulling force on the kinetochore (see the above section; Fig 1, step 2), but the opposite conversion is rare [40]. Kinetochore sliding occurs rapidly (10–50 μm/min) towards a spindle pole in vertebrate cells [77,78], but much more slowly (1–1.5 μm/min) and accompanied by transient pausing in budding yeast [8,40]. Such difference is attributed to use of different microtubule minus-end directed motors associated with kinetochores; dynein, a processive motor, is used in metazoans [78,79], whereas Kar3, a kinesin-14 family member and non-processive motor (i.e. the motor is released from microtubules after each ATPase cycle [80]), is used in budding yeast [8,40]. Dynein localizes only outside of nuclei in yeast; presumably, upon the evolution of open mitosis [81], eukaryotic cells acquired the ability to use dynein in nuclear functions.
After kinetochores are transported to the vicinity of a spindle pole in prometaphase, they move towards the spindle equator to form the metaphase plate (congression) [10]. It has been thought that congression occurs after, and depends on, sister kinetochore bi-orientation. However, in metazoan cells prior to bi-orientation, kinetochore-bound CENP-E, a kinesin-7 family plus end-directed motor, facilitates this process by transporting mono-oriented kinetochores away from a spindle pole along microtubules that are attached to other already congressed and bi-oriented kinetochores [82]. Perhaps analogously, in budding yeast, Cin8, a kinesin-5 family member, localizes at bi-oriented kinetochores and regulates kinetochore position on the metaphase spindle [83]
Mechanisms ensuring sister kinetochore bi-orientation
In addition to proteins necessary for the kinetochore-microtubule attachment, what factors are required to ensure sister kinetochore bi-orientation prior to anaphase onset? Aurora B (Ipl1 in budding yeast) kinase is a key regulator for bi-orientation [84–87] and it was suggested that this kinase promotes turnover of kinetochore-spindle pole connections and eliminates those that do not generate tension between sister kinetochores [87–90].
The Dam1 and Ndc80 complexes are crucial substrates of Aurora B/Ipl1 at kinetochores [30,36,91,92]. Dam1 is primarily phosphorylated at its C-terminus and mutants mimicking constitutive dephosphorylation show defects in bi-orientation. Recent structural studies revealed that the C-terminus of Dam1 protein could affect both oligomerization to form rings and microtubule interaction (Fig 3D) [73] (also refer to a biochemical study [93]). Intriguingly, Dam1 mutants mimicking constitutive phosphorylation reduced the efficiency of ring formation in vitro [73]. On the other hand, phosphorylation of the Ndc80 complex is clustered at the N-terminus of the Ndc80 subunit extending out of the CH domain that is important for microtubule-binding (Fig. 2A, C). Phosphorylation of this region reduced affinity of the Ndc80 complex for microtubules in vitro [30] and Ndc80 mutants mimicking constitutive dephosphorylation showed defects in bi-orientation in vivo [36]. Thus, the functional consequences of Aurora B phosphorylation on the Dam1 and Ndc80 complexes have been revealed both in vitro and in vivo.
Once bi-orientation occurs and tension is applied on kinetochores, turnover of kinetochore-spindle pole connections must stop [5,94]; otherwise bi-orientation would never be maintained. For this, sensing tension is of central importance, but which component acts as a tension sensor? Bir1 and Sli15 (Survivin and INCENP in metazoa) are binding partners of Ipl1 in yeast and regulate its kinase activity [95]. It was recently revealed that Bir1 and Sli15 form a sub-complex, which forms bridges between a microtubule and a kinetochore [96]. Bir1-Sli15 is therefore positioned ideally to sense tension and may regulate Ipl1 kinase activity accordingly. In metazoans, another good candidate for a tension sensor might be PICH, a Snf2 family member, identified in mammalian cells. PICH shows a unique thread-like localization between bi-oriented sister kinetochores and is necessary for activation of the spindle-assembly checkpoint [97]; considering its localization, PICH may work as a tension sensor to regulate both checkpoint signaling and bi-orientation.
The role of Aurora B/Ipl1 in bi-orientation was initially highlighted in budding yeast [84,85,87] where only one microtubule attaches to each kinetochore [62]. However, in metazoan cells, multiple microtubules form end-coupled attachments to each kinetochore [98]. Thus, Aurora B may have a more complex role in ensuring bi-orientation in this context. Consistent with this idea, inactivation of Aurora B leads to not only syntelic attachment defects but also frequent merotelic attachments [99,100], where a single sister kinetochore attaches to microtubules extending from opposite spindle poles. In addition to its role in promoting turnover of kinetochore-microtubule attachments, Aurora B (together with Polo kinase) facilitates resolution of sister chromatids in metazoan cells [101]; both functions may be important to avoid or correct merotelic attachment.
