Summary
The mechanisms that regulate sarcomere assembly during myofibril formation are poorly understood. In this study, we characterise the slothu45 mutant in which the initial steps in sarcomere assembly take place, but thick filaments are absent and filamentous I-Z-I brushes fail to align or adopt correct spacing. The mutation only affects skeletal muscle and mutant embryos show no other obvious phenotypes. Surprisingly we find that the phenotype is due to mutation in one copy of a tandemly duplicated hsp90a gene. The mutation disrupts the chaperoning function of Hsp90a through interference with ATPase activity. Despite being located only 2kb away from hsp90a, hsp90a2 has no obvious role in sarcomere assembly. Loss of Hsp90a function leads to down-regulation of genes encoding sarcomeric proteins and upregulation of hsp90a and several other genes encoding proteins that may act with Hsp90a during sarcomere assembly. Our studies reveal a surprisingly specific developmental role for a single Hsp90 gene in a regulatory pathway controlling late steps in sarcomere assembly.
Introduction
Myofibrils are the subcellular contractile apparatus of cardiac and striated muscle cells. Within each myofibril, Actin, Myosin and many other proteins are organised into reiterative arrays of sarcomeres. We understand how the intra- and extra-sarcomeric components of muscles fit together (Clark et al., 2002; Sellers, 2004), how muscle fibres are triggered to contract and how the sarcomere transduces force (Cooke, 2004; Rossi and Dirksen, 2006). However, despite this depth of knowledge of the structure and biophysics of the sarcomere and its constituents, our grasp of myofibrillogenesis, the process by which it takes form, remains surprisingly obscure.
During myofibrillogenesis, the first true sarcomeric components to appear are Actin filaments (Ehler et al., 1999; Van der Ven et al., 1999) which assemble into I-Z-I bodies/brushes; structures consisting of Z-discs flanked on both sides by Actin filaments (Schultheiss et al., 1990). The giant sarcomeric protein Titin integrates onto the nascent sarcomere around the stage that Z-disc and I-band epitopes are first present (Fürst et al., 1989). It is thought that thick filaments are subsequently assembled onto this scaffold.
The mechanisms by which thick filaments incorporate are controversial and three models have emerged. The first proposes that thick filaments form independently from I-Z-I brushes and these two separate sarcomeric components are integrated to form striated myofibrils (Holtzer et al., 1997; Schultheiss et al., 1990). The second holds that premyofibrils are first constructed from non-muscle Myosin which is later replaced by muscle Myosin to form functional sarcomeres (LoRusso et al., 1997; Rhee et al., 1994; Sanger et al., 2002). The third theory proposes that Titin is a scaffold upon which other sarcomeric components are assembled (Ehler et al., 1999; Gregorio et al., 1999; Trinick and Tskhovrebova, 1999; Van der Ven et al., 1999). The idea is that the N-terminal portion of Titin first associates with the Z-disc and I-band (forming I-Z-I brushes). These complexes are then brought into register, either by the unfolding of the Titin filament or further translation, and the association of m-line components (Fulton and Alftine, 1997; Fulton and L’Ecuyer, 1993). Upon this scaffold, Myosin is then integrated to form the thick filaments. In support of this model, the M-line region of Titin is important for myofibrillogenesis (Gotthardt et al., 2003; Musa et al., 2006). The current models of myofibrillogenesis have arisen from studies of cultured cardiomyocytes and in vivo studies have, to date, added little to resolve between them.
The initial formation of I-Z-I brushes is common between the myofibrillogenesis models and the controversy concerns the assembly and alignment of arrays of I-Z-I brushes and integration of thick filaments into the nascent sarcomeres. Thick filaments are elaborate structures comprised of hundreds of myosin hexamers in precise alignment with each other and other sarcomeric components. Given this inherent complexity, it is perhaps unsurprising that thick filament assembly is not well understood.
Here we describe the cloning of the slothu45 (slou45) mutation and its phenotypic consequences. Zebrafish slou45 embryos show no morphological defects and have a normal heartbeat but lack movement of skeletal muscles. Various analyses reveal that the contractility phenotype is due to a lack of assembly of thick filaments in the nascent sarcomeres of skeletal muscle fibres. Much to our initial surprise, we found that 3 slo alleles all contain mutations in the hsp90a gene.
Heat shock proteins (Hsps) are a group of proteins with transcription that is induced in response to heat or other cellular stresses. They are molecular chaperones for huge numbers of proteins and Hsp90 alone may be able to interact with more than 400 different proteins (Zhao et al., 2005). Despite extensive research upon Hsps, there has been little work addressing their developmental roles during vertebrate embryogenesis. Work from C.elegans has suggested a role for molecular chaperones during thick filament assembly and integration. The Unc45-/Cro1p-/She4p-related (UCS) protein Unc-45 functions during assembly of thick filaments (Barral et al., 1998; Barral et al., 2002; Etard et al., 2007) and Unc-45 binds stoichiometrically with Hsp90 (Barral et al., 2002). Our findings extend these studies and show that a single, developmentally regulated hsp90 gene is necessary for thick filament assembly and the construction of functional sarcomeres in skeletal myofibrils. Our data reveal unexpected specificity in the developmental role of Hsp90 and raise the possibility that other hsp genes may play similarly cell-type specific roles during vertebrate embryogenesis.
Materials and Methods
Immunohistochemistry, phalloidin and bungarotoxin staining and in situ hybridisation
Immunohistochemistry was performed with standard methods using: ZNP-1 (DSHB, University of Iowa, IA USA), anti-Actinin (EA-53; #A5044; Sigma), A4.1025 (Dan-Goor et al., 1990) which recognises all forms of sarcomeric MHC (‘pan’-MHC antibody), F59 which is specific to slow-muscle Myosin isoforms (slow-MHC), anti-Titin (T11; Sigma) which marks an eptiope in the I-band region of Titin (Fürst et al., 1988). Alexafluor conjugated phalloidin or bungarotoxin (Invitrogen) were applied after secondary antibodies. For in situ, DIG labelled antisense riboprobes were prepared using standard methods from EST constructs (Genbank refs: hspa8l: BI710554, hsp90a: CN330912, unc45b: CN318309; www.rzpd.de) or constructs made in-house.
SDS-PAGE/Western Blotting
Protein was extracted by homogenisation of deyolked embryos, 10μg of protein was seperated using 6% polyacrylamide gels, followed by transfer onto PVDF membrane (Amersham). Myosin antibodies were the same as for immunohistochemistry, rat anti-Hsp90 (16F1;Abcam) was used for Hsp90, mouse anti-gamma tubulin (Sigma) was the loading control. Peroxidase activity from HRP-conjugated secondaries (Sigma) was detected using an ECL detection kit (Amersham)..
Electron microscopy
Embryos were fixed in 2% paraformaldehyde, 2% gluteraldehyde in 0.1M sodium cacodylate buffer (pH 7.3) with 2-5% CaCl2. Tissue was post-fixed in cacodylate buffered 1% w/v osmium tetraoxide, en bloc stained in 2%w/v uranyl acetate and embedded in Agar 100 resin (Agar Scientific). Ultrathin sections (80-100nm) contrasted with saturated Uranyl acetate solution and lead citrate were examined and photographed using a JEOL1010 electron microscope.
