Abstract
Heparanase is an endoglycosidase which cleaves heparan sulfate and hence participates in degradation and remodeling of the extracellular matrix. Importantly, heparanase activity correlated with the metastatic potential of tumor-derived cells, attributed to enhanced cell dissemination as a consequence of heparan sulfate cleavage and remodeling of the extracellular matrix barrier. Heparanase has been characterized as a glycoprotein, yet glycan biochemical analysis was not performed to date. Here, we applied the Qproteome™ GlycoArray kit to perform glycan analysis of heparanase, and compared the kit results with the more commonly used biochemical analyses. We employed fibroblasts isolated from patients with I-cell disease (mucolipidosis II), fibroblasts deficient of low density lipoprotein receptor-related protein and fibroblasts lacking mannose 6-phosphate receptor, to explore the role of mannose 6-phosphate in heparanase uptake. Iodinated heparanase has been utilized to calculate binding affinity. We provide evidence for hierarchy of binding to cellular receptors as a function of heparanase concentration. We report the existence of a high affinity, low abundant (i.e., low density lipoprotein receptor-related protein, mannose 6-phosphate receptor), as well as a low affinity, high abundant (i.e., heparan sulfate proteoglycan) receptors that mediate heparanase binding, and suggest that these receptors cooperate to establish high affinity binding sites for heparanase, thus maintaining extracellular retention of the enzyme tightly regulated.
Keywords: heparanase, glycosylation, lectin array, mannose 6-phosphate, binding, receptor
Introduction
Heparanase is an endo-β-D-glucuronidase capable of cleaving heparan sulfate (HS) side chains at a limited number of sites, yielding HS fragments of still appreciable size (~4–7 kDa). Heparanase activity has long been correlated with the metastatic potential of tumor-derived cells, attributed to enhanced cell dissemination as a consequence of HS cleavage and remodeling of the extracellular matrix (ECM) barrier (Parish et al., 2001; Vlodavsky and Friedmann, 2001). Similarly, heparanase activity was implicated in cell dissemination associated with inflammation, autoimmunity and angiogenesis (Dempsey et al., 2000; Parish et al., 2001; Vlodavsky and Friedmann, 2001). This notion gained further support by employing siRNA and ribozyme methodologies, clearly depicting heparanase-mediated HS cleavage and ECM remodeling as critical requisites for inflammation, angiogenesis, and metastatic spread (Edovitsky et al., 2004; Edovitsky et al., 2005). More recently, heparanase up-regulation was documented in an increasing number of human carcinomas and hematological malignancies (Ilan et al., 2006; Vreys and David, 2007). In many cases, heparanase induction correlated with increased tumor metastasis, vascular density, and shorter post operative survival rate, thus providing a strong clinical support for the pro-metastatic and pro-angiogenic function of the enzyme and positioning heparanase as an attractive target for the development of anti-cancer drugs (Ferro et al., 2004; McKenzie, 2007; Miao et al., 2006; Sanderson et al., 2005; Vlodavsky et al., 2007). In addition, heparanase activity can liberate multitude HS-bound biological mediators such as growth factors, cytokines and chemokines, thus significantly affecting cell and tissue function. The multitude of polypeptides sequestered and regulated by HS and the ability of heparanase to convert these into bio-available molecules (Elkin et al., 2001; Folkman et al., 1988) require that these activities will be kept tightly regulated. We have shown previously that exogenously added heparanase rapidly interacts with primary human fibroblasts (Nadav et al., 2002) as well as with tumor derived cells (Gingis-Velitski et al., 2004a), followed by internalization and processing into a highly active enzyme (Zetser et al., 2004), collectively defined as heparanase uptake (Gingis-Velitski et al., 2004b). Applying HS-deficient cells, addition of heparin or xylosides, and deletion of HS-binding domains of heparanase, we have provided compelling evidence for the involvement of HS in heparanase uptake and processing (Gingis-Velitski et al., 2004b; Levy-Adam et al., 2005), maintaining extracellular retention of the enzyme tightly regulated. More recently, Verys et al have identified two additional cell surface receptors that mediate heparanase uptake, namely the low density lipoprotein receptor-related protein (LRP) and the mannose 6-phosphate receptor (MPR) as key elements in this process (Vreys et al., 2005), although a model that combine the three receptors has not been proposed. Heparanase has long been characterized as a glycoprotein and including Concanavalin A affinity chromatography in the purification scheme brought this feature into practice (Toyoshima and Nakajima, 1999; Vlodavsky et al., 1999; Zcharia et al., 2005). Six glycosylation sites were identified in the 50 kDa heparanase subunit (Hulett et al., 1999), and their role in protein secretion was established (Simizu et al., 2004), yet glycan biochemical analysis was not performed to date.
Here, we utilized a new lectin array method for studying glycan structure and composition, the Qproteome™ GlycoArray kit (Qiagen) (Rosenfeld et al., 2007), to study heparanase glycan composition and compared the lectin array results with the more common HPLC analysis. We further employed fibroblasts isolated from patients with I-cell disease (mucolipidosis II) and fibroblasts deficient of LRP and MPR, to explore the role of mannose 6-phosphate in heparanase uptake, and utilized iodinated heparanase to calculate binding affinities. We provide evidence for hierarchy of binding to cellular receptors as a function of heparanase concentration. We report the existence of high affinity, low abundant (i.e., MPR, LRP), as well as low affinity, high abundant (i.e., HSPG) receptors that mediate heparanase binding, and suggest that these receptors cooperate to establish high affinity binding sites for heparanase.
Materials and Methods
Glycan analysis by lectin array
Recombinant human heparanase was analyzed according to the manufacturer’s (Qiagene, GmbH, Germany) instructions. Briefly, Qiagene Lectin Array (Qproteome GlycoArray Kit) was incubated for 60 min with 1% BSA. Recombinant heparanase (13 μg/ml), purified from medium conditioned by heparanase-transfected CHO cells (Gingis-Velitski et al., 2004b; Zetser et al., 2003) was then applied to the array for 80 min, followed by incubation with rabbit anti heparanase 1453 antibody (Zetser et al., 2003) (60 min, diluted 1:1000) and goat anti rabbit-FITC conjugated antibody (Jackson Immunoresearch Laboratories, West Grove, PA) diluted 1:1000. Data analysis was performed using the automatic analysis software, as described (Rosenfeld et al., 2007).
Glycan preparation for HPLC analysis
Heparanase was subjected to peptide-N-gycosidase F (PNGase; 0.1U/μl) cleavage using denaturation protocol, according to the manufacturer (New-England Biolabs, Beverly, MA) instructions. Glycans were fluorescently labeled at their reducing end by 2- aminobenzamide (2AB, Merck), as described (Bigge et al., 1995).
Glycan separation by HPLC
2AB labeled glycans were separated by normal phase (NP) HPLC and weak anion exchange (WAX) HPLC columns, according to the manufacturer (GlycoSepC, Glyko-Prozyme) instructions. Briefly, glycans were separated on NP column using law salt solvent system: gradient of 35%–53% of 50 mM ammonium formate (solvent B) for 72 min, at a flow rate of 0.4 ml/min, and acetonitrile (solvent A). For WAX separation, the starting solution is 20% acetonitrile, 12 min gradient of 0–5% solvent B (20% acetonitrile, 50% 0.5 M amonium formate, pH 4.5, 30% RO water) followed by 5–21% gradient for 13 min and 21–80% gradient for the next 25 min, at a flow rate of 0.4 ml/min. Glycans were detected by fluorescence detector: excitation at λ330 nm and emmition at λ420 nm.
Enzymatic analysis of heparanase glycans
2AB labeled glycans were identified by serial enzymatic reactions using exo-glycosidases with different specificity. Sialydase (0.1 mU/μl; Roche, Mannheim, Germany), sialidase + β1,4 galactosidase (0.06 mU/μl; Calbiochem, San Diego, CA) and mannosidase (0.17 mU/μl; Calbiochem) were used in 50 mM Na2PO4, pH 6, at 37°C for 20 h. For assessment of phosphorylated glycan, alkaline phosphatase (1.3 U/ul; Calbiochem) was employed in 100 mM Tris buffer, pH 8.8, at 37°C for 20 h. Results are quantitative for peak areas above 5%.
