Abstract
The awareness that pathogens can adapt and evolve over relatively short time-scales is changing our view of infectious disease epidemiology and control. Research on the transmission dynamics of antigenically diverse pathogens is progressing and there is increasing recognition for the need of new concepts and theories. Mathematical models have been developed considering the modelling unit in two extreme scales: either diversity is not explicitly represented or diversity is represented at the finest scale of single variants. Here, we use an intermediate approach and construct a model at the scale of clusters of variants. The model captures essential properties of more detailed systems and is much more amenable to mathematical treatment. Specificities of pathogen clusters and the overall potential for transmission determine the reinfection rates. These are, in turn, important regulators of cluster dynamics. Ultimately, we detect a reinfection threshold (RT) that separates different behaviours along the transmissibility axis: below RT, levels of infection are low and cluster substitutions are probable; while above RT, levels of infection are high and multiple cluster coexistence is the most probable outcome.
Keywords: reinfection threshold, pathogen diversity, influenza drift, influenza shift
1. Introduction
Most infections induce an immune response that confers only modest protection against reinfections. This is either owing to host inefficacy in mounting specific immunity (tuberculosis, malaria) or owing to pathogen ability to generate sustainable antigenic diversity (influenza viruses, respiratory syncytial virus, malaria parasites). We have recently generated a series of simple models to explore the epidemiological consequences of different forms of immune failure (Gomes et al. 2004, 2005; Breban & Blower 2005). When immunity is partially protective, a threshold in transmission was detected above which reinfection dominates the dynamics. As the transmission coefficient is increased across this so-called ‘reinfection threshold’ (RT), the levels of infection in the population rise steeply which is accompanied by a dramatic drop in the potential effectiveness of vaccination programmes. The specification of pathogen diversity in epidemiological models is the subject of much ongoing research and debate (Andreasen et al. 1997; Gupta & Maiden 2001; Girvan et al. 2002; Gog & Grenfell 2002; Gomes et al. 2002; Boni et al. 2004; Grenfell et al. 2004). Here, we construct a relatively simple model to demonstrate that the RT can have a crucial role in regulating pathogen diversity and determining pathogen evolution. The influenza viruses provide an excellent illustration.
Influenza viruses have a segmented RNA genome, which makes them particularly prone to mutations and intratypic recombination through genetic reassortment during multiple infections. Mutation generates variants that are antigenically closely related (drift candidates) and, in the case of influenza type A, genetic reassortment involving human and avian subtypes produces antigenically distant variants (shift candidates). The conditions under which drift and shift will effectively occur can be studied by modelling the dynamics of transmission. Previous models represented the transmission dynamics of multiple variants interacting by cross-immunity and reproduced the structuring of pathogen populations into distinct clusters of antigenically similar variants (Gog & Grenfell 2002; Gomes et al. 2002; Ferguson et al. 2003). Furthermore, in the case of influenza A, the switch between clusters has been shown to occur in discrete steps (Smith et al. 2004) reinforcing the notion that the emergence of a new cluster can be modelled as a discrete event.
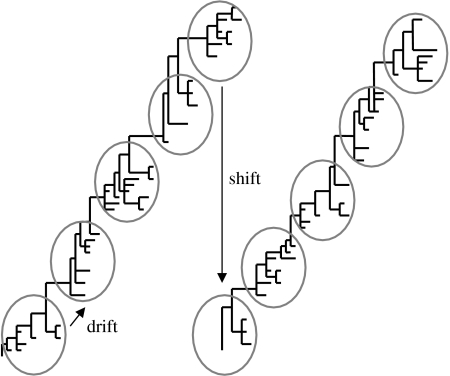
Motivated by these previous studies, we use the cluster as a modelling unit, providing a complementary approach to variant-based models. The cluster structure of two influenza A subtypes is schematically represented in figure 1. The transmission dynamics of each cluster are modelled as a SIRI (susceptible-infected-recovered-infected) model, where reinfection occurs at a reduced rate owing to partial immunity between the constituent variants. Reinfection by a variant in a different cluster is assumed to occur at a higher rate to reflect lower cross-immunity, and increases with increase in the antigenic distance between the two clusters. We describe the conditions under which a resident cluster is replaced by an invader. By varying the parameter that controls cross-immunity between clusters, we implement a range of cluster replacement scenarios and formalize the processes of drift and shift evolution within the same model framework.
