Abstract
Improving oil and protein quality for food and feed purposes is an important goal in rapeseed (Brassica napus L.) breeding programs. Rapeseed contains phytosterols, used to enrich food products, and sinapate esters, which are limiting the utilization of rapeseed proteins in the feed industry. Increasing the phytosterol content of oil and lowering sinapate ester content of meal could increase the value of the oilseed rape crop. The objective of the present study was to identify quantitative trait loci (QTL) for phytosterol and sinapate ester content in a winter rapeseed population of 148 doubled haploid lines, previously found to have a large variation for these two traits. This population also segregated for the two erucic acid genes. A close negative correlation was found between erucic acid and phytosterol content (Spearman’s rank correlation, rs = −0.80**). For total phytosterol content, three QTL were detected, explaining 60% of the genetic variance. The two QTL with the strongest additive effects were mapped on linkage groups N8 and N13 within the confidence intervals of the two erucic acid genes. For sinapate ester content four QTL were detected, explaining 53% of the genetic variance. Again, a close negative correlation was found between erucic acid and sinapate ester content (rs = −0.66**) and the QTL with the strongest additive effects mapped on linkage groups N8 and N13 within the confidence intervals of the two erucic acid genes. The results suggests, that there is a pleiotropic effect of the two erucic acid genes on phytosterol and sinapate ester content; the effect of the alleles for low erucic acid content is to increase phytosterol and sinapate ester content. Possible reasons for this are discussed based on known biosynthetic pathways.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-008-0734-2) contains supplementary material, which is available to authorized users.
Introduction
Oilseed rape (Brassica napus L.) is the major oilseed crop in temperate regions and ranks second among oilseed crops produced worldwide (http://faostat.fao.org). Interest in oilseed rape has recently increased due to its diversified utilization in food and feed production and its growing economic importance as a novel source of renewable energy, mainly as biodiesel (Mittelbach and Gangl 2001). In most parts of the world “canola” or “double low” quality type rapeseed with low seed contents of erucic acid and glucosinolates is grown. However, to some extent high erucic acid rapeseed is also cultivated and the oil is mostly used by the oleochemical industry. Besides improving its yield, yield stability and increasing the oil content, attention has recently been paid to minor seed constituents like tocopherols (Marwede et al. 2004) and sinapate esters (Hüsken et al. 2005a; Zum Felde et al. 2006). Phytosterols are another group of important, minor constituents. With concentrations ranging from 0.5 to 1% of the crude rapeseed oil, rapeseed is one of the richest natural sources of phytosterols (Piironen et al. 2000). Phytosterols have been known for more than half a century for their LDL-cholesterol-lowering properties (Best et al. 1954). Two decades ago, increased levels of plasma cholesterol have been recognized as one of the main risk factors of cardio vascular diseases—the leading cause of mortality in Western countries (Castelli 1984). One of the main mechanisms for cholesterol reduction is prevention of cholesterol absorption by its replacement with phytosterols in the intestinal-micellar phase (Nissinen et al. 2002; Trautwein et al. 2003). These observations have led to the development of a new type of “functional food”, including margarine, milk and yoghurt, enriched with phytosterols as bioactive components. Following consumption, phytosterols reduce the absorption of dietary and endogenous cholesterol by about 50% (Law 2000).
In contrast to mammals and fungi, which contain only cholesterol and ergosterol, respectively, plants have a complex mixture of different phytosterols. In rapeseed, major phytosterols are sitosterol, campesterol, brassicasterol and avenasterol, while stigmasterol and cholesterol occur only in trace amounts (Appelqvist et al. 1981). Apart from being essential membrane constituents, regulating their fluidity and permeability, phytosterols also regulate membrane-bound enzyme activities and signal transduction events (Hartmann 1998). As plant hormone-precursors they play a crucial role in plant growth and developmental processes like cell division and polarity and morphogenesis (Lindsey et al. 2003; Schaller 2004). Phytosterols are products of the isoprenoid biosynthetic pathway occurring exclusively in the cytoplasm and consisting of more than 25 enzyme-catalyzed reactions (Benveniste 2002).
Only few studies report about genetic variation in phytosterol content in seeds of oilseed rape. The analysis of phytosterol composition in ten different oil types showed that rapeseed oil had the second highest phytosterol content next to corn oil (Gordon and Miller 1997). Analysing phytosterol content of 12 different spring canola varieties, Abidi et al. (1999) reported concentrations ranging from 7,659 to 14,023 mg/kg oil. In another study, three winter rapeseed doubled haploid (DH) populations were tested in replicated field experiments (Amar 2007). Among the DH lines a twofold variation was found in the total phytosterol content, ranging from 4,475 to 9,380 mg/kg oil.
Besides being cultivated for its high oil content, the oil-extracted rapeseed meal is a valuable animal feed containing up to 40% of high nutritive value protein with near optimal amino acid composition (Yoshie-Stark et al. 2006). The utilization of rapeseed protein in food production still depends on further quality improvements (Leckband et al. 2002). In this regard, the high contents of phenolic acid esters in the meal are considered as critical. In canola seeds, phenolic compounds are predominantly sinapate esters, with sinapoylcholine (sinapine) as the most prominent one, followed by sinapoylglucose. Sinapate and other sinapate esters are found only in lower concentrations. Sinapate and the derived esters make up 1–2% of the seed dry matter (Bell 1993) and contribute to the bitter taste, astringency and low nutritive value of rapeseed meal by forming complexes with amino acids and thus inhibiting digestive enzymes (Kozlowska et al. 1990; Shahidi and Naczk 1992). Classical breeding for reduced sinapate ester content seems promising, considering the available large genetic variation along with high heritabilities (Zum Felde et al. 2006), the availability of a NIRS calibration (Zum Felde et al. 2007) and successful transgenic approaches to drastically reduce sinapate ester content in rapeseed (Hüsken et al. 2005a; Hüsken et al. 2005b).
Quantitative trait loci (QTL) for different seed quality traits such as oil content, fatty acid composition, glucosinolates and tocopherols have been identified earlier on molecular linkage maps of the rapeseed genome (Ecke et al. 1995; Uzunova et al. 1995; Marwede et al. 2005; Zhao et al. 2005, Qiu et al. 2006; Zhao et al. 2007). However, QTL for phytosterol and sinapate ester content have not been mapped so far. The objective of this study was to identify QTL for phytosterol and sinapate ester content in a winter rapeseed DH population, previously found to show a large variation for these two traits, and to segregate for the two erucic acid genes (Zum Felde et al. 2006; Amar 2007).