Cells and cell culture
Chinese hamster ovary (CHO) K1 and human cervical adenocarcinoma HeLa cells were purchased from the American Type Culture Collection (ATCC). Mouse embryonic fibroblasts (MEF) and mouse fibroblasts deficient of cell surface mannose 6-phosphate receptor (MPR300−/−) were kindly provided by Dr. Kurt von Figura (University of Göttingen, D-37075 Göttingen, Germany) (12). Mouse fibroblasts deficient of low density lipoprotein receptor related protein (LRP) (PEA-13) were kindly provided by Dr. Joachim Herz (University of Texas Southwestern, Dallas, Texas 75390, USA) (Pohlmann et al., 1995b; Willnow and Herz, 1994). Human skin and I-cell fibroblasts were kindly provided by Dr. H. Mendel (Rambam Health Care Campus, Haifa, Israel). Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FCS and antibiotics. Mutant CHO cells (pgs A-745) deficient of xylosyltransferase and unable to initiate glycosaminoglycan synthesis, were kindly provided by Dr. J. Esko (University of California, San Diego) and grown in RPMI 1640 medium supplemented with 10% FCS and antibiotics.
Adenovirus-heparanase gene constructs and infection
The chicken heparanase cDNA, in pcDNA3 plasmid (Goldshmidt et al., 2001) was sub-cloned into the shuttle vector pAdTrack-CMV using the Kpn1/Xba1 restriction sites, replacing the E1 region of the virus. The resultant plasmid was linearized by digestion with Pme1, and was subsequently co-transformed into E.coli BJ5183 bacteria with the adenoviral backbone plasmid, pAdEasy-1, kindley provided by Dr. Moshe Y. Flugelmann (Carmel Medical Center, Haifa, Israel). Following selection with kanamycin, colonies were expanded and the recombinant plasmid was transfected into adenovirus packaging 293-T cells using lipofectamine, according to the manufacturer’s (Invitrogen, Carlsbad, CA) instructions. Viruses were purified by applying 293-T cell lysates onto a cesium chloride gradient and the virus titer was determined by plaque assay, yielding a titer of ~1 × 1010 pfu/mL. For infection, cells were grown to 70% confluence and were infected (multiplicity of infection = 100) with adeno-Hepa, or control empty viral construct. Infection efficiency of 70–90% was determined by co-transfection with adenovirus carrying GFP.
Heparanase uptake
Uptake experiments were carried out essentially as described (Gingis-Velitski et al., 2004b; Zetser et al., 2004). Briefly, cells were incubated with purified 65-kDa latent heparanase (1 μg/ml) under serum-free conditions. At the indicated time points, the medium was aspirated, cells were washed three times with ice-cold phosphate-buffered saline (PBS), and cell extracts were subjected to immunoblotting with the indicated antibody, as described (Gingis-Velitski et al., 2004b; Levy-Adam et al., 2005; Zetser et al., 2004).
Heparanase binding assay
Recombinant 65 kDa latent heparanase was iodinated with Na125I (GE Healthcare, Buckinghamshire, England) and Chloramin T (Sigma), as described (Levy-Adam et al., 2005). The heparanase specific activity was 1–2 × 105 cpm/ng. Cells were grown in 24-well Nunclon multidishes and incubated with 125I-heparanase in DMEM supplemented with 10 mM HEPES and 0.2% BSA for 2 h on ice. The incubation medium was removed and the cells were washed three times with ice-cold PBS, solubilized with 1 ml of 1 M NaOH and counted by in a γ-counter. For determination of Kd and Bmax, cells were grown in 24-well Nunclon multidishes and incubated with increasing concentrations of 125I-heparanase for 2 h on ice in the absence or presence of 750 nM unlabeled heparanase. The incubation medium was removed, cells were washed with ice-cold PBS, solubilized with 200 μl of 1 M NaOH, and counted in a γ-counter. Binding data were normalized for 1 × 106 cells. Binding parameters (Kd, Bmax) were obtained by the Prism 4 software (GraphPad Software, San Diego, CA).
Heparanase activity assay
Preparation of Na235SO4-labeled ECM-coated 35-mm dishes and determination of heparanase activity were performed essentially as described in detail elsewhere (Levy-Adam et al., 2003; Vlodavsky, 1999). Briefly, cells (1 × 106) were lysed by three cycles of freeze/thaw, and the resulting cell extracts were incubated (18 h, 37°C) with 35S-labeled ECM. The incubation medium (1 ml) containing sulfate-labeled degradation fragments was subjected to gel filtration on a Sepharose CL-6B column. Fractions (0.2 ml) were eluted with PBS and their radioactivity was counted in a β-scintillation counter. Degradation fragments of HS side chains produced by heparanase are eluted at 0.5 < Kav < 0.8 (peak II, fractions 15–30). Nearly intact HSPGs released from the ECM are eluted just after the V0 (Kav < 0.2, peak I, fractions 3–15) (Vlodavsky, 1999; Vlodavsky et al., 1999). These high molecular weight products are released by proteases that cleave the HSPG core protein.
Labeling heparanase with Cy-3
Heparanase was labeled with the fluorophore Cy-3 using the Cy™ 3 Mono-Reactive Dye Pack, according to the manufecurer’s (GE Healthcare) instructions, with minor modifications. In order to protect HS-binding sites, heparanase (1 mg) was first adsorbed to heparin-Sepharose beads (GE Healthcare) and labeling was carried out while bound to the beads. Beads were washed with ice-cold 0.1 M NaHCO3 and resuspended with 1 ml ice-cold 0.1 M NaHCO3, pH 8.2. Cy-3 succinimidyl ester was added (0.5 mg in 0.2 ml 0.1 M NaHCO3) and the mixture was rotated for 1 h at 37°C. The supernatant was drained and the beads washed with 4 ml TBS and 2 ml 10 mM Tris/0.3 M NaCl, pH 7.5. Labeled heparanase was eluted with 2 ml 10 mM Tris/2 M NaCl, pH 7. Uptake of the Cy-3-labeled heparanase was examined by confocal microscopy.
Results
Analysis of heparanase glycan composition
We utilized the Qproteome™ GlycoArray kits (Rosenfeld et al., 2007) to study heparanase glycosylation. This novel method is based on binding of a glycosylated protein to a micro array that carries 25 different lectins with overlapping specificities, yielding a unique profile for each glycoprotein applied (Fig. 1) and semi-quantitative glycan composition. Application of recombinant, 65 kDa latent heparanase protein produced by CHO cells on the lectin array revealed a high level of N-linked high mannose and a low level of N-linked bi-antennary structures, while the presence of tri- and tetra- antennary structures were not detected (Table 1). The results also show low levels of sialylation and low levels of truncated antennae (terminal GlcNAc) (Table 1). O-linked glycans were not detected.
Figure 1. Heparanase glycan fingerprint profile obtained by the Qproteome GlycoArray method.
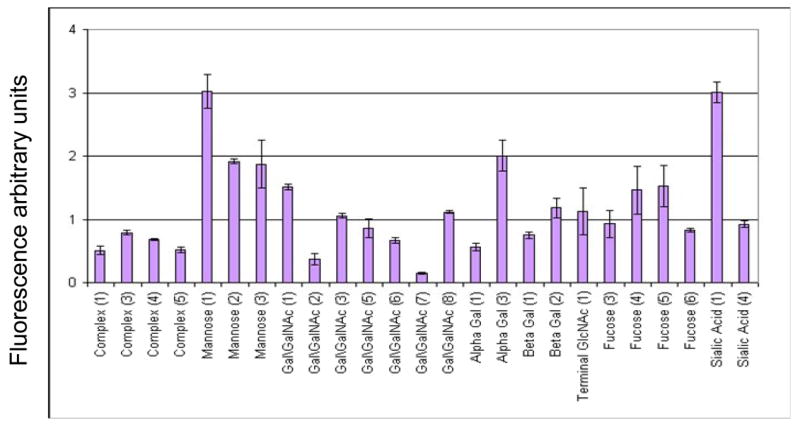
Recombinant 65 kDa heparanase purified to homogeneity from the conditioned medium of transfected CHO-K1 cells was applied onto the Qproteome GlycoArray lectin chip. Heparanase fingerprint profile was obtained, as describe under “Materials and Methods”.