Figure 1.
Schematic of clusters of antigenic variants, together with drift and shift phenomena.
When considering only one cluster, the transmission dynamics induces a RT owing to partial immunity (Gomes et al. 2004, 2005; Breban & Blower 2005), and this is associated with a steep increase in the prevalence of infection. This threshold occurs at a value of transmission such as R0=1/σ, where σ (between 0 and 1) represents the risk reduction of reinfection resulting from partial immunity in response to a primary infection. Here, we extend the model and consider more than one cluster. We show that the RT has a striking role in the regulation of pathogen antigenic diversity. Cluster replacement is identified as the preferred behaviour when the transmissibility is restricted to a window immediately below the RT. With this motivation, we propose that if the transmissibilities of influenza A viruses lie within this window, then the evolutionary pattern of drift and shift that so uniquely characterize these viruses can be accounted for by the same principles.
2. A model with multiple clusters
2.1 Cluster definition
We consider a pathogen population with the potential to accommodate antigenic diversity respecting a predefined clustered structure. The pathogen explores this potential structure in a way that is regulated by the epidemic dynamics, which in turn is largely determined by the pathogen population structure itself. Here, we consider two levels of relatedness: variants in the same cluster differ by some average within-cluster antigenic distance, σ, while different clusters are separated by some greater between-cluster antigenic distance, σx, as conceptually, greater antigenic distance represents lower cross-immunity. As such, in some appropriate normalized measure, we have 0<σ<σx≤1. Each cluster, i, is therefore responsible for three distinct rates of infection depending on the host's previous exposures: Λi for naive hosts, σΛi for hosts previously infected by the same cluster, and σxΛi for hosts previously infected by a different cluster.
2.2 Deterministic model
It is assumed that individual hosts are born with maximum susceptibility to infection, and acquire some protection as they experience sequential infections. Thus, individuals are characterized by their present infection status and history of past exposures. In analogy with previous models (Andreasen et al. 1997), and considering N={1, 2, …, n} be the set of all possible clusters, there are two types of dynamical variables: SJ is the proportion of presently uninfected individuals that have been infected by clusters in the subset J⊆N, and is the proportion of individuals presently infected with cluster i and previously infected by clusters in J. A given cluster, i, infects naive individuals at a per capita rate Λi=βiIi, where , while previously infected individuals are subject to reduced rates of infection as described above. The dynamics of transmission are described by the system of differential equations
| (2.1) |
and represented diagrammatically in figure 2 for the case of two clusters. The hierarchical structure of the pathogen population is captured by the parameters σ and σx. A crucial component in the analysis will be the difference, Δσ=σx−σ. Other parameters consist of the death rate, μ, which is assumed equal to the birth rate and takes the value μ=1/70 year−1 (representing a life expectancy at birth of 70 years), the rate of recovery from infection τ=52 year−1 (representing an average duration of infection of approximately one week) and a transmission coefficient which is the same for all clusters, β. If time is scaled to units of average duration of infection, 1/(τ+μ), the transmission coefficient scales to become the basic reproduction number, R0=β/(τ+μ). This is the average number of infections generated by a single infected individual in a totally susceptible population, and therefore it must be above 1 for a cluster to invade a susceptible population. We shall use R0 instead of β to represent the results, as this is a more universal measure, and note that the same value is assumed for all clusters.