Materials and methods
Plant material and field experiment
The mapping population consisted of 148 DH lines derived from a cross between two DH lines obtained from the ancient Dutch cultivar “Mansholt’s Hamburger Raps” (high contents of erucic acid and glucosinolates) and the modern French cultivar “Samourai” (low contents of erucic acid and glucosinolate). The DH lines were grown in a field trial during two consecutive years at two locations in a randomized block design with two replicates. Each plot consisted of a double row with around 80 plants per plot. In 1999, the two locations were two fields at Reinshof (4 km southwest of Göttingen, Germany) with different soil types. In 2000, one location was Reinshof and the other was Weende (5 km northwest of Göttingen). The two parental lines were grown only in 1999 on one of the fields at Reinshof with four replicates. Seeds harvested from three open pollinated plants per plot were bulked for the analysis (Gül 2002).
Analysis of phytosterol content and other quality traits
A gas–liquid chromatographic (GLC) method was developed and used for assessment of phytosterol content and composition as described in Amar (2007). Phytosterol extraction and preparation for the GLC was performed directly on the seeds in three major steps: alkaline hydrolysis, extraction, and derivatization to trimethyl-silyl ethers. The alkaline hydrolysis as performed allowed the quantification of the sum of free phytosterols and phytosterol fatty acid esters.
Seed oil content, expressed on seed dry matter basis, and erucic acid content were determined by using near-infrared reflectance spectroscopy (NIRS) with the calibration equation raps2001.eqa developed by Tillmann (2007). From selected samples, erucic acid content was also determined by GLC and the results were used to adjust for a systematic bias of the NIRS data. The contents of sinapine, sinapoylglucose, other sinapate esters (sinapate and minor sinapate esters grouped together) and total sinapate esters were determined using the NIRS calibration equations developed by Zum Felde et al. (2007). Samples with the highest and the lowest total sinapate ester content were also analyzed by HPLC (Dr. Baumert, Halle, Germany) and results showed conformity with the NIRS results (n = 20; R2 = 0.81).
Statistical analysis
Analysis of variance was performed with PLABSTAT software version 2N (Utz 1997) using the following model:
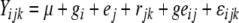 |
where Yijkwas observation of genotype i in environment j in replicate k; μ was the general mean; gi, ej and rjkwere the effects of genotype i, environment j and replicate k in the environment j, respectively; geijwas the genotype × environment interaction of genotype i with environment j and εijkwas the residual error of genotype i in environment j in replicate k. The genotypes, environments and replicates were considered as random variables. Broad-sense heritability (h2) of mean values over environments was calculated from the components of variance using PLABSTAT software version 2N (Utz 1997), as described in Hill et al. (1998):
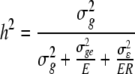 |
where 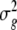 ,
, 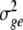 and
and 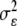 are variance components for g, ge, and ε, and E and R are number of environments and replicates, respectively. Spearman’s rank correlation coefficients were calculated using PLABSTAT software version 2N (Utz 1997). Direct and indirect path coefficients of the path coefficient analysis were calculated as described in Lynch and Walsh (1998):
are variance components for g, ge, and ε, and E and R are number of environments and replicates, respectively. Spearman’s rank correlation coefficients were calculated using PLABSTAT software version 2N (Utz 1997). Direct and indirect path coefficients of the path coefficient analysis were calculated as described in Lynch and Walsh (1998):
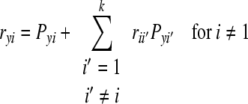 |
where ryi is the correlation coefficient between the ith causal variable (Xi) and effect variable (y), 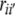 is the correlation coefficient between the ith and i′th causal variables, Pyi is the path coefficient (direct effect) of the ith causal variable (Xi),
is the correlation coefficient between the ith and i′th causal variables, Pyi is the path coefficient (direct effect) of the ith causal variable (Xi), 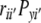 is the indirect effect of the ith causal variable via the i′th causal variable. To determine the direct effect, square matrices of the Spearman’s rank correlation coefficients between independent traits in all possible pairs were inverted and multiplied by the correlation coefficients between the independent and dependent traits. Path coefficient analyses were separately performed for phytosterol and for sinapate ester content as effect variables. In both cases oil and erucic acid content were considered as causal variables.
is the indirect effect of the ith causal variable via the i′th causal variable. To determine the direct effect, square matrices of the Spearman’s rank correlation coefficients between independent traits in all possible pairs were inverted and multiplied by the correlation coefficients between the independent and dependent traits. Path coefficient analyses were separately performed for phytosterol and for sinapate ester content as effect variables. In both cases oil and erucic acid content were considered as causal variables.
Molecular marker map
The QTL were mapped on a framework map comprising 185 evenly distributed markers selected from a previously established primary RFLP (Uzunova et al. 1995) and AFLP map (unpublished). The 185 markers were distributed over 20 linkage groups and covered 1,739 cM (Haldane) of the rapeseed genome. A table showing the markers of the framework map as well as their positions on the linkage groups is available as electronic supplementary material (ESM). Markers and more detailed information are available from the author upon request. Molecular marker genotypic data and means for phytosterol and sinapate ester phenotypic data over all environments of the segregating DH population were used for composite interval mapping performed with PLABQTL version 1.2bic (Utz 2007). Putative QTL were detected by using a LOD score threshold of 2.9 corresponding to a 5% probability of falsely declaring a QTL anywhere in the genome. The LOD score threshold has been obtained by permutation analysis with 2,000 repetitions. Cofactors for composite interval mapping were selected by stepwise regression as described in the manual of PLABQTL (Utz and Melchinger 2006). The nomenclature of Parkin et al. (1995) was used for labelling the linkage groups.
Results
Trait variation and heritability in the DH population
The DH population showed a large and significant variation for total and individual phytosterol content. Four different phytosterols were quantified: sitosterol, campesterol, brassicasterol and avenasterol (Table 1). Sitosterol was the most prominent phytosterol, accounting for 53% of the total phytosterol content, followed by campesterol (29%), brassicasterol (14%) and avenasterol (4%). Stigmasterol was present only in amounts of less than 1% (data not shown). Total phytosterol content ranged from 2,570 to 4,104 mg/kg seed. The ranges of individual phytosterol content were relatively largest for avenasterol (61–303 mg/kg seed) and campesterol (631–1,533 mg/kg seed). Large and significant variations were also found for oil, erucic acid and total sinapate ester content (Table 1). Overall high heritabilities were observed for all phytosterol traits as well as for oil, erucic acid and total sinapate ester content. The parental line “Samourai” had a higher phytosterol and total sinapate ester content compared to “Mansholt’s,” whereas “Mansholt’s” had a higher erucic acid and oil content.