Table 1.
Heparanase glycosylation pattern as evident by the Qproteome GlycoArray kit, in comparison with HPLC analyses.
| Glycan property | Qproteome GlycoArray | Verification by HPLC |
|---|---|---|
| N-linked | ||
| High mannose | High | 70% |
| Hybrid | n.a. | 1–5% |
| Complex | ||
| Bi-antennary | Low | 25% |
| Tri/tetra-antennary | Not detected | Not detected |
| Terminal GlcNAc | Low | 1–5% |
| Terminal GalNAc | Not detected | Not detected |
| Gal (α1–3) Gal | n.a. | Not detected |
| Charged glycans | ||
| Sialylated | Low | 23% |
| Phosphorylated | n.a. | 16% |
| O-linked | Not detected | n.a. |
n.a. = not analyzed
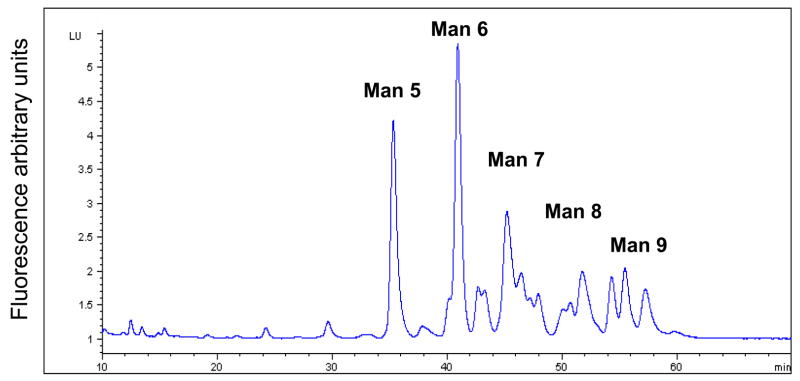
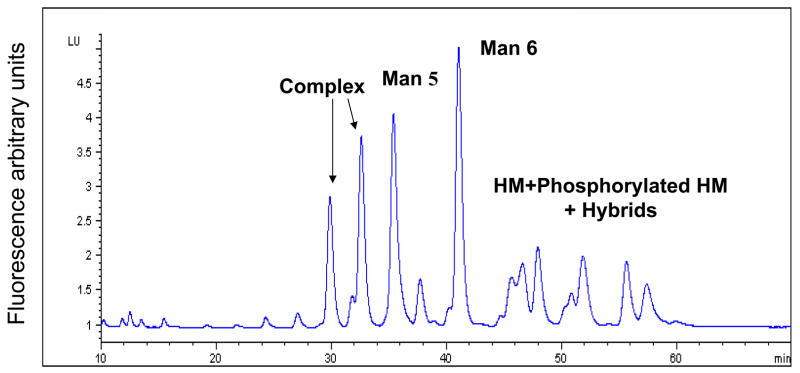
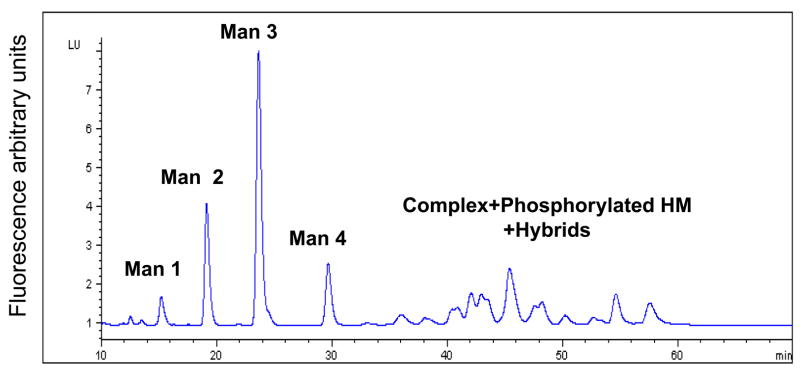
Glycoanalysis pattern obtained by the lectin array method was, next, compared with some more commonly used chromatographic and mass-spectrometric techniques, combined with enzymatic degradation. Glycan separation by NP HPLC column (Fig. 2A) indicated that most of the heparanase N-glycans are high mannose type, containing between 5–9 mannose residues (mostly 6 mannose residues). The unassigned peaks in the chromatograms can be assigned either to other (derivatized) high mannose glycans, to complex glycans, or to hybrid structures. In order to delineate between these possibilities, a series of enzymatic digests were performed on the released heparanase glycans, using sialidase, mannosidase, and galactosidase, followed by analysis of each digestion reaction by NP HPLC column. Separation of glycans treated with sialidase and β1,3 galactosidase enables the separation of complex glycans from the high mannose structures as shown in figure 2B, demonstrating that up to 25% of the glycans are complex bi-antennary type (peaks >5% were assigned). After mannosidase treatment, all neutral high mannose structures were separated from other glycans (Fig. 2C). Peaks that were not digested by these exoglucanases are concluded to be phosphorylated high mannose type and hybrid structires. In addition, weak anion exchange (WAX) chromatography was utilized for the separation of charged glycans from neutral glycans, indicating that 45% of the heparanase glycans are charged (Fig. 2D, blue line). Given the efficient trafficking of heparanase to lysosomes following cellular uptake (Goldshmidt et al, 2002; Zetser et al, 2004), and since lysosomal targeting of many lysosomal enzymes is mediated by mannose phosphorylation, we hypothesized that phosphate groups may account for the charged glycans. Glycan cleavage by alkaline phosphatase and separation by WAX HPLC column revealed a 16% increase in the neutral glycan fraction (Fig. 2D, pink line) in comparison to the untreated heparanase glycans (Fig. 2D, blue line). Subjecting glycans to desialylation prior to WAX chromatography analysis resulted in a 23% increase in the fraction of neutral glycans (data not shown). These results indicate that out of the 45% charged glycans, approximately 23% are sialylated complex type, while 16% bear phosphorylated high mannose glycans. The remaining 6% charged glycans were not identified, as they were not cleaved by either enzyme, presumably due to glycans inaccessible to the enzymes. The existence of six glycosylation sites in the heparanase molecule (Simizu et al, 2004), together with the homogenous appearance of the protein in a native polyacrylamide gel and the finding that 16% of the molecules carry phosphorylated high mannose led us to estimate that each heparanase molecule bears one phosphorylated glycan.
Figure 2. HPLC profiles of heparanase glycans.
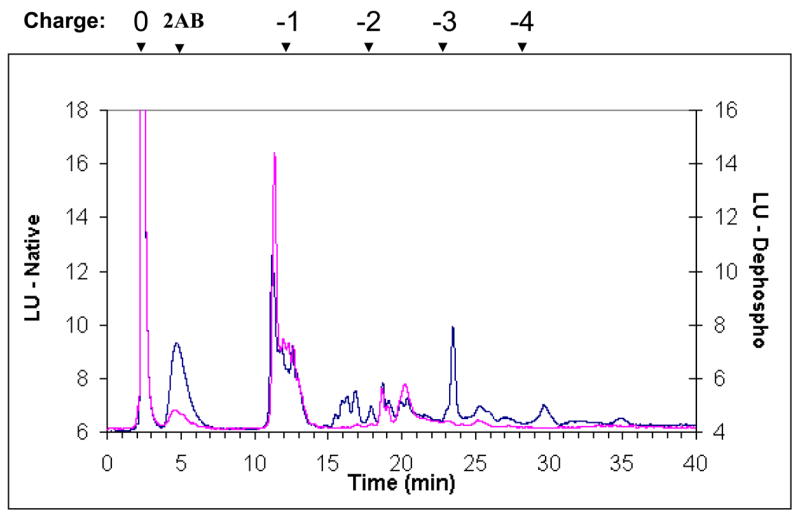
2AB labeled glycans cleaved from recombinant heparanase were separated by normal phase column before (A), and after treatment with sialidase and β1,3 galactosidase (B), or with mannosidase (C), and by weak anion exchange column, before (D, blue line) and after alkaline phosphatase treatment (D, pink line). Glycans purified from bovine Ribonuclease B (high mannose type) and fetuin (charged glycans) were used as standards. (HM = High mannose, Man = mannose).