BAC/DNA rescue/morpholino (Mo) injection
One-cell embryos were injected using capillary needles and picospritzer II (General valve corp) with 10nl of BAC DNA (rzpd/Chori; Table S2) made using a BAC kit (PSI clone). Full-length u45 mutant and wild-type hsp90a was amplified by RT-PCR, cloned into the TOPO vector (Invitrogen) and subcloned by PCR into pCS2myc+. 100-200pg of sacII-linearised constructs were injected. Mo sequences were: hsp90a: CCGACTTCTCAGGCATCTTGCTGTG, hsp90a2: TCGAGTGGTTTATTCTGAGAGTTTC (produced secondary phenotypes) or hsp90a2: CTGCTGCTCGTGAGCCTCAGGCATC (Genetools). 2pM (+/- 0.5) was injected per embryo.
Microarray analysis
Total-RNA was extracted using TRIzol from pooled slou45 and wild-type embryos. RNA and microarray processing was carried out in the ICH Gene Microarray centre (Institute of Child Health, London) according to standard methods. The MAS5 algorithm (Affymetrix) was used to compare wild-type and slou45 mutants. Genes designated as showing no change in expression or designated as absent in both wild-type and mutant were eliminated. The data was then split into two groups: genes with a slo vs wild-type signal log ratio of between 0.6 and 1 or -0.6 and -1 (1.5 to 2 fold up- or down-regulated) and genes showing a signal log ratio greater than +/-1 (greater than 2 fold up- or down-regulation)(Table S4). These groups were further sorted into genes absent in wild-type or slo mutants and those increased and decreased in slo mutants (Table S3).
Quantitative PCR (qPCR)
The following cDNA-specific primers were used for qPCR: hsp90a: F: CCACCTTAAAGAGGATCAGTCT, R: TCTTCCTCTTATTCTTGCCAT, hsp90a2 F: GCGGCGGATCAAAGAGATC, R: CACTTATCGCCATGATCGTG. cDNA was produced using superscriptII (Invitrogen) and 1μg of quality-checked Total-RNA extracted using TRIzol from pools of 10 mutants and 10 siblings. Triplicate qPCR reactions were carried out with blank controls and five standards. Copy number was determined by reference to standards normalised to a standard curve (10 triplicate 4-fold dilutions), copy number values were normalised to 1μg of total-RNA and significance determined by t-test.
Isothermal titration calorimetry, Kd determinations and Hsp90 ATPase assay
Heats of interaction were measured on a MSC system (Microcal), with a cell volume of 1.458ml, under the same buffer conditions (20mM Tris pH 7.5 containing 1mM EDTA, 5mM NaCl and 7mM MgCl2) at 30°C. 20 aliquots of 14.5μl of 1mM AMPNP were injected into 50μM yeast Hsp90, human Hsp90α or mutant protein. Heats of dilution were determined in a separate experiment by diluting protein into buffer, and the corrected data fitted using a nonlinear least square curve-fitting algorithm (Microcal Origin) with three floating variables: stoichiometry, binding constant and change in enthalpy of interaction. ATPase activity of purified mutated and normal yeast and human Hsp90 protein was measured as described previously (Panaretou et al., 1998).
Results
The motility defects of akinetou45/slothu45 mutants are due to defective skeletal muscle fibres
akinetou45 is a recessive mutation isolated in an F3 screen for ENU-induced mutations at UCL. Mutant embryos are healthy, show no obvious morphological defects and ostensibly develop as wild-type siblings for their first week. However, they do not move (VideoS1), and die by 8dpf. Pairwise breeding showed akinetou45 does not complement the tu44c and tm201 alleles of sloth (slo) (Granato et al., 1996) and we therefore re-named the novel mutation slou45. slo mutants lack spontaneous or stimulus-evoked skeletal muscle contractility but have a normal heartbeat. There is some variability in phenotype between alleles and although no movement is observed in the trunk of homozygous slou45 embryos, the fins of a minority of slou45 mutants show very slight twitching from 4dpf. In contrast, around 30% of slotu44c and slotm201 mutants show limited trunk movements at 24hpf that disappear by 48hpf, by 4dpf there is substantial, though still compromised, fin movement. Transheterozygotes (slou45/tu44c, slou45/tm201 or slotu44c/tm201) resemble the slotu44c and slotm201 phenotype, showing some trunk movement at 24hpf and more vigorous fin twitching later (compared to slou45). These results suggest that u45, tu44c and tm201 are mutations in the same gene and that u45 is the most severe allele.
The two most likely causes of the slo phenotype are defective innervation of muscle fibres and/or a defective contractile response of the muscle fibres to innervation. Examination of muscle innervation and calcium transients within muscle fibres revealed only minor differences between slou45 mutants and wild-types (Fig. S1; Table S1). The absence of neuronal or excitation-coupling problems suggested a defect intrinsic to the muscle fibres. This was corroborated by polarised microscopy, mosaic labelling and cell transplantation experiments which showed slou45 mutants had abnormally wavy muscle fibres that totally lacked birefringence (Fig. S1). This strongly suggested a defect intrinsic to the contractile apparatus of the muscle fibres.
The filamentous organisation of myofibrils is abnormal in slo mutants
Antibodies and probes for sarcomeric components elucidated the sarcomeric defects underlying the myofibrillar phenotype of slou45 embryos (Fig. 1). Although filamentous Actin is present, the myofibrils of slou45 mutants lack the organised fibrillar arrangement seen in siblings (Fig. 1A&B). Sarcomeric filamentous Actin is anchored at Z-discs composed of alpha-Actinin, capping proteins and other molecules. Double-staining for Z-discs and Actin filaments in siblings revealed evenly-spaced Z-discs in register between neighbouring fibrils and even between adjacent fibres (Fig. 1C). slou45 myofibrils lacked such regimented organisation (Fig. 1D). Much of the anti-Actinin staining was dispersed in the cytoplasm, although small structures resembling Z-discs surrounded by Actin were present. However these mutant ‘Z-discs’ were neither appropriately spaced nor in register. The sarcomeric organisation of Titin evident in siblings was absent in slou45 mutants (Fig. 1E&F) and although some Titin positive puncta were present in the slou45 mutants, they showed little evidence of organisation.
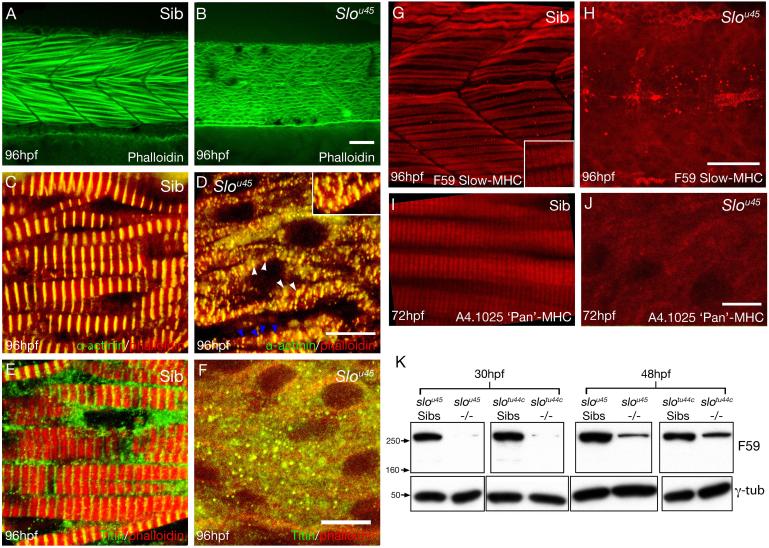
Figure 1. slou45 mutants have disorganised or missing expression of sarcomeric proteins.
Lateral views of muscle fibres in wild-type (A,C,E,G,I) and slou45 embryos (B,D,F,H,J) of ages shown bottom left and with reagents/antibodies shown bottom right.