Table 1.
Variation in phytosterol and total sinapate ester content (mg/kg seed) and in oil and erucic acid content (%) in the DH mapping population and their parents
| Sitosterol | Campesterol | Brassicasterol | Avenasterol | Total phytosterols | Total sinapate esters | Oil (%) | Erucic acid (%) | |
|---|---|---|---|---|---|---|---|---|
| Mean | 1,648 | 903 | 415 | 129 | 3,107 | 7,990 | 55 | 25 |
| Min | 1,251 | 631 | 333 | 61 | 2,570 | 6,240 | 50 | 0 |
| Max | 2,138 | 1,533 | 548 | 303 | 4,104 | 9,910 | 60 | 42 |
| F valuea | 10** | 14** | 14** | 7** | 11** | 8** | 12** | 47** |
| LSD 0.05 | 147 | 108 | 31 | 51 | 262 | 730 | 2 | 6 |
| h2 | 0.90 | 0.93 | 0.93 | 0.86 | 0.91 | 0.87 | 0.92 | 0.98 |
| “Mansholt’s” | 1,617 | 1,029 | 383 | 188 | 3,226 | 5,443 | 58 | 49 |
| “Samourai” | 1,896 | 1,552 | 469 | 207 | 4,132 | 6,942 | 55 | 0 |
** Significant at P = 0.01
aF value from analysis of variance for genetic variation in the DH lines
Correlations between traits and path coefficient analysis
Strong and positive correlations were observed between total phytosterol content and the content of all individual phytosterols (Table 2). Individual phytosterols were positively correlated with each other. A significant positive correlation was also found for phytosterols and total sinapate ester content (rs = 0.59**). The DH population segregated for erucic acid content. As expected for a trait inherited by two genes, three classes with no (eeee), medium (EEee, eeEE) and high erucic acid content (EEEE) could be distinguished (data not shown). Total phytosterol content was negatively correlated with erucic acid (rs = −0.80**) and oil content (rs = −0.58**). Strong negative correlations were also observed between total sinapate ester and oil content (rs = −0.71**) and total sinapate ester and erucic acid content (rs = −0.66**).
Table 2.
Spearman’s rank correlation (rs) between different seed quality traits in the DH mapping population
| Constituent | Sitosterol | Campesterol | Brassicasterol | Avenasterol | Total phytosterols | Oil | Erucic acid |
|---|---|---|---|---|---|---|---|
| Campesterol | 0.53** | ||||||
| Brassicasterol | 0.14 | 0.37** | |||||
| Avenasterol | 0.22** | 0.31** | 0.09 | ||||
| Total phytosterols | 0.84** | 0.86** | 0.38** | 0.40** | |||
| Oil | −0.55** | −0.44** | −0.23** | −0.36** | −0.58** | ||
| Erucic acid | −0.69** | −0.68** | −0.28** | −0.45** | −0.80** | 0.69** | |
| Total sinapate esters | 0.61** | 0.42** | 0.20* | 0.31** | 0.59** | −0.71** | −0.66** |
* Significant at P = 0.05
** Significant at P = 0.01
Since oil content was positively correlated with erucic acid content (rs = 0.69**), the question arose whether total phytosterol content was directly affected by oil content or indirectly via erucic acid content. Therefore, a path coefficient analysis was used to partition the correlations into direct and indirect effects (Table 3). Although there was a significant negative correlation between oil and phytosterol content, the results of the path coefficient analysis showed no direct effect of oil content on this trait (−0.05). Nevertheless, the observed strong direct effect of erucic acid content on total phytosterol content (−0.76) is congruent with the highly significant negative correlation between the two traits. At the same time, path analysis revealed a negative indirect effect of oil content on total phytosterol content only via erucic acid content (−0.53). A stronger direct effect of oil content on total sinapate ester content (−0.49) than of erucic acid content (−0.32) was found. Erucic acid content showed a stronger indirect effect on total sinapate ester content via oil content (−0.34) than oil content on total sinapate ester content via erucic acid content (−0.22).
Table 3.
Direct and indirect effects of oil and erucic acid content on total phytosterol and total sinapate ester content
| Seed component | Direct effect | Indirect effect | ||||
|---|---|---|---|---|---|---|
| On total phytosterols | On total sinapate esters | |||||
| On total phytosterols | On total sinapate esters | Via erucic acid | Via oil | Via erucic acid | Via oil | |
| Oil | −0.05 | −0.49 | −0.53 | – | −0.22 | – |
| Erucic acid | −0.76 | −0.32 | – | −0.04 | – | −0.34 |
| Residual effect | 0.60 | 0.66 | 0.60 | 0.66 | ||
QTL for phytosterol content
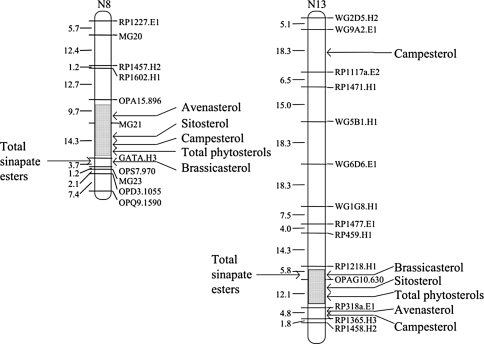
Two large and one small QTL were identified for total phytosterol content, with positive additive effects explaining 54% of the phenotypic and 60% of the genetic variance (Table 4). The two QTL with the largest effects were located on linkage groups N8 and N13 (Fig. 1). The third smaller QTL was located at the top of linkage group N18. Between four and eight QTL were detected for individual phytosterol content, together explaining between 48 and 68% of the total genetic variance of the respective trait. On N8, the confidence interval of the QTL for total phytosterol content overlapped with the confidence intervals of QTL for sitosterol, campesterol and brassicasterol content. Likewise, the QTL for total phytosterol content on N13 had a similar position as the QTL for sitosterol, brassicasterol and avenasterol content. All QTL for individual phytosterols on linkage groups N8 and N13 had positive signs indicating that the presence of the “Samourai” alleles at these QTL positions increased phytosterol content. Additional QTL for individual phytosterols with negative and positive effects were found on a number of other linkage groups (Table 4). A QTL for campesterol content on N4 mapped in 2 cM distance to a QTL for brassicasterol with an opposite effect.
Table 4.