Heparanase processing and activation by I-cell fibroblasts
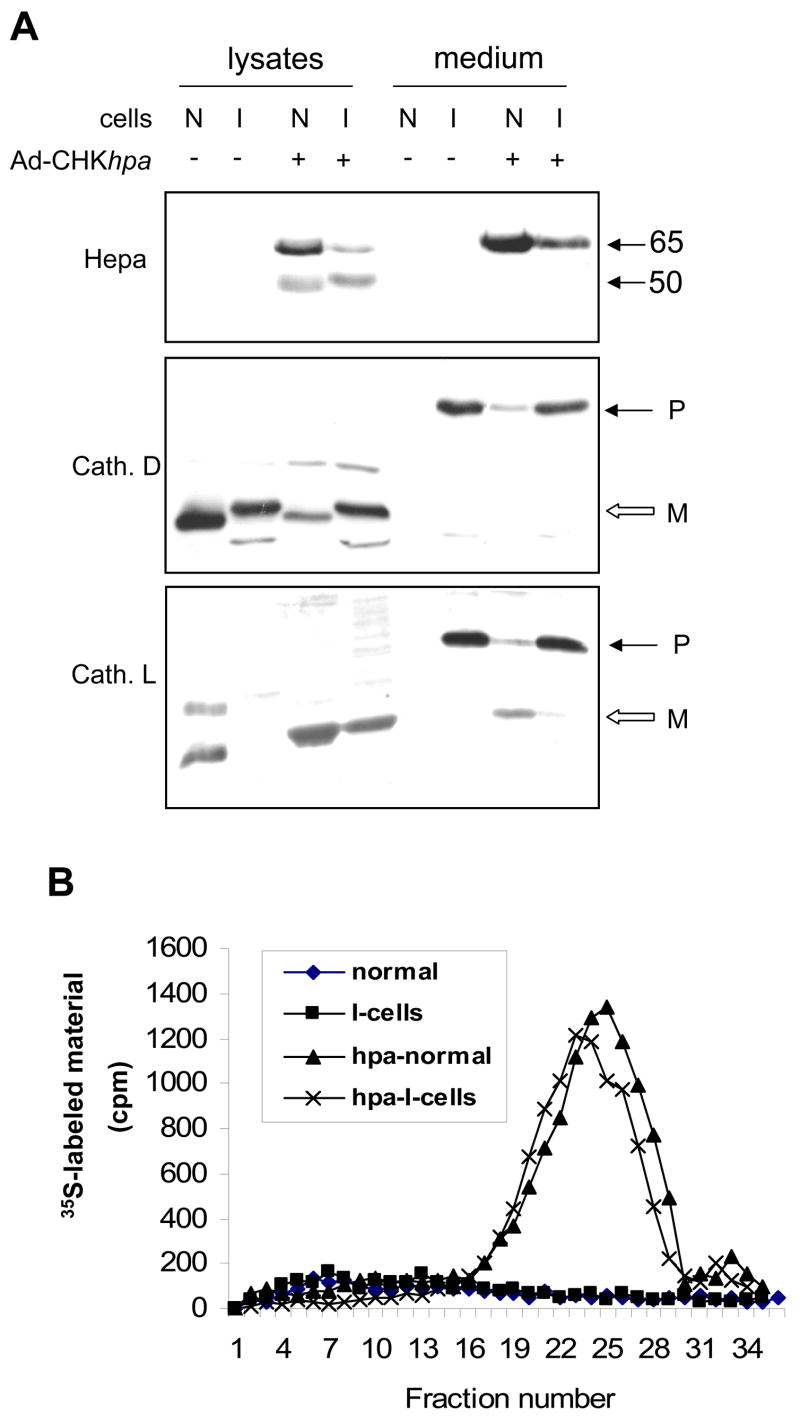
Next, we examined the biological significance of heparanase glycosylation as emerged by the glycoanalyses, focusing on glycan charge. Since mannose 6-phosphate and its receptor are considered as pre requisite for lysosomal targeting and cellular uptake of secreted lysosomal enzymes (Dittmer et al., 1998; Pohlmann et al., 1995a), we questioned whether the phosphorylated mannose moiety identified on heparanase plays a role in uptake and processing of the enzyme. To this end, we first utilized fibroblasts isolated from patients with I-cell disease (mucolipidosis II). This pathological disorder is characterized by the lack of mannose 6-phosphate modification, resulting in extracellular secretion of lysosomal enzyme at high levels, and, as a consequence, accumulation of unprocessed substrates in the lysosomal compartment (von Figura, 1991). Fibroblasts isolated from these patients are, therefore, widely used to study the role of mannose 6-phosphate residues in cellular trafficking. Fibroblasts isolated from the skin of healthy donors were used as control. We, initially, confirmed the authenticity of our I-cell fibroblasts by subjecting cell lysates and conditioned medium to immunoblotting with anti-cathepsin D (Fig. 3A, second panel) and anti-cathepsin L (Fig. 3A, lower panel) antibodies. Both cathepsins are typical lysosomal enzymes, and were found in the cell lysate of control fibroblasts (N), but not in the cell conditioned medium (Fig. 3A, lower panels, N, medium). In contrast, cathepsin D and L precursors (P) were abundantly secreted by I-cells but not by normal fibroblasts (Fig. 3A, lower panels, I, medium), thus confirming their pathological abnormality. Infection of normal control fibroblasts (N) with adeno-Hepa vector yielded readily detected, properly processed (Fig. 3A, upper panel, lysate) and active (Fig. 3B) heparanase enzyme. Noteably, subjecting heparanase infected I-cell lysates to immunoblot analysis revealed properly processed enzyme (Fig. 3A, I +, lysate) that exhibited heparanase activity almost identical to control normal fibroblasts (Fig. 3B). Thus, a role for mannose-6-phosphate modification in heparanase trafficking, processing and activation is not depicted by this model system.
Figure 3. Heparanase uptake by I-cells.
A. Immunoblotting. Control human fibroblasts (N) and fibroblasts derived from the skin of patient with I-cell disease (I) were infected with control, empty adeno-virus (−) vector, or adeno-virus vector containing the chicken heparanase cDNA (Ad-CHKhpa; +). Cell lysate and conditioned medium samples were obtained two days following infection and subjected to immunoblotting with anti-heparanase (upper panel), anti-cathepsin D (middle panel) and anti-cathepsin L (lower panel) antibodies. B. Enzymatic activity. Control (Normal) and I-cell fibroblasts were infected with control, empty adeno-virus (◆ and ■, respectively) and with the Ad-CHKhpa (▲ and x, respectively) vectors. Cells (1 × 106) were harvested two days following infection, lysed by three freeze/thaw cycles and applied onto 35 mm dishes coated with 35S-labeled ECM to determine heparanase enzymatic activity as described under “Materials and Methods”. Note heparanase processing and enzymatic activity following uptake by I-cells.