A,B) f-Actin labelling with phalloidin showing a regular arrangement of fibrils in wild-type muscle fibres whereas the slou45 mutant fibrillar organisation is disrupted.
C,D) Immunohistochemistry for alpha-Actinin (green) marks the Z-disc, here combined with phalloidin (red). Z-discs are present in the slou45 mutant (white arrowheads, D) but disordered: the distance between Z-discs is irregular (blue arrowheads) and Z-discs from neighbouring fibrils are not in register with each other (inset, D). Z-discs are flanked by Actin filaments in both siblings and mutants.
E,F) Titin labelling (green, E,F) using an antibody that marks a region of the molecule around the Z-disc, counterstained with phalloidin (red).
G-J) Immunohistochemistry for MHC (slow muscle myosin, F59, red, G,H; pan-myosin A4.1025 red, I, J). In the slou45 muscle cells anti-MHC staining is severely reduced and lacks organisation.
K) Western blots using anti-MHC antibody F59 and γ-Tubulin (γ-Tub) as a loading control on lysates of slou45 and slotu44c mutant and sibling embryos at 30hpf and 48hpf stages. MHC protein levels are reduced in mutants of both alleles.
Scale bars: (A,B,G,H=20μm; C-F=10μm; I,J=8μm)
Anti-MHC immunoreactivity was considerably reduced in slou45 mutants. Unlike in siblings, the muscle of slou45 mutants lacked strong anti-MHC labelling with no signs of striations in register between neighbouring fibres (Fig. 1G-J). Consistent with its less severe movement phenotype, MHC immunohistochemistry in the tu44c allele revealed short lengths of striation (Fig. S2A&B).
Western blots showed that MHC was almost completely absent from lysates of slo mutants at 30hpf, when myofibrils are initially forming. By 48hpf, when the majority of muscle fibres contain mature fibrils, Myosin levels had increased slightly in slo mutants although not near to levels in sibling lysates (F59:Fig. 1K; A4.1025: data not shown). These results suggest either thick filaments are not formed because of a lack of Myosin or that Myosin is degraded because thick filaments do not form correctly.
slo myofibrils are deficient in thick filaments
To examine more closely the defects in the contractile apparatus of slou45 muscle, we used transmission electron microscopy (TEM) to visualise the sarcomeric machinery at 24hpf (Fig. 2A-D), when movement is just beginning, and 48hpf (Fig. 2E-H), when coordinated movement is present.
Figure 2. Sarcomeres in slou45 mutant myofibrils lack thick filaments.
A-H) Electron micrographs of sarcomeres from wild-type (A,C,E,G) and slou45 mutant (B,D,F,H) myofibrils at the ages shown bottom left.
A,B) Caudal myotomes of 24hpf embryos. White arrowheads (A) show immature Z-discs with poorly positioned thick and thin filaments. In slou45 mutants, bundles of thin filaments are present with putative early Z-discs (B, arrowheads). Very rarely, structures with the approximate correct dimensions of thick filaments were present (box, B).
C,D) Rostral myotomes of 24hpf embryos. The wild-type has recognisable sarcomeres with almost straight Z-discs (white arrowhead, C) and more fully formed sarcoplasmic reticulum with triads (black arrowhead, C). In the slou45 mutant, I-Z-I brushes are formed (Z-discs (white arrowheads, D) flanked on either side by thin filaments), and a few putative thick filaments are evident (box, D).
E-H) 48hpf wild-type muscle fibres are packed with mature myofibrils (E&G), possessing mature triads at each Z-disc (Black arrowhead, G). Mutant muscle fibres contain no thick filaments but numerous I-Z-I brushes are evident (F,H). The Z-lines are surrounded on either side by an electron-dense region (Bracket, H; F), beyond this region the thin filaments have light striations on them. Triads are present although less common (black arrowhead, H).
I) A schematic summarising the principle differences between slou45 mutant and sibling sarcomere ultrastructure. At early stages of myofibrillogenesis (top) mutants have less numerous and malformed thick filaments (red) and I-Z-I brushes (green and blue) are present but less aligned than Siblings. By 48hpf, wild-type sarcomeres are fully formed whereas in mutants only mis-aligned and mis-spaced I-Z-I brushes with striated thin filaments are present (blue). T-tubules and sarcoplasmic reticulum are shown in yellow and purple.
Scale bars: (A-D,G,H=500nm; E,F=2μm; H inset=100nm)
In less mature caudal myotomes of wild-type 24hpf embryos, myofibrils were rare and those present were very immature. Z-discs were small in diameter and poorly aligned, thick and thin filaments were present but were few in number and were not yet organised into A and I bands (Fig. 2A). Structures resembling myofibrils were essentially absent in slou45 mutants (Fig. 2B). Where a presumptive nascent sarcomere could be found, it comprised small aggregations of thin filaments sometimes associated with densities (putative Z-discs).
In more mature rostral myotomes of 24hpf wild-types, myofibres often contained one or two almost complete myofibrils; such structures were absent in slou45 fibres (Fig. 2C&D). Although more fully constructed than their caudal counterparts, these structures were still immature in wild-type embryos. Z-discs were not straight and thick filaments were sporadic and not fully integrated. In slou45 mutants, the rostral myotomes contained some thin filament/Z-disc aggregates with Z-discs usually forming isolated ‘I-Z-I brushes’. Occasionally a few spindly thick filament-like structures were present, appearing to lace together adjacent I-Z-I brushes.
By 48hpf, wild-type myofibres contained multiple highly organised myofibrils with Z-discs in register whereas slou45 myofibres contain only disorganised I-Z-I brushes (Fig. 2E&F). These were often found in groups with the Z-discs only partially in register with their neighbours and the distance between consecutive I-Z-I brushes was also highly variable. Thick filaments were completely absent from slou45 myofibrils. Either side of the Z-disc was an electron-dense area 100-200nm wide within which the bundle of I-band thin filaments retained the same height as the Z-disc. Beyond this region, the bundles of thin filaments were slightly striated and narrower than the diameter of the Z-disc (Fig. 2 G,H). The periodicity of the striations was approximately 30nm suggesting they could be troponin complexes that would normally interact with myosin heads (Chun and Falkenthal, 1988).
The ultrastructural appearance of slotu44c muscle was broadly similar to slou45 mutants with I-Z-I brushes lacking thick filaments. However, the I-Z-I brushes were more regularly spaced and aligned with short disorganised fibrous aggregations of electron dense material between I-Z-I brushes (Fig. S2). It is probable that these aggregates correspond to the striations seen with MHC immunohistochemistry.
The heart of slou45 mutants functions normally and, unsurprisingly, the ultrastructure of the heart muscle was indistinguishable between slou45 mutants and siblings (Fig. S3).
These ultrastructural observations (summarised in Fig. 2I) reveal that the cause of the slo phenotype is a failure to assemble and integrate thick filaments into the nascent sarcomeres of myofibrils. Actin (thin) filaments and Z-lines (containing Actinin) are present, as is the excitation-coupling apparatus of T-tubules and sarcoplasmic reticulum. However, I-Z-I brushes fail to align and lack regular spacing. The lack of aligned thick filaments underlies the birefringence deficiency and lack of organisation in the Myosin immunohistochemistry. Therefore the Slo protein is not required to form a Z-disc and assemble Actin fibres, Actinin and probably other sarcomeric proteins, but is required for assembly and integration of thick filaments
slo mutations are in the hsp90a gene
Genetic mapping using SSLP markers placed the slo locus on LG20 (Fig. S4). Bespoke SNP markers for genes in the region showed tight linkage to hsp90a which is immediately adjacent to hsp90a2. Sequencing of these genes from the three mutant slo alleles revealed mutations in hsp90a (Fig. 3A and S4). A Guanine to Adenine point mutation is present in exon 3 in slou45 mutants causing a glycine to aspartic acid change in residue 94. In exon 9 of slotu44c and exon 10 of slotm201, there are point mutations that respectively change tyrosine 561 and trytophan 599 to stop codons. No mutations were found in hsp90a2 coding sequence in any slo allele.