Mapped QTL and their most likely positions for total and individual phytosterol content
| Trait | LGa | Positionb | CIc | Marker interval | LOD score | ad | Vp%e | Vg%e | Total Vp%f | Total Vg%f |
|---|---|---|---|---|---|---|---|---|---|---|
| Total phytosterols | 8 | 62 | 56–64 | MG21-GATA.H3 | 5.9 | 205 | 47 | 52 | 54 | 60 |
| 13 | 136 | 134–140 | OPAG10.630-RP318a.E1 | 22.9 | 172 | 34 | 37 | |||
| 18 | 0 | 0–0 | OPAG4.620-MG87 | 4.7 | 61 | 8 | 8 | |||
| Sitosterol | 6 | 8 | 6–12 | RP1454.E1-WG7E10.H1 | 16.7 | −70 | 30 | 33 | 61 | 68 |
| 8 | 58 | 54–60 | MG21-GATA.H3 | 32.2 | 91 | 39 | 43 | |||
| 10 | 26 | 16–30 | RP1470.H1-WG7B3.H1 | 4.0 | 22 | 4 | 4 | |||
| 13 | 134 | 130–138 | OPAG10.630-RP318a.E1 | 30.2 | 76 | 29 | 33 | |||
| Campesterol | 3 | 76 | 74–78 | RP1142.H1-RP1117a.E4 | 11.8 | 31 | 8 | 8 | 44 | 48 |
| 4 | 42 | 36–46 | RP1230.H1-RP1117a.E1 | 7.6 | 44 | 14 | 15 | |||
| 8 | 60 | 58–64 | MG21-GATA.H3 | 21.1 | 74 | 31 | 34 | |||
| 13 | 24 | 18–32 | WG9A2.E1-RP1117a.E2 | 10.6 | 28 | 6 | 7 | |||
| 13 | 146 | 142–148 | RP318a.E1-RP1365.H3 | 9.2 | 54 | 19 | 21 | |||
| Brassicasterol | 3 | 28 | 26–32 | OPAI16.1420-RP1422.E1 | 10.6 | 8 | 7 | 8 | 48 | 52 |
| 4 | 44 | 42–48 | RP117a.E1-WG4A4.H1 | 4.9 | −22 | 33 | 36 | |||
| 8 | 64 | 60–66 | GATA.H3-OPS7.970 | 8.5 | 12 | 13 | 14 | |||
| 10 | 8 | 6–10 | MG25-MG26 | 10.2 | −10 | 10 | 11 | |||
| 11 | 22 | 16–22 | RP984.H2-MG40 | 5.8 | 8 | 7 | 8 | |||
| 13 | 128 | 124–138 | RP1218.H1-OPAG10.630 | 6.1 | 11 | 11 | 12 | |||
| 15 | 48 | 40–48 | OPT9.862-RP981.H1 | 3.7 | 8 | 7 | 7 | |||
| 19 | 56 | 48–64 | RP1100.E1-RP825.H1 | 10.7 | 9 | 7 | 7 | |||
| Avenasterol | 1 | 40 | 34–44 | GATA.H1-RP1126.H1 | 9.5 | 21 | 26 | 30 | 50 | 58 |
| 3 | 102 | 100–104 | WG2D5.H1-RP793.H1 | 4.1 | −19 | 23 | 27 | |||
| 8 | 36 | 34–40 | OPA15.896-MG21 | 4.1 | 17 | 20 | 23 | |||
| 13 | 144 | 138–148 | RP318a.E1-RP1365.H3 | 3.0 | 14 | 13 | 15 | |||
| 14 | 12 | 0–26 | OPA18.600-cRT21.E1 | 3.0 | −15 | 11 | 12 | |||
| 17 | 10 | 8–12 | RP1202.H1-RP318b.E1 | 5.7 | −8 | 4 | 5 | |||
| 18 | 2 | 0–26 | MG1-RP1144.H1 | 3.3 | 9 | 6 | 7 |
aLinkage group
bPosition of the QTL expressed as the distance from the first marker in cM
cConfidence interval (0.95) expressed as the position on the linkage group in cM
dAdditive effect of the mapped QTL for phytosterol content (mg/kg seed) estimated for the substitution of a “Mansholt’s” allele by a “Samourai” allele
eProportion of phenotypic (Vp) or genetic (Vg) variance explained by the additive effect of the QTL
fProportion of total phenotypic (Vp) or total genetic (Vg) variance explained by the QTL, adjusted (Utz et al. 2000; Utz 2007)
Fig. 1.
The framework maps of linkage groups N8 and N13 with the QTL for phytosterol content. The distances between markers are given in cM (Kosambi 1944). The grey areas are representing the confidence intervals of three LOD score units around the most likely position of the erucic acid genes, according to Ecke et al. (1995)
QTL for sinapate ester content
Two large and two small QTL were identified for total sinapate ester content, with additive effects explaining 46% of the phenotypic and 53% of the genetic variance (Table 5). The two QTL with the largest additive effects were again located on linkage groups N8 and N13 (Fig. 1) at positions close to those of the QTL for total phytosterol content. They both had a positive additive main effect, indicating that the “Samourai” allele at these positions increased total sinapate ester content. Between 3 and 12 QTL were detected for the individual sinapate esters on 15 different linkage groups, which together explained between 51 and 71% of the total genetic variance. No QTL were detected on linkage groups N1, N3, N15 and N17. The QTL for total sinapate ester content on N8 mapped at 4 cM distance to a QTL for sinapoylglucose and to a QTL for other sinapate esters. The QTL for total sinapate ester content on N13 mapped at 4 and 8 cM distance to the QTL for sinapoylglucose content and for other sinapate ester content, respectively. Interestingly, no QTL for sinapine content were mapped on N8 or N13.
Table 5.