Heparanase processing and activation by MPR-deficient fibroblasts
In order to further explore the contribution of MPR to heparanase uptake, we utilized mouse embryonic fibroblasts (MEFs) deficient in cell surface MPR (MPR300−/−). Exogenously added 65 kDa latent heparanase (1 μg/ml) was subjected to rapid uptake by control fibroblasts, and the protein was detected in the cell lysate already 15 min following application (Fig. 4A, upper panel). Heparanase processing was first evident 60 min following its addition, and the amount of the 50 kDa processed enzyme further accumulated by 4 h, in agreement with the uptake kinetics reported previously (Gingis-Velitski et al., 2004a; Gingis-Velitski et al., 2004b; Zetser et al., 2004). Interestingly, heparanase uptake by MPR300−/− cells resembled its uptake by control fibroblasts (Fig. 4A, lower panel), as judged by the appearance of a 65 kDa protein band 15 min following heparanase addition. In contrast, heparanase processing was significantly delayed, and the 50 kDa processed enzyme was first detected 4 h following addition (Fig. 4A, lower panel). The amount of the 50 kDa subunit significantly accumulated thereafter, yielding enzymatic activity equivalent to heparanase activity detected in lysates of control fibroblasts (Fig. 4B). We suspected that the delay in heparanase processing is due to reduced cathepsin activity in the lysosomal compartment of MPR300−/− cells, rather than to impaired internalization. Indeed, Cy3-labeled heparanase appeared to be mainly confined within endocytic vesicles, most likely endosomes and lysosomes, 1 h following its exogenous addition to control cells (Fig. 4C, left panel), and a similar localization of the labeled enzyme was noted in MPR300−/− fibroblasts (Fig. 4C, right panel). Since heparanase processing by MPR300−/− cells was not evident at this time point (Fig. 4A, lower panel), we concluded that heparanase binding and internalization by MPR300−/− cells is not impaired, and that under these experimental conditions heparanase is directed to endocytic vesicles even in the absence of MPR. The delay in heparanase processing is most likely due to a reduced cathepsin activity in these cells.
Figure 4. Heparanase uptake by MPR-deficient cells.
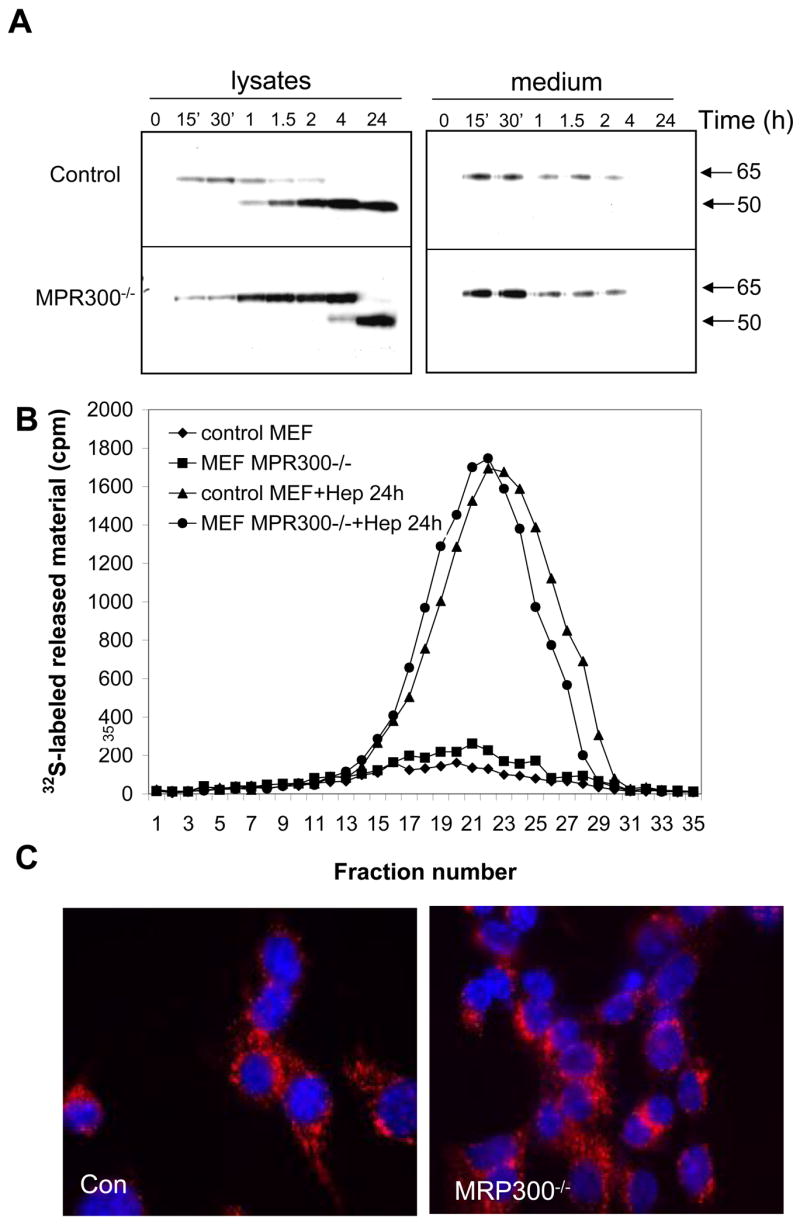
A. Immunoblotting. Control mouse fibroblasts (MEF, upper panel) and fibroblasts deficient of MPR (MPR300−/−, lower panel) were left untreated (0) or incubated with purified latent 65 kDa heparanase (1 μg/ml, ~15 nM) for the time indicated. Cell lysate and medium samples were subjected to immunoblotting with anti-heparanase antibody. B. Enzymatic activity. Control (▲, ◆) and MPR-deficient fibroblasts (■, ●) were left untreated (◆, ■) or incubated with heparanase (1μg/ml) for 24 h (▲ and ●, respectively). Cells (1 × 106) were then harvested, lysed and heparanase enzymatic activity was evaluated as described above. C. Immunofluorecence staining. Control and MPR-deficient fibroblasts were incubated with Cy3-labeled heparanase for 2 h. Cells were then washed with PBS, fixed, mounted and examined by confocal microscopy. Note heparanase internalization, processing, and activity by MPR-deficient cells.
Hierarchal uptake of heparanase by cell-surface receptors
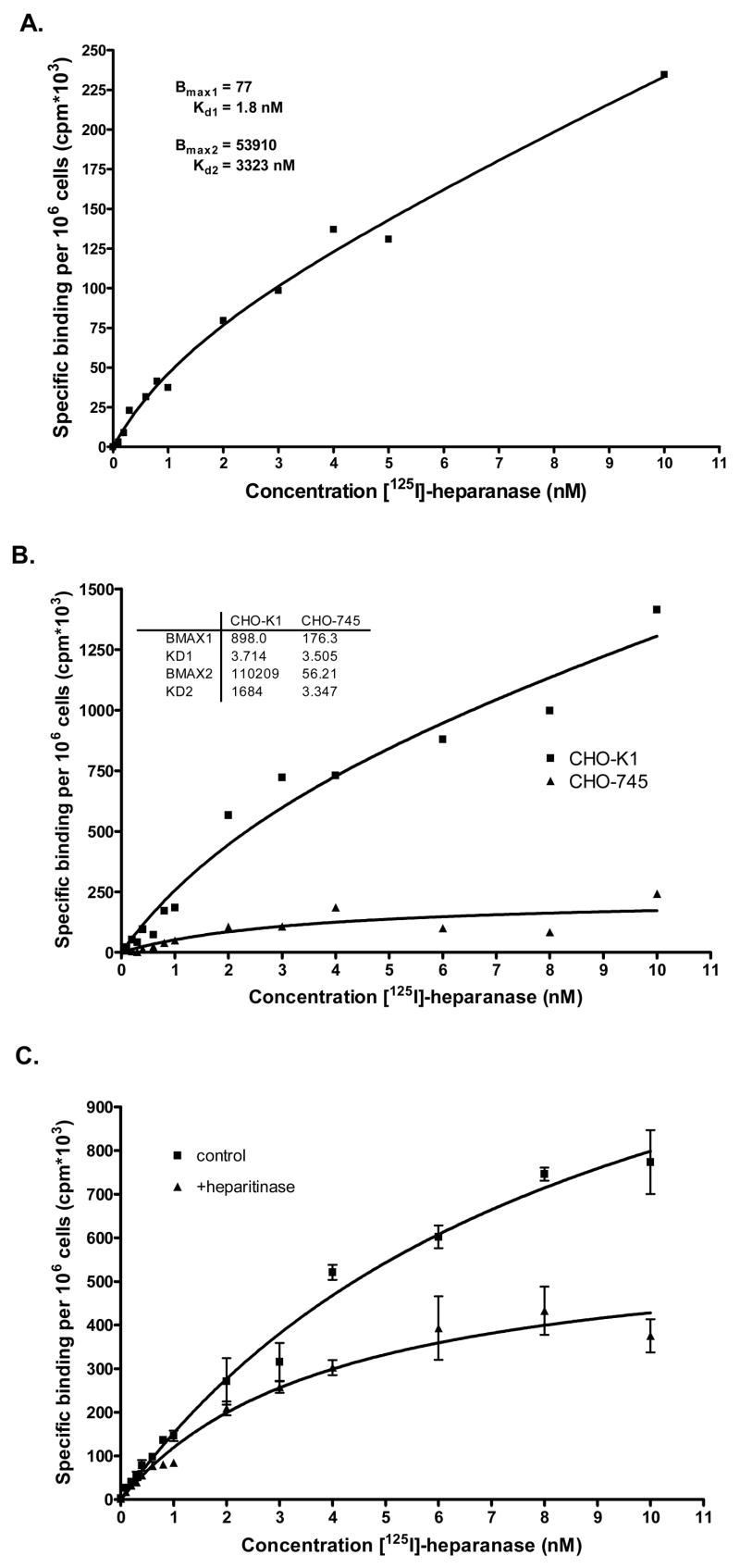