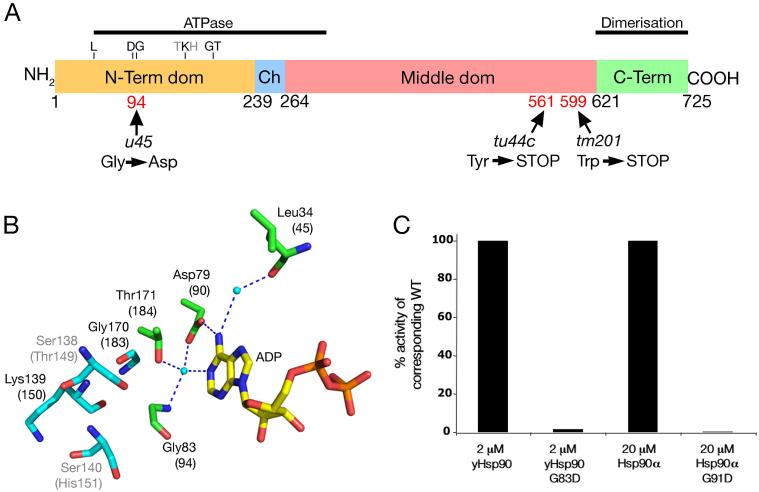
Figure 3. The ATPase activity of Hsp90 is abrogated by the slou45 mutation.
A) Hsp90a domain structure (amino acid numbers of domain boundaries are shown below; known functions of regions indicated by black bars). The positions and consequences of mutations in three slo alleles: u45, tu44c and tm201 are shown below. The equivalent residue locations of yeast Hsp90 represented in B are shown above.
B) Pymol diagram showing critical binding interactions of ADP with yeast Hsp90. Dotted blue lines are hydrogen bonds and amino acid residues (corresponding residue numbers in zebrafish in parentheses - also indicated in A) involved are in green, water molecules are cyan coloured balls and residues packed against the cα-atom of Gly83 are shown in cyan. Illustrated residues are conserved between yeast and zebrafish except Ser138/Thr149 and Ser140/His151 (illustrated in grey), which are conservative substitutions unlikely to cause major structural changes. Binding of ATP/ADP to the Hsp90 N-terminal domain involves highly conserved interactions including the carboxylate side-chain of Asp79 and main-chain carboxyl of Leu34, via a tightly bound water molecule, to the exocyclic N6 of adenine. The same Asp79 also interacts via another tightly bound water to the N1 imino nitrogen of the adenine. This same water is bound by an interaction with the side-chain hydroxyl of Thr171 and main-chain amide of Gly83. An aspartic acid residue substitution at Gly83 (mimicking the u45 mutation) would lead to steric clashes likely to disrupt critical hydrogen bonding interactions with ADP/ATP.
C) ATPase activity of yeast Hsp90, human Hsp90α and their u45-mimick mutants. ATPase activity of the yeast G83D and human G91D mutants is negligible relative to corresponding wild-type.
The nonsense slotu44c and slotm201 mutations would result in truncated molecules missing the C-terminus domain which is important for both homo- and hetero-dimerisation (Ali et al., 2006); such truncations would have severe functional consequences for the molecule.
The molecular consequence of the slou45 missense mutation was initially less obvious but bioinformatic analysis and further experiments demonstrated it abrogates the ATPase function of the Hsp90a protein. The N-terminal region of Hsp90 harbouring the mutation is required for ATP hydrolysis (Panaretou et al., 1998) and a ClustalW alignment of available Hsp90a sequences demonstrated the glycine mutated in slou45 to be universally conserved (Fig. S5). To predict the functional consequences of the Gly to Asp change, we assessed the probable consequence of this change on the protein structure of yeast Hsp90 for which the crystallographic structure has previously been determined ((Ali et al., 2006);Fig. 3B). The C-α atom of Gly83 sits in a confined space that is 3.4 Å from the hydroxyl of Ser138, 3.8 Å from the main-chain carbonyl of Lys139, 3.9 Å from the main-chain carbonyl of Gly170, and 4.4 and 4.8 Å from the main-chain amides of Gly170 and Lys139, respectively (Fig. 3B). The large side-chain of an aspartic acid residue would not be easily accommodated and would disrupt the local folding of the protein. As the glycine is involved in critical interactions with bound ATP/ADP, it is highly likely that the side-chain change would severely affect the ability of mutant Hsp90 to bind nucleotide.
To test directly whether a Gly to Asp mutation at this site affects ATPase function we recapitulated the mutation in yeast and human forms and tested their ATPase and ATP-binding properties in vitro. The mutations completely abrogated the ability of the molecules to hydrolyse ATP (Fig. 3C). Binding between the ATP analogue AMPPNP and mutant yeast and human Hsp90 was also negligible (Fig. S6). Functional ATPase activity is essential for chaperoning by Hsp90 (Panaretou et al., 1998; Pearl and Prodromou, 2006). The slou45 mutation is therefore catastrophic for the function of zebrafish Hsp90a as a chaperone.
The slo phenotype is rescued by exogenous Hsp90a
To confirm that the mutations identified above were indeed the cause of the muscle defects in slo mutants, we attempted to rescue the phenotype. Several BACs that covered the genomic region (Fig. S4; also Table S2 for clone names) were injected into batches of embryos from crosses of slou45 or slotu44c heterozygotes. At 3dpf, injected embryos showed normal movement, partial movement or paralysis (Video 2; Table 1). Examination of muscles from partially moving embryos with polarised light revealed a mosaic pattern of birefringence in the myotomes (Fig S4) indicating some cells, most likely those containing mosaically inherited BAC DNA, had functional myofibrillar assemblies. Although hsp90a and hsp90a2 are separated by only 2Kb we found 2 BACs (8 & 9) with ends in this 2Kb region facing in opposite directions, thus separating the two genes. Injection of BAC 9 (hsp90a only) produced mosaically rescued embryos, as expected. Surprisingly, BAC 8 (hsp90a2 only) also rescued some embryos (Table 1), albeit less efficiently, suggesting that exogenous Hsp90a2 can partially compensate for loss of Hsp90a.
Table 1.