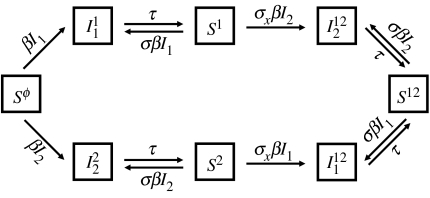
Figure 2.
Diagram representing the model with two clusters.
2.3 Stochastic model
We construct a continuous time stochastic model by replacing the differential equation system (2.1) with corresponding Poisson processes with state-dependant rates and exponential waiting time between state transitions. These stochastic models are investigated with Monte Carlo simulation experiments using standard techniques (Renshaw 1991). All stochastic simulations are performed with two clusters only, with the additional assumption that both clusters have the same transmission coefficient. The transition rates of the specific model with two clusters are given in table 1.
Table 1.
Transition rates.
| transition | rate |
|---|---|
| τ | |
| τ | |
| τ | |
| τ | |
3. Results
3.1 Endemic equilibria
According to the deterministic model, a single cluster invades a naive population and establishes an endemic state given that its transmissibility is above the epidemic threshold (R0>1). The endemic state is reached in a damped oscillatory manner if transmissibility is below the RT (R0<1/σ) and characterized by low prevalence level. Above the RT, equilibrium prevalences are about two orders of magnitude higher and oscillations are no longer observed (see Gomes et al. 2004). When two clusters are introduced, the long-term equilibrium is characterized by coexistence where both clusters maintain the same endemic level (Nunes et al. 2006). This work is focused on invasion by new clusters and the dynamics on shorter time-scales. The first insight into the associated behaviour is obtained by simulation of the deterministic model and the results are then consolidated by its stochastic counterpart.
3.2 Cluster replacement
Simulations consider that a resident cluster (cluster 1) is approaching equilibrium in a host population. An invading cluster (cluster 2) is then introduced. The reduced risk of infection owing to within-cluster acquired immunity, σ, was set at 0.25 for every simulation, unless stated otherwise. The antigenic distance between clusters is represented by a weaker risk reduction here modelled by varying σx>σ. Therefore, the same framework can be used to model either antigenic drift or antigenic shift, depending on whether Δσ is small or large, respectively. Given the increasing evidence for the existence of heterosubtypic immunity between different subtypes of influenza (Yetter et al. 1980; Liang et al. 1994; Heinen et al. 2001), we assume that σx<1 in all settings.
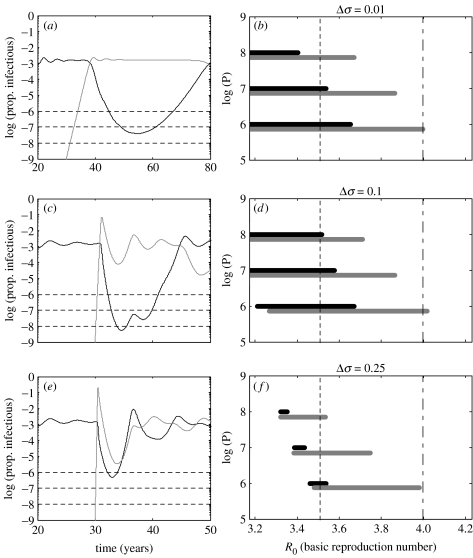
Figure 3 illustrates the behaviour of the deterministic model, when the RT is placed at 4 (σ=0.25). When the invading cluster is introduced, we observe a transient effect: the invader generates an epidemic peak and the resident goes through a trough before initiating convergence towards a new equilibrium. This transient effect, leading to the extinction of one or both clusters, depends essentially on two factors: the position of R0 relative to the RT and the antigenic distance between the two clusters. Regarding the first factor, as transient oscillations are only observed below the RT, the potential for cluster extinctions is also confined to this parameter regime. Regarding the second factor, by fixing R0 below the RT (e.g. R0=3.5) and varying Δσ, the resident cluster always goes through a deep trough associated with probable extinction (figure 3a,c,e, black line), while the invading cluster induces an initial epidemic followed by a trough whose amplitude increases with Δσ, facilitating the extinction of both clusters (figure 3a,c,e, grey line).