Utilizing recombinant heparanase at 1 μg/ml (~15 nM), we have reported that heparanase uptake is mediated by cell surface HSPG (Gingis-Velitski et al., 2004b). More recently, by using serum-free medium conditioned by heparanase transfected 293 cells, yielding an estimated heparanase concentration of ~5 nM, Vreys et al have shown that, in addition to HSPG, MPR and LRP receptors play a critical role in heparanase uptake (Vreys et al., 2005). We, therefore, questioned whether heparanase binding to cell surface molecules is affected by the applied concentration. Binding of iodinated heparanase to HSPG-deficient CHO-745 cells was significantly reduced compared with wild type CHO-K1 cells (Fig. 5A). Interestingly, however, no differences in heparanase binding were observed between CHO-K1 and CHO-745 cells upon lowering heparanase concentration from 5 nM (Fig. 5A) to 0.5 nM (Fig. 5B), suggesting that at low concentrations, cell surface molecules other than HS dominate its binding. Indeed, binding of 125I-heparanase (~0.5 nM) to MPR-deficient (MRP300−/−, Fig. 5C), and LRP-deficient (PEA-13, Fig. 5C) cells was reduced 4.5-, and 7-folds, respectively, compared with control fibroblasts (MEF). These results suggest a hierarchy of binding to cell surface molecules as a function of heparanase concentration, and suggest the existence of high affinity, low abundant (i.e., MPR, LRP), and low affinity, high abundant (i.e., HSPG) receptors that mediate heparanase binding.
Figure 5. Hierarchy of heparanase binding.
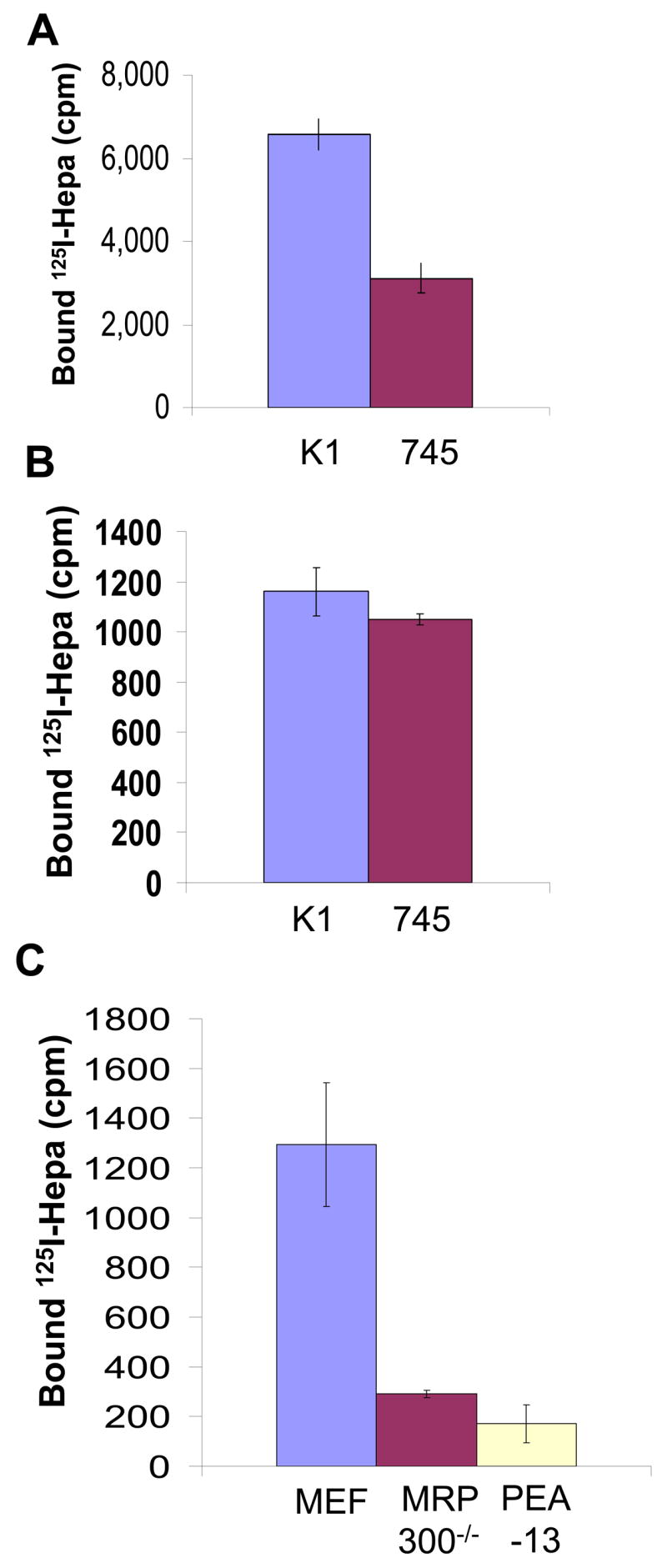
Control CHO K1 and HS-deficient CHO-745 cells were incubated for 30 min with 125I-heparanase at 5 nM (A) and 0.5 nM (B). Cells were then washed, lysed and counted in a γ-counter. Control fibroblasts (MEF) and fibroblasts deficient of MPR (MPR300−/−) or LRP (PEA 13) were incubated for 30 min with 125I-heparanase at 0.5 nM and heparanase binding was examined as above.
We approached this possibility by performing binding assays and calculating Bmax and Kd values for heparanase interactions (Fig. 6). Binding of 125I-heparanase to HeLa cells exhibited linearity at low concentrations that was not saturated even at high levels of heparanase (Fig. 6A). Further analyses of heparanase binding by the Prism 4 software suggest the existence of low affinity (Kd = 3 mM) and highly abundant (Bmax = 1 × 108) receptors, and of high affinity (Kd = 2 nM), low abundant (Bmax = 1.7 × 104) receptors. We suspected that the low affinity, high abundant binding sites are HSPG, as was shown, for example, for bFGF (Ornitz, 2000), and examined this possibility by comparing heparanase binding to CHO-K1 cells and their HS-deficient CHO-745 counterpart cells. Binding of 125I-heparanase to its high-affinity binding sites appeared almost identical, and Kd values of 3.7 and 3.5 nM were calculated for CHO-K1 and 745 cells, respectively (Fig. 6B), in agreement with the high affinity Kd value calculated for HeLa cells (Fig. 6A). In contrast, the number of low-affinity sites was significantly reduced and Bmax values of 1 × 108 and 5.6 × 105 were calculated for CHO K1 and 745 cells, respectively. Moreover, similar results were obtained following treatment of CHO K1 cells with bacterial heparitinase (Fig. 6C), clearly implying that HSPG serve as low affinity binding sites, while MPR and LRP likely represent high affinity heparanase receptors.
Figure 6. Binding assay reveals the existence of low-, and high-affinity heparanase receptors.