Exogenous hsp90 rescues the slo phenotype. Percentages of paralysed embryos and those showing normal and partial movement after injection with BACs, BACs with MOs and in non-manipulated populations
| slo | Injected embryos | Non-injected control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| construct | allele | normal | partial | paralysed | n | normal | partial | paralysed | n |
| BAC 2 | u45 | 73.3 | 14.4 | 12.3 | 146 | 76.2 | 0 | 23.8 | 126 |
| BAC 4 | u45 | 74.7 | 16.9 | 8.4 | 83 | 80.5 | 0 | 19.5 | 113 |
| BAC 5 | u45 | 77.2 | 21.5 | 1.3 | 79 | 77.2 | 0 | 22.8 | 127 |
| BAC 6 | u45 | 75 | 21.7 | 3.3 | 60 | 81.9 | 0 | 18.1 | 72 |
| BAC 8 (hsp90a2) | u45 | 83.1 | 9.6 | 7.2 | 83 | 73 | 0 | 27 | 74 |
| BAC 8 (hsp90a2) | tu44c | 75.7 | 6.5 | 17.8 | 185 | 78.3 | 0 | 21.7 | 230 |
| BAC 8 + hsp90a2MO | tu44c | 71.6 | 0 | 28.4 | 81 | 75 | 0 | 25 | 88 |
| BAC 9 (hsp90a) | u45 | 72.7 | 25.5 | 1.8 | 55 | 76.3 | 0 | 23.7 | 59 |
| BAC 9 (hsp90a) | tu44c | 79 | 11.2 | 9.8 | 143 | 77.6 | 0.6 | 21.8 | 174 |
| BAC 9 + hsp90aMO | tu44c | 0 | 2.9 | 97.1 | 35 | - | - | - | - |
A caveat of these rescue experiments is that BACs contain more than one gene and so one cannot be certain that the rescue is due to the activity of a single gene. To address this issue, we co-injected Mos against hsp90a or hsp90a2 along with the corresponding BAC. In both cases, the ability of the BACs to rescue movement of slou45 embryos was blocked, confirming it is the activity of the Hsp90 proteins that mediates rescue. Additionally, hsp90a Mo produces immotile embryos lacking birefringence (Table1); conversely, neither of two hsp90a2 Mos disrupted muscle function (data not shown and see Etard et al (2007)).
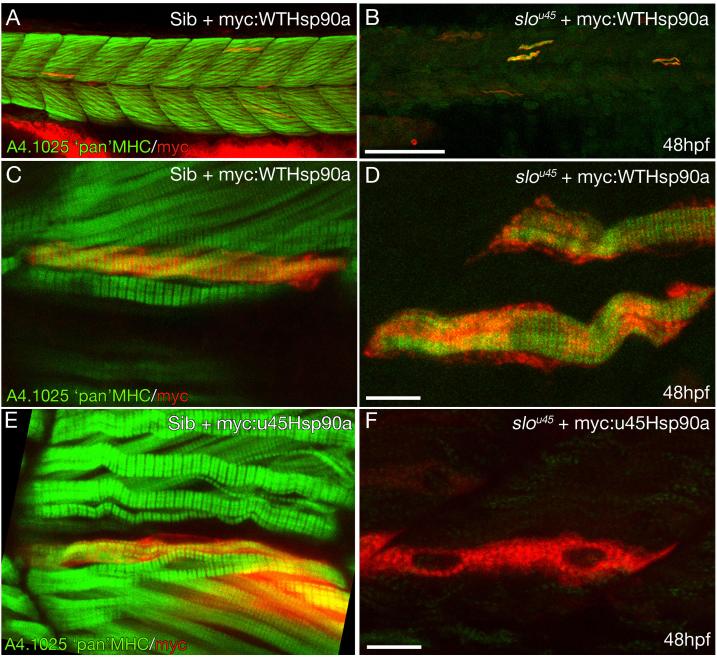
The slou45 allele is a missense mutation and, despite our study of the yeast and human mutant proteins, we could not be certain first that it is an amorphic (or weakly antimorphic) mutation in the zebrafish and secondly that the mutation is responsible for the lack of thick filaments. Therefore, to determine if the mutant protein has any residual activity in myofibrillogenesis, we assayed if exogenous mutant protein could rescue movement and thick filament generation. Constructs encoding N-terminal myc-tagged wild-type (myc:WThsp90a) and u45 mutant (myc:u45Hsp90a) Hsp90a were injected into slou45 mutants. Only the myc:WThsp90a construct rescued movement in slou45 mutants. Mosaically distributed myc:WTHsp90a expressing muscle cells in slou45 mutants possessed apparently normal levels of organised Myosin and resembled wild-type muscle fibres transplanted into slou45 embryos (Fig. 4A-D cf. Fig. S1K) indicating a rescue of thick filament formation. Muscle cells in slou45 mutants expressing the myc:u45Hsp90a protein had neither higher levels of Myosin immunoreactivity nor organisation in the Myosin present at low level (Fig. 4F). Sibling muscle fibres expressing myc:u45hsp90a retain the ability to express Myosin in orderly myofibrils suggesting that u45 mutant Hsp90a does not have detectable antimorphic activity (Fig. 4E).
Figure 4. Wildtype Hsp90a rescues thick filament formation in slou45 muscle cells whereas Hsp90au45 does not.
A-D) Muscle fibres from a slou45 mutant (B,D) and sibling (A,C) injected with DNA encoding myc:WTHsp90a double stained for myc (red) and MHC (MHC;green). MHC staining in siblings decorates the cells with organised bands (C) indicative of mature A-bands. In the slou45 mutant, striations in the MHC staining are only evident in myc+ cells (D).
E,F) Muscle fibres in sibling embryos injected with myc-tagged hsp90au45 DNA show organised MHC staining of myc+ cells (E). Myc+ cells in slou45 mutants injected with the same construct do not show organised myofibrils (F).
Scale bars: (A,B=100μm; C-F=10μm)
The slou45 missense allele gives a slightly stronger contractility phenotype and more severe disruption to myofibrillogenesis than the truncated slotu44c allele (Figs 1, 2 & S2) and we assayed if this might be due to enhanced activity of the hsp90a2 gene in the tu44c background. However, Mo-based abrogation of Hsp90a2 activity in slotu44c mutants failed to block muscle twitches. This suggests that the slotu44c mutant Hsp90a protein retains residual ability to mediate myofibrillogenesis. This is probably due to function of the intact ATPase region and since the dimerisation domain is missing in this allele, it suggests the Hsp90 monomer retains some activity. All together, these results confirm that hsp90a is the gene affected by the slo mutations and that Hsp90a has a unique and specific developmental role in the assembly and integration of thick filaments onto nascent sarcomeres of skeletal muscle.
hsp90a and genes encoding proteins that may interact with Hsp90a to mediate sarcomere assembly are up-regulated in slo mutants
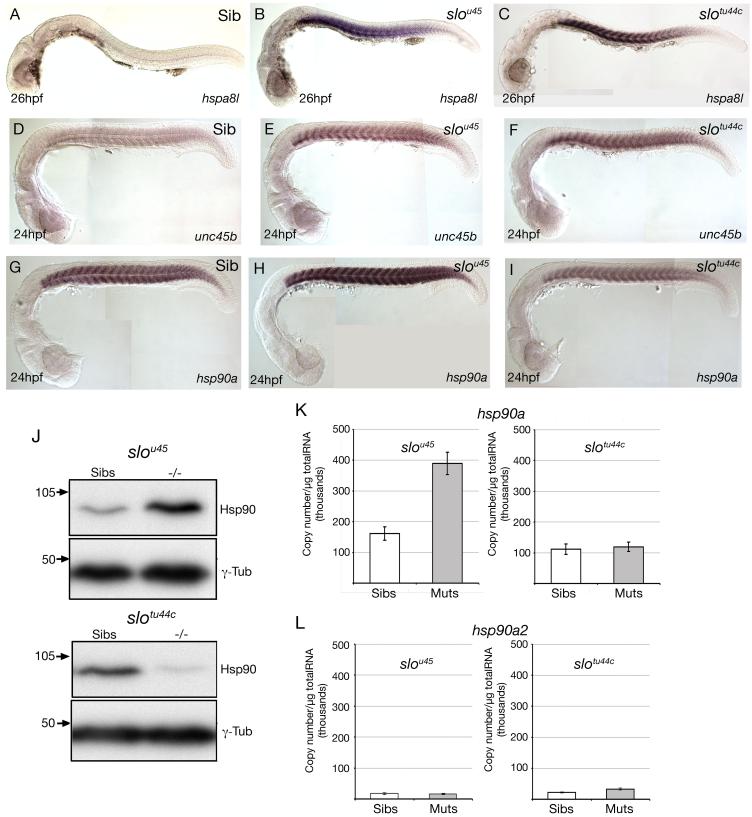
Using Affymetrix arrays, we compared mRNA expression profiles of wild-type and slou45 embryos (Tables S1&S2). Amongst the genes with lowered levels of expression were several encoding sarcomeric proteins including Myosin light chains, Titin and Troponins. Notably, several genes encoding heat-shock proteins and heat-shock protein interacting factors were upregulated. hspa8l and a similar gene homologous to human HS7C showed the largest slou45 versus wild-type differences with 150-160x upregulation in mutants. Both hsp90a and unc45b, a gene proposed to interact with Hsp90 during myofibrillogenesis (Barral et al., 2002; Etard et al., 2007) were 4x upregulated in mutants. For selected genes, in situ hybridisation (ISH) analysis corroborated the expression changes found using the microarray. hspa8l was massively upregulated in the myotomes of both slou45 and slotu44c mutants (Fig. 5A-C) while both unc45b and hsp90a showed more modest increases in ISH signal in slou45 mutants (Fig. 5D-I). In surprising contrast to the upregulation of hsp90a in slou45 embryos and hsp90a morphants (Fig. 5M), hsp90a transcript levels were reduced in slotu44c mutants (for which no microarray analysis was carried out) despite upregulation of both unc45b and hspa8l in the morphant and both mutant alleles (Fig. 5C,F,I, S7). Complimenting the changes at the mRNA level, western blot analysis revealed that Hsp90 protein levels are increased in slou45 mutants and decreased in slotu44c mutants when compared to siblings.