Mapped QTL and their most likely positions for sinapate ester content
| Trait | LGa | Positionb | CIc | Marker interval | LOD score | ad | Vp%e | Vg%e | Total Vp%f | Total Vg%f |
|---|---|---|---|---|---|---|---|---|---|---|
| Total sinapate esters | 5 | 64 | 62–66 | OPC19.1090-WG4C5.H1 | 6.9 | −148 | 7 | 9 | 46 | 53 |
| 6 | 76 | 54–82 | WG1F6.H1-OPB5.910 | 3.8 | −184 | 8 | 10 | |||
| 8 | 64 | 58–68 | GATA.H3-OPS7.970 | 15.6 | 398 | 36 | 41 | |||
| 13 | 128 | 126–130 | RP1218.H1-OPAG10.630 | 17.5 | 305 | 23 | 27 | |||
| Sinapine | 4 | 0 | 0–6 | WG6F10.H1-RP1230.H1 | 10.9 | 107 | 10 | 11 | 62 | 71 |
| 5 | 64 | 62–66 | OPC19.1090-WG4C5.H1 | 15.7 | −214 | 27 | 33 | |||
| 7 | 0 | 0–4 | RP1146.H3-RP830.E1 | 12.9 | 173 | 21 | 24 | |||
| 7 | 50 | 42–56 | MG11-RP1457.H1 | 3.0 | −158 | 17 | 20 | |||
| 9 | 4 | 0–10 | WG3F7.H1-RP1175.H1 | 4.7 | −79 | 5 | 5 | |||
| 9 | 72 | 64–76 | RP1253.E1-TG2F9.H1 | 6.7 | 105 | 8 | 10 | |||
| 10 | 6 | 4–8 | OPA18.900-MG25 | 4.2 | 77 | 5 | 6 | |||
| 11 | 22 | 18–22 | RP984.H2-MG40 | 7.6 | −213 | 28 | 32 | |||
| 12 | 64 | 58–70 | RP1565.E1-OPA18.820 | 3.1 | 133 | 13 | 15 | |||
| 12 | 124 | 120–128 | WG7A8.H1-WG4E12.H1 | 18.0 | −186 | 22 | 25 | |||
| 16 | 74 | 64–86 | MG9-RP1087.H1 | 3.5 | 103 | 8 | 9 | |||
| 19 | 36 | 34–44 | RP1100.E1-RP825.H1 | 13.7 | −135 | 14 | 16 | |||
| Sinapoylglucose | 2 | 68 | 62–74 | RP1146.H1-MG16 | 3.6 | −68 | 4 | 5 | 57 | 68 |
| 6 | 38 | 30–40 | RP1068.E1-WG1F6.H1 | 5.1 | −108 | 10 | 12 | |||
| 7 | 10 | 6–16 | RP1122.H1-OPD3.1190 | 8.3 | −120 | 14 | 17 | |||
| 8 | 60 | 56–64 | MG21-GATA.H3 | 27.9 | 263 | 39 | 47 | |||
| 13 | 132 | 126–136 | OPAG10.630-RP318a.E1 | 20.2 | 251 | 36 | 43 | |||
| 14 | 6 | 0–24 | OPA18.600-cRT21.E1 | 3.0 | −79 | 5 | 6 | |||
| Other sinapate esters | 8 | 60 | 56–64 | MG21-GATA.H3 | 29.3 | 142 | 34 | 41 | 43 | 51 |
| 13 | 136 | 132–140 | OPAG10.630-RP318a.E1 | 13.6 | 112 | 22 | 27 | |||
| 18 | 0 | 0–0 | OPAG4.620-MG87 | 6.7 | 74 | 14 | 17 |
aLinkage group
bPosition of the QTL expressed as the distance from the first marker in cM
cConfidence interval (0.95) expressed as the position on the linkage group in cM
dAdditive effect of the mapped QTL for sinapate ester content (mg/kg seed) estimated for the substitution of a “Mansholt’s” allele by a “Samourai” allele
eProportion of phenotypic (Vp) or genetic (Vg) variance explained by the additive effect of the QTL
fProportion of total phenotypic (Vp) or total genetic (Vg) variance explained by the QTL, adjusted (Utz et al. 2000; Utz 2007)
Discussion
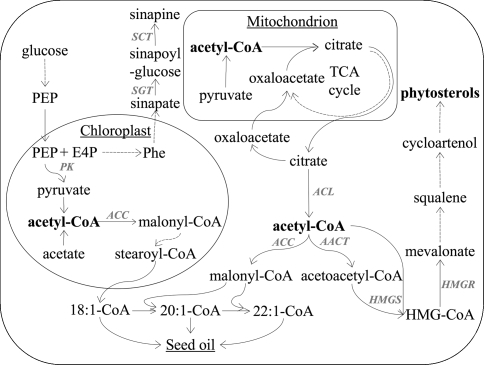
Traditional rapeseed contains around 50% erucic acid in the seed oil. The detection of low erucic acid mutants led to a complete conversion to low erucic acid cultivars in the 1970s in Canada and in Europe (Shahidi 1990). Nowadays, the “Canola” or double zero (“00”) quality type cultivars with low erucic acid content in the seed oil and low glucosinolate content in the seed meal are exclusively being cultivated in most rapeseed growing areas of the world. The conversion to low erucic acid seed oil quality has not been reported to affect any other seed constituents than the fatty acid composition and the oil content (Ecke et al. 1995). This is rather surprising, since erucic acid is formed via consecutive elongation of oleoyl-CoA with malonyl-CoA (Fig. 2). Unlike the synthesis of fatty acids with chain lengths shorter than 18 carbons that occurs in plastids, the elongation of oleoyl-CoA occurs in cytoplasm. There, malonyl-CoA is generated through carboxylation of acetyl-CoA. Cytoplasmic acetyl-CoA is also required for the biosynthesis of a plethora of other compounds, e.g. isoprenoids, phenolics, flavonoids, stilbenoids, alkaloids, anthocyanins, etc. (Fatland et al. 2005). Among these, phytosterols are present in the highest concentration in the seed (0.3%; Table 1).
Fig. 2.
Scheme of putative phytosterol, fatty acid and sinapate ester biosynthetic pathways in plants (adapted from Milkowski et al. 2004; Fatland et al. 2005; Ruuska et al. 2002; Fischer et al. 1997). Solid and dashed arrows represent single and multiple enzymatic reactions, respectively. TCA tricarboxylic acid cycle, HMG-CoA 3-hydroxy-3-methylglutaryl-CoA, PEP phosphoenolpyruvate, E4P erythrose-4-phosphate, Phe phenylalanine, ACL ATP-citrate lyase, ACC acetyl-CoA carboxylase, AACT acetoacetyl-CoA thiolase, HMGS HMG-CoA synthase, HMGR HMG-CoA reductase, PK pyruvate kinase; SGT sinapate glucosytransferase, SCT choline sinapoyltransferase
Considering the fact, that up to 25% of the seed may consist of erucic acid and that two molecules of acetyl-CoA are required for the synthesis of one molecule of erucic acid, it can be anticipated that a major change in erucic acid content could have an effect on contents of other seed constituents synthesized from cytoplasmic acetyl-CoA. Indeed, the results of the present study show a close negative association between total phytosterol content and erucic acid content (Table 2). The results from the path coefficient analysis confirmed the direct negative effect of erucic acid on total phytosterol content (Table 3). Further evidence for the association between erucic acid content and total phytosterol content can be drawn from the fact that the two largest QTL for total phytosterol content on linkage groups N8 and N13 mapped within the confidence intervals of the two erucic acid genes (Fig. 1). Their additive effects were positive, indicating that the alleles increasing the total phytosterol content were coming from the low erucic acid parent “Samourai”. The two erucic acid genes were mapped earlier in the present population on linkage groups LG6 and LG12 by Ecke et al. (1995), which correspond to linkage groups N8 (B. rapa) and N13 (B. oleracea) mapped by Parkin et al. (1995). Obviously, there is a pleiotropic effect of the erucic acid genes on phytosterol content, which may be explained by the competition for cytoplasmic acetyl-CoA—an early essential precursor for both compounds (Fig. 2) and presumably available only in limited quantities (Fatland et al. 2005). Otherwise, it could be also possible that major genes for phytosterol content are closely linked to the erucic acid genes. This could be checked easily by analyzing transgenic rapeseed expressing the erucic acid fae1-gene (James et al. 1995) in an otherwise low erucic acid background (Han et al. 2001).