A. HeLa cells were incubated with the indicated concentrations of 125I-heparanase in the absence or presence of cold heparanase (200 nM) for 2 h on ice. Following three washes with ice cold PBS cells were lyased and counted. Binding curve was obtained by the Prism 4 software. Similar analyses were performed for CHO K1 vs. CHO-745 cells (B), and CHO K1 cells pre-treated with bacterial heparitinase (C).
Discussion
Applying a highly purified recombinant heparanase on the Qproteome GlycoArray, revealed high levels of N-linked mannose, in agreement with its high affinity to Concanavalin A lectin (Vlodavsky et al., 1999; Zcharia et al., 2005), and low levels of bi-antennary structures and sialic acid (Table 1). Glycoanalysis obtained by the lectin array was confirmed by biochemical analysis (Table 1). Among the parameters included, detection of charged glycans is the major limitation of the lectin array. While the array is capable of detecting sialic acid, charge contributed by phosphorylation is not detected. Thus, mannose 6-phosphate modification, although anticipated, was only revealed by subjecting heparanase glycans to alkaline phosphatase. In order to further explore the significance of heparanase glycosylation, we utilized several model systems focusing on mannose 6-phosphate modification. Mannose 6-phosphate is present on oligosaccharides of most lysosomal enzymes and serves as the major recognition signal for lysosomal targeting, functioning as a high-affinity ligand for mannose 6-phosphate receptors (MPRs) at the trans-Golgi network (TGN) (Ghosh et al., 2003; Pohlmann et al., 1995b). In this manner, lysosomal enzymes are distinguished from secretory and membrane proteins and are selectively delivered to lysosomes. In spite of this unique and efficient targeting mechanism, a small fraction of acid hydrolases may be released from cells. Cell surface MPRs (MPR300/IGFRII) are responsible for recapturing and uptake of secreted acid hydrolases, thus ensuring a tightly regulated extracellular retention of these enzymes (Ghosh et al., 2003). Heparanase seems to primarily utilize this alternate pathway in its trafficking route, utilizing a set of cell surface receptors for its uptake. We have previously reported that heparanase is subjected to rapid and efficient cellular uptake (Nadav et al., 2002), mediated by cell surface HSPG of the syndecan family (Gingis-Velitski et al., 2004b). More recently, MPR and LRP have also been implicated in heparanase uptake, possibly dominating the role of HSPG (Vreys et al., 2005). Since Vreys et al have used a lower concentration of heparanase in their studies (~5 nM), we suspected that binding preferences may be dictated by the applied concentration. Indeed, binding of iodinated heparanase (5 nM) by HS-deficient CHO-745 cells was reduced two folds compared to CHO-K1 cells, 30 min following application, in agreement with the observations of Vreys et al (Vreys et al., 2005). In contrast, binding to wild type K1 and HS-deficient CHO-745 cells appeared unchanged upon a ten-fold lowering of heparanase concentration (0.5 nM, Fig. 5B). Under the same conditions, binding to MPR-deficient, and even more so to LRP-deficient cells (PEA13) was significantly reduced (Fig. 5C), suggesting a hierarchy between the different types of heparanase receptors. According to this notion, HSPG, due to their high abundance, are more critical at high heparanase concentrations, while MPR and LRP dominate cellular uptake at low heparanase levels. This concept is supported by binding experiments, clearly revealing the existence of two types of heparanase binding sites, exhibiting high (Kd = 2–3 nM) and low (Kd = 3 mM) affinities (Fig. 6). The latter appear to be HSPG as indicated by a marked decrease in low affinity binding sites upon heparitinase treatment, or heparanase binding to HS-deficient CHO-745 cells (Fig. 6). While MPR specifically captures mannose 6-phosphate-containing client proteins, LRP mediates the clearance of over 30 different ligands belonging to a variety of protein families. By far, the largest group of ligands recognized by LRP is proteinases. Certain serine proteases and metalloproteinases (MMP) bind directly to LRP, while others preferentially bind to LRP once complexed to their specific inhibitor (Herz and Strickland, 2001). Examples are MMP-9-TIMP-1 (Kd = 20 nM) (Hahn-Dantona et al., 2001), uPA-PAI-1 (Kd = 3 nM) (Croucher et al., 2006), and factor VIIa-TFPI complexes (Herz and Strickland, 2001). The results described here (Fig. 5) and by Vreys et al (Vreys and David, 2007; Vreys et al., 2005) add heparanase to the growing list of enzymes that bind LRP, and further support a critical role for the LRP system in the uptake and clearance of proteases and glucuronidases from the extracellular environment. Furthermore, the calculated affinity of heparanase for LRP (Kd = 2–3.5 nM) is in the range, or even higher than that found for most other LRP ligands (Croucher et al., 2006; Hahn-Dantona et al., 2001; Muramatsu et al., 2000; Page et al., 2006; Rohlena et al., 2003), supporting its biological relevance.
The concept of low affinity, high abundant vs. high affinity, low abundant receptors is well established. In the case of bFGF, for example, HSPG are considered low-affinity receptors, co-operating with the high-affinity bFGF receptor to enhance receptor dimerization and exert potent biological effects (Eswarakumar et al., 2005; Ornitz, 2000; Yayon et al., 1991). Several lines of evidence suggest that HSPG and MPR/LRP co-operate in the course of heparanase uptake. Stable transfection of heparanase into HS-deficient CHO-745 (Gingis-Velitski et al., 2004b) and CHO-677 (Vreys et al., 2005) cells resulted in accumulation of the 65 kDa latent heparanase in the culture medium, accompanied by reduced levels of the active 50 kDa enzyme in the cell lysate (Gingis-Velitski et al., 2004b). In these cells, extracellular levels of heparanase are suspected to be lower than 5 nM, calculated for transiently-transfected 293 cells (Vreys et al., 2005), thus expected to primarily utilize MPR/LRP receptors for efficient uptake. Co-operation is further supported by studies utilizing heparanase gene constructs deficient, or mutated in the heparin/HS binding domain (HBD). As already mentioned, a deletion mutant lacking the HBD sequence Gln270-Lys280 (65Δ10) accumulates at high levels extracellularly, is not internalized and lacks enzymatic activity (Levy-Adam et al., 2005). In contrast, heparanase mutant deleted at a second HBD, Lys158-Asn171 (65Δ15), failed to get secreted (Levy-Adam et al., 2005), likely due to removal of N-glycosylation sites required for proper secretion (Simizu et al., 2004). This protein variant appeared to accumulate in the ER/Golgi apparatus, is not subjected to proteolytic processing and, hence, lacks enzymatic activity (Levy-Adam et al., 2005). Thus, in the absence of HBD, MPR by itself appears not sufficient for proper lysosomal targeting of heparanase, unlike, for example, the mechanism utilized by cathepsins.