Figure 5. Abrogation of hsp90a but not hsp90a2 function leads to upregulation of genes encoding proteins likely to be involved in sarcomere assembly.
A-I) Lateral views at the stage shown bottom left showing expression of genes indicated bottom right in wild-type and mutant embryos (indicated top right). Note that although hspa8l and unc45b are upregulated in the slotu44c allele, hsp90a itself is not.
J) Western blots using lysates of slou45 and slotu44c mutant and sibling embryos. More Hsp90 protein was present in slou45 mutant lysates then the corresponding siblings. In contrast, slotu44c mutant lysates contained less Hsp90 protein than siblings.
K,L) Quantitative PCR (qPCR) analyses of levels of hsp90a and hsp90a2 expression in wild-type, slou45 and slotu44c mutants. Absolute expression levels of hsp90a (normalised to μg of total RNA) were significantly increased (K; p=0.002, n=5) in slou45 mutants compared to siblings; levels of expression of hsp90a in slotu44c mutants were unchanged compared to siblings (K; p=0.75, n=3); and, levels of expression of hsp90a2 were not significantly different between slou45 mutants and siblings nor slotu44c mutants and siblings (L; p=0.813 n=5; p=0.074 n=3 respectively)
In summation, these results show that when Hsp90a function is compromised as in slou45 mutants, levels of transcription of genes encoding certain sarcomeric proteins are reduced; whereas expression levels of genes that encode several chaperone and co-chaperone proteins (hspa8l; hsp90a; unc45b and others) are upregulated (with the allele specific exception of hsp90a in slotu44c mutants).
Hsp90a2 has no role in myofibrillogenesis
Our data have revealed a requirement for hsp90a in myofibrillogenesis but not for its neighbouring hsp90a2 gene. The reasons why hsp90a2 appears to have little or no role in muscle were not clear and so we designed experiments to assess expression of hsp90a2 and further explore its function (or lack of function). ISH analysis suggests that, like hsp90a, hsp90a2 is expressed in muscle tissue but expression levels are low (data not shown and (Etard et al., 2007)). However, as expression was low and to strengthen this data, quantitative RT-PCR (qPCR) was performed to examine the differences in levels of mRNA for hsp90a and hsp90a2 in wild-type and slo embryos (u45 and tu44c alleles).
hsp90a expression levels were significantly increased in slou45 mutants compared to siblings (Fig. 5K), whereas expression levels of hsp90a2 were unaffected by either mutation (Fig. 5L). This indicates that although both genes are adjacent to each other, they are subject to differential transcriptional regulation. This conclusion is supported by analysis of the absolute levels of transcription of the two genes in wild-type embryos with hsp90a expression nearly 8 times greater than hsp90a2 (Fig. 5K&L; Fig. S7). This offers a likely explanation for why endogenous hsp90a2 does not compensate for mutations in hsp90a. Presumably, the low expression level of hsp90a2 that fails to be upregulated as a consequence of mutation in hsp90a means that there is very little Hsp90a2 activity in developing muscle of either wild-type or slo embryos. In contrast, we assume that the expression level of hsp90a2 following BAC injection is sufficiently high to mediate partial rescue. Supporting the conclusion that hsp90a2 has no role in muscle development, hsp90a2 morphants show normal birefringence of muscle fibres and fail to upregulate hsp90a, unc45b or hspa8l and hsp90a/hsp90a2, double morphants exhibit no more severe phenotype than the hsp90a and hsp90a2 single mutants/morphants (Fig. S8 and (Etard et al., 2007)).
Discussion
slo mutants exhibit a developmental phenotype specifically affecting skeletal muscle myofibrillogenesis. Within slo mutant myofibres, the initial steps in construction of sarcomeres are relatively normal, leading to formation of Actin filament decorated Z-bands. However, these I-Z-I brushes fail to align or become properly spaced and nascent sarcomeres lack properly formed thick filaments. The level of MHC protein is reduced in mutants suggesting Myosin protein is degraded. Four main lines of evidence indicate that this phenotype is due to mutations in hsp90a: mutations are found in three slo alleles; the phenotype is rescued by exogenous wild-type Hsp90 and phenocopied by hsp90a Mo; and the transcriptional upregulation of hsp90a and hspa8l (a hsp70-related gene) is a conserved response to the loss of Hsp90a activity (Blagg and Kerr, 2006). Moreover, structural and functional analysis of the U45 mutation indicate that it is the ATPase-dependent chaperoning activity of Hsp90 that is important for thick filament formation and integration.
The developmental requirement for hsp90a is restricted to skeletal muscle cells
Given the vast literature on roles for Hsp90 in many cellular events, we were initially surprised to find such a specific role for hsp90a during embryonic development. Little has been published on the requirements for Hsps during normal developmental processes in vertebrates, so the issue of tissue and cell-type specificity of function has not previously been addressed.
Two possible explanations for the specificity of the hsp90a mutant phenotype are either that skeletal muscle is the only embryonic cell type to require Hsp90 activity or that other hsp90 genes function in other developing cells. In favour of a specific requirement in skeletal muscle cells, construction of the myofibrillar apparatus is an enormous challenge to the machinery of the cell, and muscle cells probably have the greatest load of any developing cell type in terms of protein folding and construction of multimeric protein complexes. To build a sarcomere requires balancing of the transcription, translation and folding of proteins and their incorporation into reiterative sarcomeric units. In this context, the demands upon protein chaperones will be high so Hsp90a may have been recruited for this specific developmental role. Additionally, given the large numbers of hsp genes, it is almost certain that others will have developmental roles. With respect to hsp90a genes, in zebrafish there are at least three, hsp90a, hsp90a2 and hsp90ab1 (formerly hsp90b: (Krone et al., 1997)) raising the possibility that different paralogues function in different developmental events.