Figure 3.
Simulations with different cut-offs (a,c,e) and associated replacement windows (b,d,f). (a,c,e) Simulation of model (2.1) with two clusters and R0=3.5, just below the RT, set at 4 (σ=0.25). Black and grey lines correspond to clusters 1 and 2, respectively. Cut-offs for extinction marked at 10−6, 10−7 and 10−8 represent population sizes P=106, 107 and 108, respectively. (b,d,f) Range of R0 where replacement is expected as determined from the deterministic model with cut-offs as above (black bars) and the stochastic model with corresponding population sizes (grey bars: range of R0 where replacement occurs in at least one of 100 simulations). Three values of Δσ are represented: (a,b) Δσ=0.01; (c,d) Δσ=0.1; (e,f) Δσ=0.25.
The infinite host population size inherent to the deterministic model cannot, however, accommodate cluster extinctions, as infection can survive the most infinitesimal troughs. To analyse the probable outcome in a finite population, extinction cut-offs are placed at frequency 1/P, depending on the population size P considered. A cluster is considered extinct when it crosses the assumed cut-off (figure 3a,c,e, dashed lines).
Depending on the parameters and the population size, we observe three possible scenarios: extinction of both clusters, replacement of the resident by the invading cluster, and coexistence of both clusters. These are, hereinafter, referred simply as ‘extinction’, ‘replacement’ and ‘coexistence’, and predominate in this order as R0 increases, except for small Δσ (e.g. Δσ=0.01), where extinction is never observed. Replacement is typically confined to a window between extinction and coexistence (figure 3b,d,f, black bars) and this window becomes narrower as we increase the antigenic distance between clusters, Δσ. The replacement window is always contained below the RT, determining an important basis for the occurrence of antigenic drift and antigenic shift, as these require the previous cluster to become extinct and the invading one to survive. These results are confronted with their stochastic counterpart (figure 3b,d,f, grey bars) obtained for R0>3.2. Below this value, stochastic fluctuations make persistence of the resident cluster alone virtually impossible owing to sustained oscillations inherent to the stochastic model (Nåsell 2002). Stochasticity appears to extend the replacement window towards higher R0, reflecting a stronger tendency for cluster replacement.
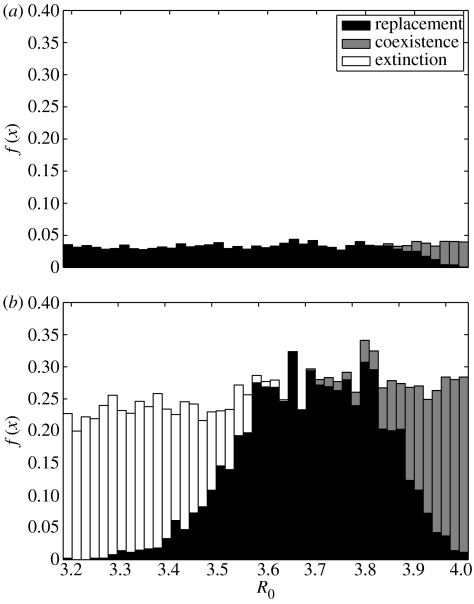
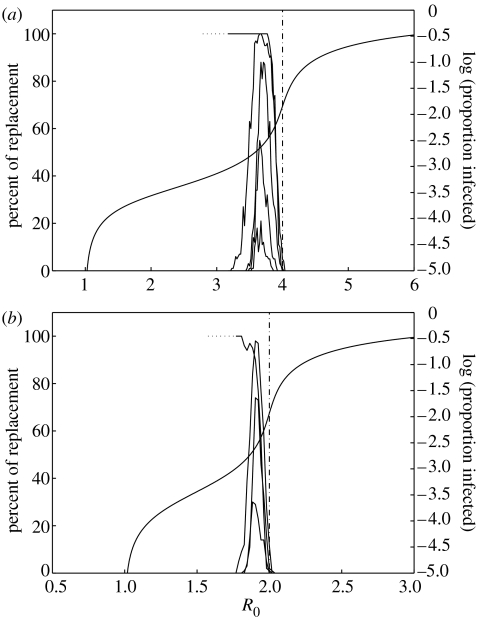
It is important to quantify the probability of these different events according to the values of the epidemiological parameters. Figures 4 and 5 show detailed results for the stochastic model, with a population of size P=106. There are two main differences when comparing with the deterministic model: for the same R0 and Δσ, several outcomes are now possible owing to stochasticity and the second cluster may not succeed to invade. For each set of parameter values, simulations were carried out to obtain 100 invasions. Of these successful invasions, the numbers of extinction, replacement and coexistence events were recorded. The results for the probability of each of these events, given that invasion occurs, are depicted in figure 4 for Δσ=0.01 and 0.25. Figure 5a,b shows the probabilities of replacement, conditional on invasion, for σ=0.25 and 0.5, along with the context provided by the equilibrium prevalence curve for a single cluster and the associated RT. The RT at R0=1/σ and the nested replacement windows located below this threshold are consistent across different values of sigma. Furthermore, when considering influenza A, the values of R0 encountered in the literature (Oxford 2000; Mills et al. 2004; Ferguson et al. 2005) are well accommodated within the parameter region associated with our replacement windows.