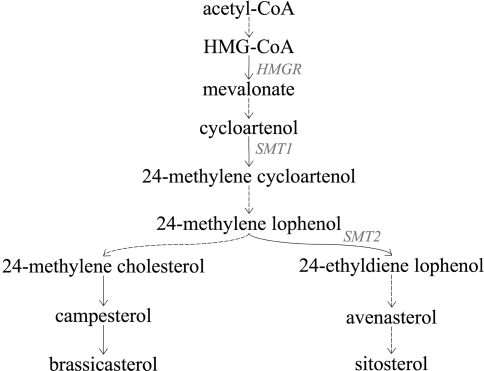
The two major QTL for total phytosterol content on N8 and N13 were at the same time QTL for individual phytosterols. Confidence intervals were overlapping, except for the QTL for avenasterol on N8 and the QTL for campesterol on N13. This was not the case for the third minor QTL for total phytosterol content on N18; only a QTL for avenasterol was detected at a distance of 2 cM. It could be that the effect of this QTL for total phytosterol content was too small to be detected in QTL mapping for the other individual phytosterols. QTL for sitosterol and campesterol content mapped at the same position on N18, but were below the critical LOD score threshold (data not shown). At this time, it can only be speculated about the gene behind the QTL on N18 and its function. Results from transgenic plants show, that over expression or inhibition of the key biosynthetic enzymes 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR; Fig. 3) and cycloartenol C-24-methyltransferase (SMT1), influence the amount of total phytosterol content in Nicotiana tabacum and Arabidopsis thaliana seed and leaf tissue (Holmberg et al. 2003; Harker et al. 2003a; Schaller 2004). Hence, these genes are candidates for the QTL on N18. Transgenic approaches have been successful in modifying phytosterol composition and accumulation in seeds. Ectopic expression of the HMGR gene in transgenic tobacco led to 3.2-fold and 10-fold increased phytosterol levels in seed and leaf tissue, respectively (Harker et al. 2003b). Overexpression of the SMT1 gene in tobacco seed tissue increased the amount of total phytosterols up to 44% and modulated the phytosterol composition; sitosterol proportion was increased by up to 50% and levels of isofucosterol (sitosterol precursor) and campesterol increased by up to 80% (Holmberg et al. 2002).
Fig. 3.
Scheme of phytosterol biosynthetic pathway in plants (adapted from Holmberg et al. 2003; Benveniste 2002). Solid and dashed arrows represent single and multiple enzymatic reactions, respectively. HMG-CoA 3-hydroxy-3-methylglutaryl-CoA, HMGR HMG-CoA reductase, SMT1 cycloartenol C-24-methyltransferase, SMT2 SAM-24-methylene lophenol C-24-methyltransferase
The correlations among individual phytosterols were all positive (Table 2), suggesting that most of the variation is caused by genetic differences before the separation of the two pathways leading to brassicasterol and sitosterol (Fig. 3). On N4, there was a QTL for campesterol that mapped at a distance of only 2 cM to a QTL for brassicasterol. Those QTL may be identical. However, so far little is known about the gene(s) responsible for the conversion of campesterol to brassicasterol (Schaller 2004).
The close negative association between total sinapate ester and erucic acid content (Table 2) were confirmed by the results from the QTL analysis, which showed that the two major QTL for total sinapate ester content mapped at very similar positions as the erucic acid genes (Table 5). The confidence intervals of the erucic acid genes (Ecke et al. 1995) and of the two QTL for total sinapate ester content were overlapping (data not shown). This also explains the negative association between total sinapate ester content and oil content (indirect effect of oil via erucic acid content). However, path coefficient analysis also revealed a direct effect of oil content on total sinapate ester content (Table 3). From known biochemical pathways there is no obvious connection between the biosynthesis of sinapate esters and erucic acid. Starting from phenylalanine, sinapate esters are synthesized in the cytoplasmic cell compartment via the phenylalanine/hydroxycinnamate pathway (Fig. 2; Milkowski et al. 2004). It can be only speculated that plastidic phosphoenolpyruvate (PEP), as a common precursor for de novo fatty acid and phenylalanine biosynthesis (Fischer et al. 1997), is limiting and hence may explain the negative association between sinapate esters and oil content. Amongst other enzyme activities, pyruvate kinase (PK; Fig. 2) has been found to be enhanced in a B. napus line with high oil content compared to a near-isogenic line with a low oil content, corroborating the importance of this step in storage lipid biosynthesis (Li et al. 2006). Zum Felde et al. (2006) analyzed the same DH population as in the present study, but reported only a slightly negative, non-significant correlation between oil content and total sinapate ester content. This may be due to the fact, that the seed material used in the present study was derived from the field trial with no additional N-fertilizers, which led to overall higher contents of oil. Even so, for two additional DH populations analyzed by Zum Felde et al. (2006), no significant association between oil and sinapate ester content were reported; both populations were low in erucic acid content. Further studies are needed to elucidate the association between total sinapate ester content and oil content as well as erucic acid content. The effects of the two major QTL on N8 and on N13 were limited to sinapoylglucose and to the other sinapate esters; for sinapine content no significant QTL were found in these regions. Sinapoylglucose is the direct precursor for sinapine. This step is catalyzed by the choline sinapoyltransferase (SCT, Fig. 2; Milkowski et al. 2004). The QTL for sinapine at the top of N7 with a positive effect mapped 10 cM away from a QTL for sinapoylglucose with a negative effect, indicating that they may be identical. A candidate for this QTL could be the SCT gene (Milkowski et al. 2004).