Hsp90a may co-operate with Unc45 in the chaperoning of Myosin during myofibril assembly
Hsp90s are well-characterised molecular chaperones with diverse and wide-ranging roles in cellular physiology (Pearl and Prodromou, 2006). The current hypothesis for the chaperoning role of the Hsp90 homodimer is that the N-terminus ATPase domain flexes upon binding of ATP and this facilitates the maturation of client proteins (Ali et al., 2006; Pearl and Prodromou, 2006).
Although this hypothesis suggests how Hsp90 chaperones, it does not explain how client specificity is managed (Pearl and Prodromou, 2006). Unlike many chaperones, Hsp90 shows much specificity for the proteins it binds although these client proteins are very diverse and may number over 400 (Zhao et al., 2005). With this specific, yet potentially wide-ranging network of interactions, it is perhaps all the more surprising that slo mutants only show defective skeletal thick filament formation. By what mechanisms might Hsp90a specifically act to chaperone Myosin during thick filament formation?
Substrate-specific binding sites in the middle domain of the Hsp90 molecule, in addition to recruitment of various co-chaperone molecules, may contribute to the client specificity of the chaperoning activity (Pearl and Prodromou, 2006). Included among co-chaperones is the UCS-factor Unc45 identified through a C. elegans screen as being required for muscle development (Barral et al., 1998; Venolia et al., 1999). These studies led to the proposition that Unc45 acts in concert with Hsp90 to chaperone Myosin. In a recent study Etard et al (2007) demonstrated that the steif zebrafish motility mutant is due to mutations in the unc-45b gene and that Steif/Unc-45b binds Hsp90. These results point to a highly conserved mechanism whereby Unc-45 provides specificity with Hsp90 for chaperoning Myosin during myofibrillogenesis.
The regulation of hsp90a during skeletal muscle development is likely to be independent of the normal heat-shock response pathway
During the course of our studies, we made several observations regarding the regulation of mRNAs and proteins in wild-type and slo muscles: we find hsp90a is strongly expressed in wild-type skeletal muscle; both hsp90a mRNA and Hsp90 protein and hspa8l and unc45 mRNA levels are increased in slou45 mutants whereas hsp90a2 is not; hsp90a mRNA is not upregulated and Hsp90 protein levels are lower in slotu44c mutants; Myosin protein levels are severely reduced in slou45 mutants; and genes encoding several sarcomeric proteins are down-regulated in slou45 mutants. Do these observations make sense and shed any light on the transcriptional and post-translational mechanisms that work during sarcomere assembly?
There are two obvious possibilities to explain the strong, tissue specific expression of hsp90a during normal skeletal muscle development. The first is that the stress levels that occur in developing myofibres induce the “heat-shock response”, a well-established trigger for upregulation of hsp90 transcription. The second is that the high level expression of hsp90a in skeletal muscle cells is developmentally regulated, independent of the heat-shock regulation of hsp90a transcription. We favour this second possibility as our data suggest that the heat-shock response is not triggered during normal muscle development whereas it is triggered in slo mutant muscle cells.
A well-established signature for reduced Hsp90 function is the upregulation of hsp90 and hsp70 related genes and the ubiquitination and proteosome-mediated destruction of Hsp90 client proteins (Proisy et al., 2006). In such situations, it is thought that the depletion of Hsp90 (through cellular stress induced client binding, mutations or other reasons) frees the transcription factor Hsf which triggers the heat shock response through up-regulation of various Hsp genes. Thus the upregulation of chaperone genes and depletion of client proteins (Myosin) in slo mutants is entirely consistent with the muscle cells mounting a stress response due to loss of Hsp90a function.
In contrast to the situation in slo mutants, there is no indication that the expression of hsp90a during normal development is a direct consequence of the cell mounting a stress response. Perhaps most notably, hspa8l expression is virtually undetectable inwild-type developing muscle in contrast to slo mutant muscle. Thus it seems much more likely that an alternative, developmentally regulated, transcriptional mechanism leads to upregulation of hsp90a during normal myogenesis. This would, in principle, not be difficult to achieve as various muscle-specific transcription factors are active during the period of myofibrillogenesis (Hinits and Hughes, 2007).
The reduction in Myosin protein levels in slou45 and slotu44c mutants could be explained by the well-established proteosome mediated degradation of Hsp90 client proteins in the absence of Hsp90 function (Blagg and Kerr, 2006). We have no explanation for the reduced transcription of other genes involved in myofibrillogenesis although it suggests a feedback mechanism balancing transcription levels with the translation and further processing of sarcomeric proteins.
It is curious that slotu44c mutants show upregulation of unc45 and hspa8l but not hsp90a itself. The fact that tu44c is a nonsense mutation may lead to nonsense-mediated mRNA decay and, if this happens, it might lead to lowering of transcript and protein levels. However, it does not seem likely that mRNA decay would completely mask the strong constitutive expression that should be induced by the stress response. The milder phenotype of slotu44c and slou45/tu44c mutants suggests that some translation does occur and that the Slotu44c Hsp90a protein retains some function, presumably in its N-terminal portion. It may still retain the ability to sequester HSF and hence a milder stress response may be mounted in the mutant muscle cells. However this does not really explain why unc45 and hspa8l respond similarly in both mutant alleles. Finally, one should also consider that the truncated Hsp90tu44c protein may in some unknown way suppress the hsp90a transcriptional upregulation response.
It is also curious that despite being located adjacent to hsp90a, hsp90a2 is subject to different transcriptional regulation and has no obvious role in muscle formation. Given their proximity and similarity of sequence, it seems likely that the two genes arose through a tandem duplication event. Hsp90a2 still encodes a functional protein and so it must retain a function in zebrafish, though this may not be evident during embryogenesis.
Implications for myofibrillogenesis models
How do our data impact upon the three current models of myofibrillogenesis outlined in the introduction? The core of all three models is a sequential deposition of the myofibrillar components from Z-line to M-line, with I-Z-I brushes forming first, followed by the integration of thick filaments to complete sarcomeric assembly. Our data support the notion that I-Z-I brushes form first but these do not align or become properly spaced in slo mutants leaving no well-structured scaffold onto which thick filaments can integrate. Thus, our data suggest a more active role for the assembly and integration of thick filaments in the linking together, alignment and spacing of Z-discs during sarcomere maturation.
Both Sanger’s model (2002) and the “Titin” model (Trinick and Tskhovrebova, 1999) of myofibrillogenesis suggest that a well-formed scaffold of I-Z-I brushes is established prior to thick filament integration, with either non-muscle Myosin or Titin fulfilling the role of linking the I-Z-I brushes. In both cases, the lack of assembly and integration of thick filaments should not significantly impact upon the construction of the sarcomeric scaffold. In slou45 mutants, scaffold formation is compromised as I-Z-I brushes do not properly align, are not correctly spaced and probably do not correctly lace up with adjacent I-Z-I brushes. This suggests either that Hsp90a has unsuspected roles in the assembly of non-muscle Myosin or Titin or that the models may need some revision. Indeed, Titin immunohistochemistry in slou45 mutants suggests that the molecule is not even correctly integrated into the I-Z-I brushes that are present, raising the possibility that sarcomeric Titin integration might lie parallel to, or even downstream of, thick filament assembly and integration. The third model of myofibrillogenesis (Ehler et al., 1999) holds that I-Z-I brushes are loaded with thick filaments at specific cellular locations and subsequently assembled into full sarcomeres. The slou45 phenotype is not inconsistent with this idea but neither does it provide strong support.
The mutants described in this and related papers enhance our understanding of myofibrillogenesis through study of the process in vivo. We are hopeful that further genetic studies of sarcomere formation will refine existing models and help build a more complete picture of myofibrillogenesis.