Mps1 is another evolutionarily conserved protein kinase, required for the spindle assembly checkpoint and, in some organisms, for duplication of microtubule-organizing centres [102]. Separately from these functions, however, Mps1 has an important role in chromosome segregation [103]. It was recently shown that, in budding yeast, Mps1 has a crucial role in establishing sister kinetochore bi-orientation on the mitotic spindle [104]. Similarly to Ipl1, Mps1 promotes re-orientation of kinetochore-spindle pole connections and eliminates those that do not generate tension between sister kinetochores. Intriguingly, both Ipl1 and Mps1 phosphorylate the Dam1 subunit of the Dam1 complex, but at different sites [91,105]. The role of Mps1 in bi-orientation needs to be investigated further.
Conclusion and perspectives
Over the last couple of years, the kinetochore-microtubule attachment interface has been revealed in increasing detail, in particular, through studies of the Ndc80 and Dam1 complexes. These discoveries have shed new light on the mechanisms underlying kinetochore motion and bi-orientation. Biochemical reconstitutions, structural analysis, genetics, and cell biology have all contributed to these discoveries and will continue to advance research in this field.
Acknowledgments
We thank Lesley Clayton and Karen Oegema for helpful comments on the manuscript; Stephen C. Harrison and Eva Nogales for providing high-resolution versions of their figures. Work in Tanaka lab has been supported by the funding from Cancer Research UK, the Wellcome Trust, Human Frontier Science Program, Lister Research Institute Prize and Association for International Cancer Research. Work in the Desai lab has been supported by the Ludwig Institute for Cancer Research, the National Institute of Health (R01-GM074215), and the Human Frontiers Science Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Tomoyuki U. Tanaka, Email: t.tanaka@lifesci.dundee.ac.uk.
Arshad Desai, Email: abdesai@ucsd.edu.
References
- 1.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Maiato H, Deluca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman I, Desai A. Molecular Architecture of the Kinetochore-Microtubule Interface. Nat Rev Mol Cell Biol. doi: 10.1038/nrm2310. in press. [DOI] [PubMed] [Google Scholar]
- 4.Kline-Smith SL, Sandall S, Desai A. Kinetochore-spindle microtubule interactions during mitosis. Curr Opin Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka TU, Stark MJ, Tanaka K. Kinetochore capture and bi-orientation on the mitotic spindle. Nat Rev Mol Cell Biol. 2005;6:929–942. doi: 10.1038/nrm1764. [DOI] [PubMed] [Google Scholar]
- 6.Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayden JH, Bowser SS, Rieder CL. Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. J Cell Biol. 1990;111:1039–1045. doi: 10.1083/jcb.111.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 9.Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 10.Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 12.Rogers GC, Rogers SL, Sharp DJ. Spindle microtubules in flux. J Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- 13.Kwok BH, Kapoor TM. Microtubule flux: drivers wanted. Curr Opin Cell Biol. 2007;19:36–42. doi: 10.1016/j.ceb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Mellone B, Erhardt S, Karpen GH. The ABCs of centromeres. Nat Cell Biol. 2006;8:427–429. doi: 10.1038/ncb0506-427. [DOI] [PubMed] [Google Scholar]
- 15.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 16.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 17.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14:R797–805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 18.Kline-Smith SL, Walczak CE. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Hauf S, Watanabe Y. Kinetochore orientation in mitosis and meiosis. Cell. 2004;119:317–327. doi: 10.1016/j.cell.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 22.Ciferri C, Musacchio A, Petrovic A. The Ndc80 complex: hub of kinetochore activity. FEBS Lett. 2007;581:2862–2869. doi: 10.1016/j.febslet.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 25.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 26.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- 29•.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. The authors quantify the copy number of each kinetochore component at a single centromere in budding yeast. They compared the signal intensity of GFP-tagged kinetochore components with that of GFP-tagged Cse4, a centromeric histone H3 variant. In budding yeast, a centromere is specified by a single nucleosome [106] containing two copies of Cse4. The authors also perform photobleaching studies which revealed that the Dam1 complex and the Ndc80 complex are stably bound in metaphase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. The authors biochemically reconstitute KNL-1, the Ndc80 complex, and the Mis12 complex, and show that they directly associate to form a larger complex, named the KMN network, which possesses two microtubule-binding domains: one in the globular Ndc80/Nuf2 heterodimer and the other in KNL-1. Phosphorylation of the N-terminal extension of the Ndc80 subunit by Aurora B reduced affinity of the Ndc80 complex for microtubules. Reconstitution of the network, resulted in a significant enhancement of microtubule-binding affinity relative to the individual microtubule-binding domains. [DOI] [PubMed] [Google Scholar]