Figure 4.
Detailed results characterizing the probabilities of invasion as three possible outcomes: extinction (white), replacement (black) and coexistence (grey). (a) Δσ=0.01 and (b) Δσ=0.1. f(x) represents the frequency of each event, relative to the total number of simulations performed. The RT is set at 4 (σ=0.25).
Figure 5.
Summary of the results from the stochastic simulations for σ=0.25 and 0.5 (RT set at 4 and 2, respectively). The nested curves depict the percentage of replacement events, conditional to invasion of the second cluster, for five values of Δσ (from top to bottom): 0.01, 0.1, 0.25, 0.4 and 0.6. The upper curves (Δσ=0.01) are discontinued at R0=3.2 (in (a)) and R0=1.8 (in (b)) as stochastic simulations are unfeasible at low R0. Deterministic simulations suggested the trend indicated by the dotted segments. Equilibrium curve calculated from the deterministic model (black).
The results presented here show that the RT marks an upper bound on R0 for the occurrence of successive replacements of antigenic clusters that compete by cross-immunity. Above this threshold, the trend is for cluster coexistence and accumulation of antigenic diversity. Based on this, we propose that the RT has a potential role as a regulator of pathogen diversity.
4. Discussion
Supported by empirical evidence (Plotkin et al. 2002; Levin et al. 2004; Smith et al. 2004) and theoretical studies (Gog & Grenfell 2002; Gomes et al. 2002), a novel approach to the modelling of antigenically diverse pathogens, centred on the cluster of antigenic variants, is presented here, with an application to influenza. This approach intends to capture essential properties of the epidemiology resulting from what is now demonstrated as the emergence of an intermediate scale of aggregation for pathogen variants, the cluster. This is not contradictory to previous models centred on the individual strain, showing the emergence of these clusters, but in fact complementary.
Antigenically distinct clusters are introduced sequentially into circulation in a host population and the outcome of competition monitored. Cluster replacement is a phenomenon restricted to a range of transmissibility values that becomes narrow as we increase the antigenic distance between the clusters. This fundamental region is located just below the RT moving with it in an attached manner. Furthermore, it separates two other types of behaviour: extinction of both clusters at lower transmissibility and coexistence at higher transmissibility. Above the RT, accumulation of antigenic diversity is thus the expected result of any succession of cluster introductions. We expect influenza A to lie within the parameter region where replacement occurs, as the accumulation of diversity is limited for this pathogen, whose evolutionary pattern is characterized by a sequence of drift and shift events.