Conclusions
The present study provides some evidence that the two erucic acid genes exert a pleiotropic effect on phytosterol and total sinapate ester content in seeds of oilseed rape. Canola quality type rapeseed has a higher total sinapate ester and phytosterol content than high erucic acid rapeseed material. These results have recently been confirmed by using a set of intervarietal substitution lines derived from the same parents as in this study (Kebede 2007). The present findings have consequences when screening Brassica genetic resources for reduced contents of sinapate esters or increased phytosterol contents (Zum Felde et al. 2007). In this case some adjustment should be performed depending on the erucic acid content of the seed oil. It is likely that the two erucic acid genes exert a pleiotropic effect also on other secondary compounds synthesized from cytoplasmic acetyl-CoA. Beside the two major QTL, additional QTL for total and individual phytosterol and sinapate ester content were identified, which may be useful in future candidate gene mapping projects. The utilization of an additional DH population not segregating for erucic acid content for mapping QTL for phytosterol and sinapate ester content should increase the power of detection for QTL with smaller effects. Alternatively, conditional QTL mapping, taking the variation in the erucic acid content into account could be performed with the present DH population (Zhao et al. 2006).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors express gratitude to Uwe Ammerman for his professional technical assistance, to Dr. Ujjal Nath for his help with the path coefficient analysis, to Dr. Mladen Radoev for his help with the QTL mapping and to Dr. Kemal Gül for providing the seed samples. The authors are also grateful to Dr. Alfred Baumert (IPB Halle, Germany) for performing HPLC reference analysis of sinapate ester content. Samija Amar gratefully appreciates the financial support of the Hans-Böckler-Stiftung.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-008-0734-2) contains supplementary material, which is available to authorized users.
References
- Abidi SL, List GR, Rennick KA (1999) Effect of genetic modification on the distribution of minor constituents in canola oil. J Am Oil Chem Soc 76:463–467 [DOI]
- Amar S (2007) Genetic variation and inheritance of phytosterol content in Brassica napus L. Ph.D. Thesis, Faculty of Agriculture, Georg-August-Universität Göttingen, Germany. http://webdoc.sub.gwdg.de/diss/2007/amar/amar.pdf
- Appelqvist LAD, Kornfeldt AK, Wennerholm JE (1981) Sterols and steryl esters in some Brassica and Sinapis seeds. Phytochemistry 20:207–210 [DOI]
- Bell JM (1993) Factors affecting the nutritional value of canola meal. Can J Anim Sci 73:679–697
- Benveniste P (2002) Sterol metabolism. In: Meyerowitz EM, Somerville C (eds) The Arabidopsis book. American Society of Plant Biologists, pp 1–31. Available via http://www.aspb.org/publications/arabidopsis/. Accessed 12 June 2007
- Best MM, Duncan CH, van Loon EJ, Wathens JD (1954) Lowering of serum cholesterol by the administration of plant sterol. Circulation 10:201–206 [DOI] [PubMed]
- Castelli WP (1984) Epidemiology of coronary heart disease: The Framingham Study. Am J Med 76:4–12 [DOI] [PubMed]
- Ecke W, Uzunova M, Weißleder K (1995) Mapping the genome of rapeseed (Brassica napus L.). II. Localisation of genes controlling erucic acid synthesis and seed oil content. Theor Appl Genet 91:972–977 [DOI] [PubMed]
- Fatland BL, Nikolau BJ, Syrkin WE (2005) Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell 17:182–203 [DOI] [PMC free article] [PubMed]
- Fischer K, Kammerer B, Gutensohn M, Arbinger B, Weber A, Häusler RE, Flügge U-I (1997) A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9:453–462 [DOI] [PMC free article] [PubMed]
- Gordon MH, Miller LAD (1997) Development of steryl ester analysis for the detection of admixtures of vegetable oils. J Am Oil Chem Soc 74:505–510 [DOI]
- Gül K (2002) QTL-Kartierung und Analyse von QTL × Stickstoff Interaktionen beim Winterraps (Brassica napus L.). Cuvillier Verlag, Göttingen
- Han J, Lühs W, Sonntag K, Zähringer U, Borchardt DS, Wolter FP, Heinz E, Frentzen M (2001) Functional characterization of β-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol Biol 46:229–239 [DOI] [PubMed]
- Harker M, Hellyer A, Clayton JC, Duvoix A, Lanot A, Safford R (2003a) Co-ordinate regulation of sterol biosynthesis enzyme activity during accumulation of sterol in developing rape and tobacco seed. Planta 216:707–715 [DOI] [PubMed]
- Harker M, Holmberg N, Clayton JC, Gibbaard CL, Wallace AD, Rawlins S (2003b) Enhancement of seed phytosterol levels by expression of N-terminal truncated Hevea brasiliensis (rubber tree) 3-hydroxy-3methylgutaryl-CoA reductase. Plant Biotech J 1:113–121 [DOI] [PubMed]
- Hartmann M-A (1998) Plant sterols and membrane environment. Trends Plant Sci 3:170–175 [DOI]
- Hill J, Becker HC, Tigerstedt PMA (1998) Quantitative and ecological aspects of plant breeding. Chapman & Hall, London, pp 119–120
- Holmberg N, Harker M, Wallace AD, Clayton JC, Rawlins S, Hellyer A, Safford R (2002) Sterol C-24 methyltransferase type 1 controls the flux of carbon into sterol biosynthesis in tobacco seed. Plant Physiol 130:303–311 [DOI] [PMC free article] [PubMed]
- Holmberg N, Harker M, Wallace AD, Clayton JC, Gibbard CL, Hellyer A, Safford R (2003) Co-expression of N-terminal truncated 3-hydroxy-3-methylglutaryl CoA reductase and C24-sterol methyltransferase type 1 in transgenic tobacco enhances carbon flux towards end-product sterol. Plant J 36:12–20 [DOI] [PubMed]
- Hüsken A, Baumert A, Strack D, Becker HC, Möllers C, Milkowski C (2005a) Reduction of sinapate ester content in transgenic oilseed rape (Brassica napus L.) by dsRNAi-based suppression of BnSGT1 gene expression. Mol Breed 16:127–138 [DOI]
- Hüsken A, Baumert A, Milkowski C, Becker HC, Strack D, Möllers C (2005b) Resveratrol glucoside (Piceid) synthesis in seeds of transgenic oilseed rape (Brassica napus L.). Theor Appl Genet 111:1553–1562 [DOI] [PubMed]