- 31••.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. The authors report the crystal structure of the most conserved region of the Ndc80 globular domain and conclude that it forms a calponin-homology domain. They further show that an Ndc80-Nuf2 heterodimer binds the microtubule lattice in vitro. The N-terminal 80 amino acids of Ndc80, not present in the structure, contribute significantly to the binding affinity. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi I, Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J Biol Chem. 2003;278:36430–36434. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- 33.Djinovic Carugo K, Banuelos S, Saraste M. Crystal structure of a calponin homology domain. Nat Struct Biol. 1997;4:175–179. doi: 10.1038/nsb0397-175. [DOI] [PubMed] [Google Scholar]
- 34•.Sandblad L, Busch KE, Tittmann P, Gross H, Brunner D, Hoenger A. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. The authors report that Mal3 (fission yeast EB1), which is a calponin-homology domain microtubule-binding protein, binds along one of the microtubule grooves between protofilaments. The alignment of Mal3 along a microtubule groove seems to coincide with the microtubule lateral seam, whose stabilization is dependent on Mal3. [DOI] [PubMed] [Google Scholar]
- 35.Nogales E, Wang HW. Structural intermediates in microtubule assembly and disassembly: how and why? Curr Opin Cell Biol. 2006;18:179–184. doi: 10.1016/j.ceb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 36•.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. The authors report that microinjection of an antibody against the N-terminus of Ndc80 suppresses the dynamic behaviour of microtubules at kinetochores in PtK1 cells. They also find that the N-terminus of Ndc80 is phosphorylated by Aurora B kinase in vitro. Expression of non-phosphorylatable Ndc80 causes frequent merotelic attachment and chromosome mis-segregation. [DOI] [PubMed] [Google Scholar]
- 37.Kerres A, Jakopec V, Fleig U. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol Biol Cell. 2007;18:2441–2454. doi: 10.1091/mbc.E06-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. Using electron tomography, the authors visualized the kinetochore-microtubule interface in high resolution, in metaphase of PtK1 cells. Within the kinetochore outer plate, distinct unit motifs are not found; instead, several fibers either embed the microtubule plus-end in a radial mesh or extend out to bind microtubule walls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. The authors reveal two distinct mechanisms, by which kinetochores are transported by microtubules towards spindle poles after they are captured by microtubule lateral surface in prometaphase of budding yeast. Kar3, a kinesin-14 member, drives kinetochore sliding along the microtubule lateral surface, while the Dam1 complex promotes motion of kinetochores, tethered at microtubule ends (end-coupled pulling). Sliding is often converted to end-coupled pulling but the opposite conversion is rare. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. Embo J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong J, Nakajima Y, Westermann S, Shang C, Kang JS, Goodner C, Houshmand P, Fields S, Chan CS, Drubin D, et al. A Protein Interaction Map of the Mitotic Spindle. Mol Biol Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 45.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a Dynamic Kinetochore- Microtubule Interface through Assembly of the Dam1 Ring Complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 46•.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. Using in vitro reconstitution, the authors show that Dam1 complexes accumulate at the ends of depolymerising microtubules, processively follow a shrinking microtubule end and can couple movement of a bead to the depolymerizing microtubule. When Dam1 complexes bound along a microtubule, they exhibited one-dimensional diffusion. [DOI] [PubMed] [Google Scholar]
- 47.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, Onuchic JN. A driving and coupling “Pac-Man” mechanism for chromosome poleward translocation in anaphase A. Proc Natl Acad Sci U S A. 2006;103:18432–18437. doi: 10.1073/pnas.0608962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. In these two papers, using optical tweezers, the authors measured the amount of tension that can be resisted by a Dam1-complex-coated beads localizing at the microtubule plus ends [49]. Higher tension decreases the frequency of microtubule catastrophe and increases that of microtubule rescue [50], similarly to the effects on kinetochore microtubules during metaphase in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. Embo J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franco A, Meadows JC, Millar JB. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. [DOI] [PubMed] [Google Scholar]