We find that the crucial factors determining replacement are:
the characteristic cluster size (factor affecting reinfection by a variant in the same cluster, σ) as this determines the position of the RT;
the distance between competing clusters (factor affecting reinfection by a variant in the competing cluster, σx>σ) as this determines the width of the replacement window; and
the position of R0 relative to the RT (located at R0=1/σ) and the replacement window determined above.
Reinfection is indeed central to the mechanism of replacement as a simple SIR (susceptible-infected-recovered) model with no reinfection, equivalent to setting σ=0, exhibits no substitution for any given set of parameters. Our proposal is that the evolutionary pattern of influenza A viruses is largely determined by a characteristic aggregation into clusters of antigenic variants and the subsequent reinfection dynamics regimented by this aggregation.
Although no explicit phylogeny is extracted from the model, a ‘slender-trunked’ phylogenetic tree, such as obtained for the influenza A hemagglutinin, would be most probable if transmissibility takes place in the replacement regime. This extrapolation from antigenic to genetic diversity is supported by the parallel between the two measures in the case of influenza A established by Smith et al. (2004). Furthermore, the hemagglutinin gene encodes the most important target of the immune system (the hemagglutinin protein) implying that patterns of genetic diversity will reflect antigenic constraints.
The drift of influenza A as a single lineage and the tendency for subtype replacement has stimulated other theoretical investigations. A highly elaborate epidemiological model produces this behaviour under the hypothesis that infection induces some variant-transcending immunity, which completely prevents reinfections for some time (Ferguson et al. 2003). The authors assume this immunity to apply between variants of the same subtype and between subtypes, decaying exponentially with a half-life period of six months. This variant-transcending (hetero- and homosubtypic) immunity was included as a means to enhance competition between lineages and subtypes, and presented as a necessary condition to obtain the patterns of drift as a single lineage and shift evolution characteristic of influenza A.
The cluster-based model proposed here also requires some sort of hetero- and homosubtypic immunity. However, we implement a long-lasting partially protective effect in agreement with immunological studies (Yetter et al. 1980; Liang et al. 1994; Heinen et al. 2001). The only manifestation of a short-lasting total protection in our model is the impediment of co-infection. This protective effect can be attributed not only to immunity during the course of infection but also to other phenomena such as behavioural change associated with disease. As observed by Ferguson et al. (2003), some form of temporary variant-transcending protective effect appears necessary to oppose trends of coexistence and accumulation of diversity. However, our framework permits a considerable relaxation of the assumption by reducing the average duration of protection from six months to one week. Despite the lack of more definite experimental results, it is important that different assumptions are explored by modelling studies.
Acknowledgments
We thank Jorge Carneiro, Ben Cooper and Graham Medley for their valuable comments. This research was funded by the Calouste Gulbenkian Foundation (FCG), the Portuguese Research Council (FCT) and the European Commission, grant MEXT-CT-2004-14338. J.B.O.M. had a studentship from PRODEP III.