Taken together, the results presented here and by Vreys and David (Vreys and David, 2007) suggest a model by which heparanase uptake is mediated by co-operation between low-affinity HSPG and high-affinity MPR/LRP cell surface receptors, that can be interrupted by the deletion of HBD, or by competition with soluble heparin, RAP, or mannose 6-phosphate (Gingis-Velitski et al., 2004b; Levy-Adam et al., 2005; Vreys and David, 2007; Vreys et al., 2005).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230:229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- Croucher D, Saunders DN, Ranson M. The urokinase/PAI-2 complex: a new high affinity ligand for the endocytosis receptor low density lipoprotein receptor-related protein. J Biol Chem. 2006;281:10206–10213. doi: 10.1074/jbc.M513645200. [DOI] [PubMed] [Google Scholar]
- Dempsey LA, Brunn GJ, Platt JL. Heparanase, a potential regulator of cell-matrix interactions. Trends Biochem Sci. 2000;25:349–351. doi: 10.1016/s0968-0004(00)01619-4. [DOI] [PubMed] [Google Scholar]
- Dittmer F, Hafner A, Ulbrich EJ, Moritz JD, Schmidt P, Schmahl W, Pohlmann R, Figura KV. I-cell disease-like phenotype in mice deficient in mannose 6-phosphate receptors. Transgenic Res. 1998;7:473–483. doi: 10.1023/a:1008823315416. [DOI] [PubMed] [Google Scholar]
- Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase Gene Silencing, Tumor Invasiveness, Angiogenesis, and Metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- Edovitsky E, Lerner I, Zcharia E, Peretz T, Vlodavsky I, Elkin M. Role of endothelial heparanase in delayed-type hypersensitivity. Blood. 2005;107:3609–3616. doi: 10.1182/blood-2005-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. Faseb J. 2001;15:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ferro V, Hammond E, Fairweather JK. The development of inhibitors of heparanase, a key enzyme involved in tumour metastasis, angiogenesis and inflammation. Mini Rev Med Chem. 2004;4:693–702. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- Folkman J, Klagsbrun M, Sasse J, Wadzinski M, Ingber D, Vlodavsky I. A heparin-binding angiogenic protein--basic fibroblast growth factor--is stored within basement membrane. Am J Pathol. 1988;130:393–400. [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase Induces Endothelial Cell Migration via Protein Kinase B/Akt Activation. J Biol Chem. 2004a;279:23536–23541. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- Gingis-Velitski S, Zetser A, Kaplan V, Ben-Zaken O, Cohen E, Levy-Adam F, Bashenko Y, Flugelman MY, Vlodavsky I, Ilan N. Heparanase Uptake Is Mediated by Cell Membrane Heparan Sulfate Proteoglycans. J Biol Chem. 2004b;279:44084–44092. doi: 10.1074/jbc.M402131200. [DOI] [PubMed] [Google Scholar]
- Goldshmidt O, Zcharia E, Aingorn H, Guatta-Rangini Z, Atzmon R, Michal I, Pecker I, Mitrani E, Vlodavsky I. Expression pattern and secretion of human and chicken heparanase are determined by their signal peptide sequence. J Biol Chem. 2001;276:29178–29187. doi: 10.1074/jbc.M102462200. [DOI] [PubMed] [Google Scholar]
- Hahn-Dantona E, Ruiz JF, Bornstein P, Strickland DK. The low density lipoprotein receptor-related protein modulates levels of matrix metalloproteinase 9 (MMP-9) by mediating its cellular catabolism. J Biol Chem. 2001;276:15498–15503. doi: 10.1074/jbc.M100121200. [DOI] [PubMed] [Google Scholar]
- Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Levy-Adam F, Abboud-Jarrous G, Guerrini M, Beccati D, Vlodavsky I, Ilan N. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J Biol Chem. 2005;280:20457–20466. doi: 10.1074/jbc.M414546200. [DOI] [PubMed] [Google Scholar]
- Levy-Adam F, Miao HQ, Heinrikson RL, Vlodavsky I, Ilan N. Heterodimer formation is essential for heparanase enzymatic activity. Biochem Biophys Res Commun. 2003;308:885–891. doi: 10.1016/s0006-291x(03)01478-5. [DOI] [PubMed] [Google Scholar]
- McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br J Pharmacol. 2007;151:1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao HQ, Liu H, Navarro E, Kussie P, Zhu Z. Development of heparanase inhibitors for anti-cancer therapy. Curr Med Chem. 2006;13:2101–2111. doi: 10.2174/092986706777935230. [DOI] [PubMed] [Google Scholar]
- Muramatsu H, Zou K, Sakaguchi N, Ikematsu S, Sakuma S, Muramatsu T. LDL receptor-related protein as a component of the midkine receptor. Biochem Biophys Res Commun. 2000;270:936–941. doi: 10.1006/bbrc.2000.2549. [DOI] [PubMed] [Google Scholar]
- Nadav L, Eldor A, Yacoby-Zeevi O, Zamir E, Pecker I, Ilan N, Geiger B, Vlodavsky I, Katz BZ. Activation, processing and trafficking of extracellular heparanase by primary human fibroblasts. J Cell Sci. 2002;115:2179–2187. doi: 10.1242/jcs.115.10.2179. [DOI] [PubMed] [Google Scholar]
- Ornitz DM. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Page S, Judson A, Melford K, Bensadoun A. Interaction of lipoprotein lipase and receptor-associated protein. J Biol Chem. 2006;281:13931–13938. doi: 10.1074/jbc.M600995200. [DOI] [PubMed] [Google Scholar]
- Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- Pohlmann R, Boeker MW, von Figura K. The two mannose 6-phosphate receptors transport distinct complements of lysosomal proteins. J Biol Chem. 1995a;270:27311–27318. doi: 10.1074/jbc.270.45.27311. [DOI] [PubMed] [Google Scholar]
- Pohlmann R, Boeker MWC, von Figura K. The Two Mannose 6-Phosphate Receptors Transport Distinct Complements of Lysosomal Proteins. J Biol Chem. 1995b;270:27311–27318. doi: 10.1074/jbc.270.45.27311. [DOI] [PubMed] [Google Scholar]
- Rohlena J, Kolkman JA, Boertjes RC, Mertens K, Lenting PJ. Residues Phe342-Asn346 of activated coagulation factor IX contribute to the interaction with low density lipoprotein receptor-related protein. J Biol Chem. 2003;278:9394–9401. doi: 10.1074/jbc.M209097200. [DOI] [PubMed] [Google Scholar]
- Rosenfeld R, Bangio H, Gerwig GJ, Rosenberg R, Aloni R, Cohen Y, Amor Y, Plaschkes I, Kamerling JP, Maya RB. A lectin array-based methodology for the analysis of protein glycosylation. J Biochem Biophys Methods. 2007;70:415–426. doi: 10.1016/j.jbbm.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Sanderson RD, Yang Y, Kelly T, Macleod V, Dai Y, Theus A. Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: Growth regulation and the prospect of new cancer therapies. J Cell Biochem. 2005;96:897–905. doi: 10.1002/jcb.20602. [DOI] [PubMed] [Google Scholar]
- Simizu S, Ishida K, Wierzba MK, Osada H. Secretion of Heparanase Protein Is Regulated by Glycosylation in Human Tumor Cell Lines. J Biol Chem. 2004;279:2697–2703. doi: 10.1074/jbc.M300541200. [DOI] [PubMed] [Google Scholar]
- Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274:24153–24160. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I. Preparation of extracellular matrices produced by cultured corneal endothelial and PF-HR9 endodermal cells. In: Bonifacino MDJS, Hartford JB, Lippincott-Schwartz J, Yamada KM, editors. Protocols in Cell Biology. Vol. 1. New York: John Wiley & Sons; 1999. pp. 10.4.1–10.4.14. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: Structure, Biological Functions, and Inhibition by Heparin-Derived Mimetics of Heparan Sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- von Figura K. Molecular recognition and targeting of lysosomal proteins. Curr Opin Cell Biol. 1991;3:642–646. doi: 10.1016/0955-0674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–52. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreys V, Delande N, Zhang Z, Coomans C, Roebroek A, Durr J, David G. Cellular Uptake of Mammalian Heparanase Precursor Involves Low Density Lipoprotein Receptor-related Proteins, Mannose 6-Phosphate Receptors, and Heparan Sulfate Proteoglycans. J Biol Chem. 2005;280:33141–33148. doi: 10.1074/jbc.M503007200. [DOI] [PubMed] [Google Scholar]
- Willnow T, Herz J. Genetic deficiency in low density lipoprotein receptor-related protein confers cellular resistance to Pseudomonas exotoxin A. Evidence that this protein is required for uptake and degradation of multiple ligands. J Cell Sci. 1994;107:719–726. [PubMed] [Google Scholar]
- Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- Zcharia E, Zilka R, Yaar A, Yacoby-Zeevi O, Zetser A, Metzger S, Sarid R, Naggi A, Casu B, Ilan N, et al. Heparanase accelerates wound angiogenesis and wound healing in mouse and rat models. Faseb J. 2005;19:211–221. doi: 10.1096/fj.04-1970com. [DOI] [PubMed] [Google Scholar]
- Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase Affects Adhesive and Tumorigenic Potential of Human Glioma Cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- Zetser A, Levy-Adam F, Kaplan V, Gingis-Velitski S, Bashenko Y, Schubert S, Flugelman MY, Vlodavsky I, Ilan N. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci. 2004;117:2249–2258. doi: 10.1242/jcs.01068. [DOI] [PubMed] [Google Scholar]