- 53•.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore-microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007 doi: 10.1101/gad.449407. in press. The authors show that in budding yeast, which shows closed mitosis, DNA replication of centromeres causes their detachment from microtubules, thus displacing them away from a spindle pole for 1–2 min. Soon afterwards kinetochores are reassembled, leading to their recapture by microtubules and subsequent transport towards a spindle pole. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 56.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McIntosh JR. Rings around kinetochore microtubules in yeast. Nat Struct Mol Biol. 2005;12:210–212. doi: 10.1038/nsmb0305-210. [DOI] [PubMed] [Google Scholar]
- 59.Liu X, McLeod I, Anderson S, Yates JR, He X. Molecular analysis of kinetochore architecture in fission yeast. Embo J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. Embo J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wordeman L. Microtubule-depolymerizing kinesins. Curr Opin Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 64.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 65.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 66.Ohi R, Sapra T, Howard J, Mitchison TJ. Differentiation of Cytoplasmic and Meiotic Spindle Assembly MCAK Functions by Aurora B-dependent Phosphorylation. Mol Biol Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Emanuele MJ, Stukenberg PT. Xenopus Cep57 is a novel kinetochore component involved in microtubule attachment. Cell. 2007;130:893–905. doi: 10.1016/j.cell.2007.07.023. Using Xenopus egg extracts, the authors find that, Cep57 localizes at both kinetochores and centrosomes. Cep57 is required to strengthen kinetochore-microtubule attachment and to maintain microtubule anchorage at spindle poles. [DOI] [PubMed] [Google Scholar]
- 70.Tan D, Asenjo AB, Mennella V, Sharp DJ, Sosa H. Kinesin-13s form rings around microtubules. J Cell Biol. 2006;175:25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71•.Moores CA, Cooper J, Wagenbach M, Ovechkina Y, Wordeman L, Milligan RA. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5:1812–1815. doi: 10.4161/cc.5.16.3134. In these two papers, the authors show that kinesin-13 proteins are associated with microtubule protofilaments and facilitate their curling, leading to formation of rings and spirals around microtubules in vitro, in the presence of non-hydrolysable ATP analogue. [DOI] [PubMed] [Google Scholar]
- 72.Davis TN, Wordeman L. Rings, bracelets, sleeves, and chevrons: new structures of kinetochore proteins. Trends Cell Biol. 2007;17:377–382. doi: 10.1016/j.tcb.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73••.Wang HW, Ramey VH, Westermann S, Leschziner AE, Welburn JP, Nakajima Y, Drubin DG, Barnes G, Nogales E. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. Using electron microscopy, the authors determined the structures of the free Dam1 complex and of the microtubule-encircling ring formed by oligomerization of Dam1 complexes. Ring formation was facilitated by a conformational change upon microtubule binding. The C-terminus of the Dam1 subunit is at a position that could affect both protein oligomerization and microtubule interaction. The direct docking between the Dam1 complexes in the ring and the tubulin subunits of the microtubule was not observed; this may facilitate sliding of the entire ring when coupling to depolymerizing microtubules. [DOI] [PubMed] [Google Scholar]
- 74•.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. Embo J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. The authors identified the Ska1/2 complex in mammals, which associates with kinetochores following microtubule attachment in prometaphase. Depletion of Ska1/2 shows metaphase arrest due to spindle-checkpoint activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75•.Porter IM, Swedlow JR. Bod1, a novel kinetochore protein required for chromosome biorientation. J Cell Biol. 2007;179 doi: 10.1083/jcb.200704098. in press. The authors identified Bod1 protein through proteomic analysis of chromosomes. Bod1 depletion causes frequent syntelic kinetochore-microtubule attachment and reduces MCAK phosphorylation without an appreciable change in Aurora B activity or localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 77.Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78•.Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. By inhibiting ZW10 that is required for dynein association with kinetochores, the authors studied the roles of dynein at kinetochores in mammalian cells. Dynein at kinetochores is required for rapid poleward chromosome motion in prometaphase and chromosome congression in metaphase. Moreover, ZW10 depletion leads to a reduced velocity of anaphase chromosome motion without affecting microtubule poleward flux. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.King JM, Hays TS, Nicklas RB. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J Cell Biol. 2000;151:739–748. doi: 10.1083/jcb.151.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Endow SA. Kinesin motors as molecular machines. Bioessays. 2003;25:1212–1219. doi: 10.1002/bies.10358. [DOI] [PubMed] [Google Scholar]