References
- Andreasen V, Lin L, Levin S.A. The dynamics of cocirculating strains conferring partial cross-immunity. J. Math. Biol. 1997;35:825–842. doi: 10.1007/s002850050079. [DOI] [PubMed] [Google Scholar]
- Boni M.F, Gog J.R, Andreasen V, Christiansen F.B. Influenza drift and epidemic size: the race between generating and escaping immunity. Theor. Popul. Biol. 2004;65:179–191. doi: 10.1016/j.tpb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Breban R, Blower S. The reinfection threshold does not exist. J. Theor. Biol. 2005;235:151–152. doi: 10.1016/j.jtbi.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Galvani A.P, Bush R.M. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Cummings D.A.T, Cauchemez S, Fraser C, Riley S, Meeyai A, Iamsirithaworn S, Burke D.S. Strategies for containing an emerging influenza pandemic in southeast Asia. Nature. 2005;8:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- Girvan M, Callaway D, Newman M, Strogatz S. A simple model of epidemics with pathogen mutation. Phys. Rev. E. 2002;65:031915. doi: 10.1103/PhysRevE.65.031915. [DOI] [PubMed] [Google Scholar]
- Gog J.R, Grenfell B.T. Dynamics and selection of many-strain pathogens. Proc. Natl Acad. Sci. USA. 2002;99:17 209–17 214. doi: 10.1073/pnas.252512799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes M.G.M, Medley G.F, Nokes D.J. On the determinants of population structure in antigentically diverse pathogens. Proc. R. Soc. B. 2002;269:227–233. doi: 10.1098/rspb.2001.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes M.G.M, White L.J, Medley G.F. Infection, reinfection, and vaccination under suboptimal immune protection: epidemiological perspectives. J. Theor. Biol. 2004;228:539–549. doi: 10.1016/j.jtbi.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Gomes M.G.M, White L.J, Medley G.F. The reinfection threshold. J. Theor. Biol. 2005;236:111–113. doi: 10.1016/j.jtbi.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Grenfell B.T, Pybus O.G, Gog J.R, Wood J.L.N, Daly J.M, Mumford J.A, Holmes E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- Gupta S, Maiden M.C.J. Exploring the evolution of diversity in pathogen populations. Trends Microbiol. 2001;9:181–185. doi: 10.1016/S0966-842X(01)01986-2. [DOI] [PubMed] [Google Scholar]
- Heinen P, deBoer-Luijtze E.A, Bianchi T.J. Respiratory and systemic humoral and cellular immune of pigs to a heterosubtypic influenza A virus infection. J. Gen. Virol. 2001;82:2697–2707. doi: 10.1099/0022-1317-82-11-2697. [DOI] [PubMed] [Google Scholar]
- Levin S.A, Dushoff J, Plotkin J.B. Evolution and persistence of influenza A and other diseases. Math. Biosci. 2004;188:17–28. doi: 10.1016/j.mbs.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Liang S, Mozdzanowska K, Palladino G, Gerhard W. Heterosubtypic immunity to influenza type A in mice—effector mechanisms and their longevity. J. Immunol. 1994;152:1653–1661. [PubMed] [Google Scholar]
- Mills C.E, Robins J.M, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nåsell I. Stochastic models of some endemic infections. Math. Biosci. 2002;179:1–19. doi: 10.1016/S0025-5564(02)00098-6. [DOI] [PubMed] [Google Scholar]
- Nunes A, Telo da Gama M.M, Gomes M.G.M. Localised contacts between hosts reduce pathogen diversity. J. Theor. Biol. 2006;241:477–487. doi: 10.1016/j.jtbi.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Oxford J.S. Influenza A pandemics of the 20th century with special reference to 1918: virology, pathology and epidemiology. Rev. Med. Virol. 2000;10:119–133. doi: 10.1002/(SICI)1099-1654(200003/04)10:2%3C119::AID-RMV272%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Plotkin J.B, Dushoff J, Levin S.A. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc. Natl Acad. Sci. USA. 2002;99:6263–6268. doi: 10.1073/pnas.082110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw E. Cambridge University Press; Cambridge, UK: 1991. Modelling biological populations in space and time. [Google Scholar]
- Smith D.J, Lapedes A.S, de Jong J.C, Bestebroer T.M, Rimmelzwaan G.F, Osterhaus A.D, Fouchier R.A. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Yetter R.A, Barber W.H, Small P.A. Heterosubtypic immunity to influenza in ferrets. Infect. Immun. 1980;29:650–653. doi: 10.1128/iai.29.2.650-653.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]