- James DW, Lim E, Keller J, Plooy I, Ralston E, Dooner HK (1995) Direct tagging of the Arabidopsis fatty acid elongation (fae1) gene with the maize transposon activator. Plant Cell 7:309–319 [DOI] [PMC free article] [PubMed]
- Kebede B (2007) Development of intervarietal substitution lines in Brassica napus L. using marker assisted selection and mapping of QTL for agronomically important traits. Ph.D. Thesis, Faculty of Agriculture, Georg-August-Unveristät Göttingen, Germany. http://webdoc.sub.gwdg.de/diss/2007/kebede/kebede.pdf
- Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
- Kozlowska H, Naczk M, Shahidi F, Zadernowski R (1990) Phenolic acids and tannins in rapeseed and canola. In: Shahidi F (ed) Canola and rapeseed. Production, chemistry, nutrition and processing technology. Van Nostrand Reinhold, New York, pp 193–210
- Law M (2000) Plant sterol and stanol margarines and health. Br Med J 320:861–864 [DOI] [PMC free article] [PubMed]
- Leckband G, Frauen M, Friedt W (2002) NAPUS 2000. Rapeseed (Brassica napus) breeding for improved human nutrition. Food Res Int 35:273–278 [DOI]
- Li R-J, Wang H-Z, Mao H, Lu Y-T, Hua W (2006) Identification of differentially expressed genes in seeds of two near-isogenic Brassica napus lines with different oil content. Planta 224:952–962 [DOI] [PubMed]
- Lindsey K, Pullen ML, Topping JF (2003) Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci 8:1360–1385 [DOI] [PubMed]
- Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates Inc., Sunderland, pp 823–831
- Marwede V, Schierholt A, Möllers C, Becker HC (2004) Genotype × environment interactions and heritability of tocopherols content in canola. Crop Sci 44:728–731
- Marwede V, Gül KM, Becker HC, Ecke W (2005) Mapping of QTL controlling tocopherol content in winter oilseed rape. Plant Breed 124:20–24 [DOI]
- Milkowski C, Baumert A, Schmidt D, Nehlin L, Strack D (2004) Molecular regulation of sinapate ester metabolism in Brassica napus: expression of genes, properties of the encoded proteins and correlation of enzyme activities with metabolite accumulation. Plant J 38:80–92 [DOI] [PubMed]
- Mittelbach M, Gangl S (2001) Long storage stability of biodiesel made from rapeseed and used frying oil. J Am Oil Chem Soc 78:573–577 [DOI]
- Nissinen M, Gylling H, Vuoristo M, Miettinen TA (2002) Micellar distribution of cholesterol and phytosterols after duodenal plant stanol ester infusion. Am J Physiol Gastrointest Liver Physiol 282:1009–1015 [DOI] [PubMed]
- Parkin IAP, Sharpe AG, Keith DJ, Lydiate DJ (1995) Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38:1122–1131 [DOI] [PubMed]
- Piironen V, Lindsay DG, Miettinen TA, Toivo J, Lampi A-M (2000) Plant sterols: biosynthesis, biological function and their importance to human nutrition. J Sci Food Agric 80:939–966 [DOI]
- Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, Zhuang X, Wang Y, Tan X, Dietrich E, Weihmann T, Everett C, Vanstraelen S, Beckett P, Fraser F, Trick M, Barnes S, Wilmer J, Schmidt R, Li J, Li D, Meng J, Bancroft I (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114:67–80 [DOI] [PubMed]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14:1191–1206 [DOI] [PMC free article] [PubMed]
- Schaller H (2004) New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol Biochem 42:465–476 [DOI] [PubMed]
- Shahidi F (1990) Rapeseed and canola: global production and distribution. In: Shahidi F (eds) Canola and rapeseed. Production, chemistry, nutrition and processing technology. Van Nostrand Reinhold, New York, pp 3–13
- Shahidi F, Naczk M (1992) An overview of the phenolics of canola and rapeseed: chemical, sensory and nutritional significance. J Am Oil Chem Soc 69:917–924 [DOI]
- Tillmann P (2007) http://www.vdlufa.de/nirs. Accessed 12 June 2007
- Trautwein EA, Duchateau G, Lin Y, Melcnikov SM, Molhuizen H, Ntanios FY (2003) Proposed mechanisms of cholesterol-lowering action of plant sterols. Eur J Lipid Sci Technol 105:171–185 [DOI]
- Utz HF (1997) PLABSTAT: a computer program for the computation of variances and covariances. Universität Hohenheim, Germany
- Utz HF (2007) PLABQTL: a program for the composite interval mapping of QTL. http://www.uni-hohenheim.de/ipspwww/soft.html. Accessed 12 June 2007
- Utz FH, Melchinger AE (2006) PLABQTL: a computer program to map QTL. Institute of Plant Breeding, Seed Science, and Population Genetics, University of Hohenheim. http://www.uni-hohenheim.de/ipspwww/soft.html. Accessed 19 December 2007
- Utz FH, Melchinger AE, Schön CC (2000) Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154:1839–1849 [PMC free article] [PubMed]
- Uzunova M, Ecke W, Weissleder K, Röbbelen G (1995) Mapping the genome of rapeseed (Brassica napus L.). I. Construction of an RFLP linkage map and localisation of QTLs for seed glucosinolate content. Theor Appl Genet 90:194–204 [DOI] [PubMed]
- Yoshie-Stark Y, Wada Y, Schott M, Wäsche A (2006) Functional and bioactive properties of rapeseed protein concentrates and sensory analysis of food application with rapeseed protein concentrates. LTW Food Sci Technol 39:503–512
- Zhao J, Becker HC, Zhang D, Zhang Y, Ecke W (2005) Oil content in a European × Chinese rapeseed population: QTL with additive and epistatic effects and their genotype–environment interactions. Crop Sci 45:51–59
- Zhao J, Becker HC, Zhang D, Zhang Y, Ecke W (2006) Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor Appl Genet 113:33–38 [DOI] [PubMed]
- Zhao J, Dimov Z, Becker HC, Ecke W, Möllers C (2007) Mapping QTL controlling fatty acid composition in a doubled haploid rapeseed population segregating for oil content. Mol Breed. doi:10.1007/s11032-007-9113-y
- Zum Felde T, Becker HC, Möllers C (2006) Genotype × environment interactions, heritability and trait correlations of sinapate ester content in winter rapeseed (Brassica napus L.). Crop Sci 46:2195–2199 [DOI]
- Zum Felde T, Baumert A, Strack D, Becker HC, Möllers C (2007) Genetic variation for sinapate ester content in winter rapeseed (Brassica napus L.) and development of NIRS calibration equation. Plant Breed 126:291–296 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.