- 81.Sazer S. Nuclear envelope: nuclear pore complexity. Curr Biol. 2005;15:R23–26. doi: 10.1016/j.cub.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 82••.Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. By using live-cell imaging and correlative electron microscopy, the authors found that mono-oriented chromosomes could glide towards the spindle equator (metaphase plate) along the kinetochore microtubule bundles of other, already bi-oriented chromosomes. This gliding is dependent on the kinetochore-associated plus end-directed motor protein CENP-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83•.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. The authors show that Kip1/Cin8 (kinesin-5) and Kip3 (kinesin-8) are associated with kinetochores during metaphase in budding yeast. Kar3 (kinesin-14) is at kinetochores in prometaphase [40] but its amount decreases in metaphase. Cin8 and Kip1 regulate kinetochore positions on the metaphase spindle while Kip3 regulates poleward kinetochore motion in anaphase A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 86.Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis. Curr Biol. 2002;12:798–812. doi: 10.1016/s0960-9822(02)00820-5. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 88.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, Van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- 90.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 91.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, 3rd, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 92.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. The Set1 Methyltransferase Opposes Ipl1 Aurora Kinase Functions in Chromosome Segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miranda JJ, King DS, Harrison SC. Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol Biol Cell. 2007;18:2503–2510. doi: 10.1091/mbc.E07-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 95.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 96•.Sandall S, Severin F, McLeod IX, Yates JR, 3rd, Oegema K, Hyman A, Desai A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. Using a biochemical reconstitution approach, the authors found that a Bir1-Sli15 complex links budding yeast centromeres to microtubules. This linkage does not require Ipl1 kinase, whose targeting and activation is regulated by Bir1 and Sli15. Elimination of the Bir1-Sli15 linkage, for example, by deletion of the microtubule-binding domain of Sli15, leads to a defect in bi-orientation, similarly to ipl1 mutants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97•.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. The authors identify PICH as a Plk1 (Polo-like kinase 1) binding protein and its substrate. Depletion of PICH abrogated the spindle checkpoint. PICH localizes along threads between sister kinetochores during metaphase, which diminish during anaphase. The PICH-positive threads are sensitive to DNase and exacerbated by inhibition of topoisomerase II. [DOI] [PubMed] [Google Scholar]
- 98.McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in PTK cells. J Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 100•.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. These two papers report that inhibition of Aurora B kinase activity leads to frequent occurrence of merotelic kinetochore-microtubule attachments in vertebrate cells. In prometaphase, Aurora B inhibition causes suppression of kinetochore-microtubule turnover [99]. Aurora B is enriched at merotelic attachment sites, independently of its kinase activity, and Aurora B enriches MCAK at these sites using its kinase activity [100] [DOI] [PubMed] [Google Scholar]
- 101.Losada A, Hirano M, Hirano T. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 2002;16:3004–3016. doi: 10.1101/gad.249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Winey M, Huneycutt BJ. Centrosomes and checkpoints: the MPS1 family of kinases. Oncogene. 2002;21:6161–6169. doi: 10.1038/sj.onc.1205712. [DOI] [PubMed] [Google Scholar]
- 103.Jones MH, Huneycutt BJ, Pearson CG, Zhang C, Morgan G, Shokat K, Bloom K, Winey M. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 104•.Maure J-F, Kitamura E, Tanaka TU. Mps1 kinase promotes sister kinetochore bi-orientation by a tension-dependent mechanism. Curr Biol. 2007 doi: 10.1016/j.cub.2007.11.032. The authors show that Mps1 kinase has a crucial role in establishing sister kinetochore bi-orientation on the mitotic spindle in budding yeast. This role of Mps1 is separate from its function in the spindle-assembly checkpoint and spindle pole body duplication. Similarly to Ipl1, Mps1 facilitates bi-orientation in a tension-dependent mechanism by eliminating wrong kinetochore-spindle pole connections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Yates JR, 3rd, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.King JM, Nicklas RB. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J Cell Sci. 2000;113:3815–3823. doi: 10.1242/jcs.113.21.3815. [DOI] [PubMed] [Google Scholar]