Supplementary Material
Acknowledgements
The study was supported by the BBSRC, Wellcome Trust and EC ZF-models integrated project (zf-models.org). Hsp90 work at ICR is supported by the Wellcome Trust (LHP). A-PH was supported by an EC Marie-Curie Fellowship and RA by a BBSRC David Phillips Fellowship. TH received a UCL Bogue Fellowship to visit Stanford University. We thank Mike Hubank and Nipurna Jina (ICH Gene Microarray Centre) for microarray processing, Phil Ingham, Sarah Baxendale, Elizabeth Busch and Derek Stemple for sharing data and information prior to publication; Rodrigo Young and Mark Turmaine for practical assistance; Hans-Georg Frohnhöfer and Carole Wilson and her team for fish care.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–7. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Bauer CC, Ortiz I, Epstein HF. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol. 1998;143:1215–25. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45as a molecular chaperone for myosin. Science. 2002;295:669–71. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- Blagg BS, Kerr TD. Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med Res Rev. 2006;26:310–38. doi: 10.1002/med.20052. [DOI] [PubMed] [Google Scholar]
- Chun M, Falkenthal S. Ifm(2)2 is a myosin heavy chain allele that disrupts myofibrillar assembly only in the indirect flight muscle of Drosophila melanogaster. J Cell Biol. 1988;107:2613–21. doi: 10.1083/jcb.107.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. STRIATED MUSCLE CYTOARCHITECTURE: An Intricate Web of Form and Function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Cooke R. The sliding filament model: 1972-2004. J Gen Physiol. 2004;123:643–56. doi: 10.1085/jgp.200409089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan-Goor M, Silberstein L, Kessel M, Muhlrad A. Localization of epitopes and functional effects of two novel monoclonal antibodies against skeletal muscle myosin. J Muscle Res Cell Motil. 1990;11:216–26. doi: 10.1007/BF01843575. [DOI] [PubMed] [Google Scholar]
- Ehler E, Rothen BM, Hammerle SP, Komiyama M, Perriard JC. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J Cell Sci. 1999;112(Pt 10):1529–39. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- Etard C, Behra M, Fischer N, Hutcheson D, Geisler R, Strahle U. The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev Biol. 2007;308:133–43. doi: 10.1016/j.ydbio.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Fulton AB, Alftine C. Organization of protein and mRNA for titin and other myofibril components during myofibrillogenesis in cultured chicken skeletal muscle. Cell Struct Funct. 1997;22:51–8. doi: 10.1247/csf.22.51. [DOI] [PubMed] [Google Scholar]
- Fulton AB, L’Ecuyer T. Cotranslational assembly of some cytoskeletal proteins: implications and prospects. J Cell Sci. 1993;105(Pt 4):867–71. doi: 10.1242/jcs.105.4.867. [DOI] [PubMed] [Google Scholar]
- Fürst DO, Osborn M, Nave R, Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J Cell Biol. 1988;106:1563–72. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst DO, Osborn M, Weber K. Myogenesis in the mouse embryo: differential onset of expression of myogenic proteins and the involvement of titin in myofibril assembly. J Cell Biol. 1989;109:517–27. doi: 10.1083/jcb.109.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt M, Hammer RE, Hubner N, Monti J, Witt CC, McNabb M, Richardson JA, Granzier H, Labeit S, Herz J. Conditional expression of mutant M-line titins results in cardiomyopathy with altered sarcomere structure. J Biol Chem. 2003;278:6059–65. doi: 10.1074/jbc.M211723200. [DOI] [PubMed] [Google Scholar]
- Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Granzier H, Sorimachi H, Labeit S. Muscle assembly: a titanic achievement? Curr Opin Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134:2511–9. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzer H, Hijikata T, Lin ZX, Zhang ZQ, Holtzer S, Protasi F, Franzini-Armstrong C, Sweeney HL. Independent assembly of 1.6 microns long bipolar MHC filaments and I-Z-I bodies. Cell Struct Funct. 1997;22:83–93. doi: 10.1247/csf.22.83. [DOI] [PubMed] [Google Scholar]
- Krone PH, Lele Z, Sass JB. Heat shock genes and the heat shock response in zebrafish embryos. Biochem Cell Biol. 1997;75:487–97. [PubMed] [Google Scholar]
- LoRusso SM, Rhee D, Sanger JM, Sanger JW. Premyofibrils in spreading adult cardiomyocytes in tissue culture: evidence for reexpression of the embryonic program for myofibrillogenesis in adult cells. Cell Motil Cytoskeleton. 1997;37:183–98. doi: 10.1002/(SICI)1097-0169(1997)37:3<183::AID-CM1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Mishra M, D’Souza V,M, Chang KC, Huang Y, Balasubramanian MK. Hsp90 protein in fission yeast Swo1p and UCS protein Rng3p facilitate myosin II assembly and function. Eukaryot Cell. 2005;4:567–76. doi: 10.1128/EC.4.3.567-576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa H, Meek S, Gautel M, Peddie D, Smith AJ, Peckham M. Targeted homozygous deletion of M-band titin in cardiomyocytes prevents sarcomere formation. J Cell Sci. 2006;119:4322–31. doi: 10.1242/jcs.03198. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. Embo J. 1998;17:4829–36. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–94. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Proisy N, Sharp SY, Boxall K, Connelly S, Roe SM, Prodromou C, Slawin AM, Pearl LH, Workman P, Moody CJ. Inhibition of hsp90 with synthetic macrolactones: synthesis and structural and biological evaluation of ring and conformational analogs of radicicol. Chem Biol. 2006;13:1203–15. doi: 10.1016/j.chembiol.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Rhee D, Sanger JM, Sanger JW. The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil Cytoskeleton. 1994;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- Rossi AE, Dirksen RT. Sarcoplasmic reticulum: the dynamic calcium governor of muscle. Muscle Nerve. 2006;33:715–31. doi: 10.1002/mus.20512. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Chowrashi P, Shaner NC, Spalthoff S, Wang J, Freeman NL, Sanger JM. Myofibrillogenesis in skeletal muscle cells. Clin Orthop. 2002:S153–62. doi: 10.1097/00003086-200210001-00018. [DOI] [PubMed] [Google Scholar]
- Schultheiss T, Lin ZX, Lu MH, Murray J, Fischman DA, Weber K, Masaki T, Imamura M, Holtzer H. Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J Cell Biol. 1990;110:1159–72. doi: 10.1083/jcb.110.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. Fifty years of contractility research post sliding filament hypothesis. J Muscle Res Cell Motil. 2004;25:475–82. doi: 10.1007/s10974-004-4239-6. [DOI] [PubMed] [Google Scholar]
- Trinick J, Tskhovrebova L. Titin: a molecular control freak. Trends Cell Biol. 1999;9:377–80. doi: 10.1016/s0962-8924(99)01641-4. [DOI] [PubMed] [Google Scholar]
- Van der Ven PF, Ehler E, Perriard JC, Furst DO. Thick filament assembly occurs after the formation of a cytoskeletal scaffold. J Muscle Res Cell Motil. 1999;20:569–79. doi: 10.1023/a:1005569225773. [DOI] [PubMed] [Google Scholar]
- Venolia L, Ao W, Kim S, Kim C, Pilgrim D. unc-45 gene of Caenorhabditis elegans encodes a muscle-specific tetratricopeptide repeat-containing protein. Cell Motil Cytoskeleton. 1999;42:163–77. doi: 10.1002/(SICI)1097-0169(1999)42:3<163::AID-CM1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–27. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.