Abstract
The reproduction number, R, defined as the average number of secondary cases generated by a primary case, is a crucial quantity for identifying the intensity of interventions required to control an epidemic. Current estimates of the reproduction number for seasonal influenza show wide variation and, in particular, uncertainty bounds for R for the pandemic strain from 1918 to 1919 have been obtained only in a few recent studies and are yet to be fully clarified. Here, we estimate R using daily case notifications during the autumn wave of the influenza pandemic (Spanish flu) in the city of San Francisco, California, from 1918 to 1919. In order to elucidate the effects from adopting different estimation approaches, four different methods are used: estimation of R using the early exponential-growth rate (Method 1), a simple susceptible–exposed–infectious–recovered (SEIR) model (Method 2), a more complex SEIR-type model that accounts for asymptomatic and hospitalized cases (Method 3), and a stochastic susceptible–infectious–removed (SIR) with Bayesian estimation (Method 4) that determines the effective reproduction number at a given time t. The first three methods fit the initial exponential-growth phase of the epidemic, which was explicitly determined by the goodness-of-fit test. Moreover, Method 3 was also fitted to the whole epidemic curve. Whereas the values of R obtained using the first three methods based on the initial growth phase were estimated to be 2.98 (95% confidence interval (CI): 2.73, 3.25), 2.38 (2.16, 2.60) and 2.20 (1.55, 2.84), the third method with the entire epidemic curve yielded a value of 3.53 (3.45, 3.62). This larger value could be an overestimate since the goodness-of-fit to the initial exponential phase worsened when we fitted the model to the entire epidemic curve, and because the model is established as an autonomous system without time-varying assumptions. These estimates were shown to be robust to parameter uncertainties, but the theoretical exponential-growth approximation (Method 1) shows wide uncertainty. Method 4 provided a maximum-likelihood effective reproduction number 2.10 (1.21, 2.95) using the first 17 epidemic days, which is consistent with estimates obtained from the other methods and an estimate of 2.36 (2.07, 2.65) for the entire autumn wave. We conclude that the reproduction number for pandemic influenza (Spanish flu) at the city level can be robustly assessed to lie in the range of 2.0–3.0, in broad agreement with previous estimates using distinct data.
Keywords: Spanish flu, pandemic, influenza, reproduction number, San Francisco
1. Introduction
The present study aims at assessing different approaches to the estimation of the transmissibility of the influenza pandemic of 1918–1919. To perform this comparison, we estimate epidemiological parameters for daily case notification (i.e. morbidity) time-series for the autumn wave of the 1918 influenza pandemic in the city of San Francisco, California using four different methods. These approaches include the estimation of the initial intrinsic growth rate of the epidemic followed by its substitution into a formula derived from the linearization of the deterministic epidemic model (e.g. Anderson & May 1991; Nowak et al. 1997; Lloyd 2001; Lipsitch 2003), trajectory matching (least-square fitting) of epidemic models to epidemic curve data (examples of recent work include Riley et al. 2003; Chowell et al. 2006) and sequential Bayesian inference to estimate the effective reproduction number at a given time t, from a stochastic formulation of a SIR model (Bettencourt & Ribeiro 2006).
The presence of the highly pathogenic A/(H5N1) influenza virus in avian populations in several regions of the world has highlighted the urgent need to prepare for the next influenza pandemic. While the great majority of transmission events (236 confirmed human cases as of 9 August 2006 (The World Health Organization 2006)) have resulted from direct contact with birds, a limited number of human-to-human transmission events have been reported as probable (Ungchusak et al. 2005). Should this virus become adapted for efficient human-to-human transmission, an influenza pandemic could develop with devastating consequences.
Genetic drift in viral populations leads to annual seasonal epidemics of influenza worldwide (Webster et al. 1992). Much more rarely, major changes in the influenza virus antigenic structure (genetic shifts) have the potential to cause major pandemics, which are associated with high morbidity and mortality rates because the population is immunologically naive to the new pathogen. The 1918 influenza pandemic (Spanish flu) has been the most devastating among these in recent history, with a death toll estimated at over 20 million worldwide (Johnson & Mueller 2002). The 1918–1919 pandemic strain probably originated from an avian virus that adapted its tropism to humans (Taubenberger et al. 2005), but this conclusion is currently under debate (Antonovics et al. 2006; Gibbs & Gibbs 2006).
In the advent of a next influenza pandemic, the accurate and early estimation of the number of secondary cases generated by a primary infectious case (known as the reproduction number) is of high priority for public health management. The reproduction number associated with the pandemic provides a measure of the intensity of interventions required to achieve control. In the context of a completely susceptible population, this quantity is referred to as the basic reproduction number and denoted by (Anderson & May 1991). When a fraction p of the population is effectively protected from infection, this quantity is known as the reproduction number (and often denoted by R) and is related to by , assuming a well-mixed population (Diekmann & Heesterbeek 2000). For the case of pandemics we can expect .
Parameter estimations of the epidemiology of influenza have been of great concern to modellers for sometime (Longini et al. 1982, 1984; Cauchemez et al. 2004). The evaluation of potential intervention strategies using detailed mathematical frameworks has become an important tool towards mitigating future outbreaks in different parts of the world (Flahault et al. 1988; Longini & Halloran 2005; Longini et al. 2005; Ferguson et al. 2006), but evaluation of these actions suffers at present from uncertainty resulting from the scarcity of empirical estimates obtained from past pandemics. In addition, to date, only a small number of estimates exist for the reproduction number of the pandemic strain that circulated during 1918–1919 (Mills et al. 2004; Gani et al. 2005; Chowell et al. 2006; Bettencourt & Ribeiro 2006), and these were achieved via different dynamical models and estimation procedures, as well as over distinct datasets, organized at different levels of temporal and regional aggregation. As a consequence, there is still insufficient information to fully clarify the transmission dynamics of the great 1918–1919 pandemic. In addition, previously suggested values of R for seasonal influenza varied widely with some studies assuming (Dushoff et al. 2004) and (Gog et al. 2003), while others argue that it should only be slightly above unity (Gani et al. 2005). Different methods and assumptions as well as the absence of critical analyses regarding the robustness and validity of these estimates have contributed to this large uncertainty, which has lead to substantial confusion, even among specialists (Koopman 2004). This situation is owing, at least to a large extent, to the limited amount and type of available data, so that few estimates from incidence time-series have been performed to date. Indeed, the sources of information for the 1918 pandemic influenza completely differed from one study to the next. Moreover, since the available epidemiological information is not sufficient to validate a detailed (e.g. agent-based) model for the transmission of pandemic influenza, estimation and analysis procedures must rely on simpler methods within broader model assumptions (Arino et al. 2006). Here, we explore several of these methods and associated parameter estimation procedures to help settle the uncertainty bounds on R for San Francisco in 1918–1919.
2. Materials and methods
2.1 Historical background
The 1918 influenza pandemic known as the ‘Spanish flu’ was caused by the influenza virus A (H1N1). In San Francisco, California (United States), 28 310 cases including 1908 deaths were reported during the autumn wave (September–November) comprising 63 epidemic days, giving a case fatality of 6.7%. The city of San Francisco, California is located on the tip of the San Francisco Peninsula and covers an area of 121 km2. In 1918, the city of San Francisco had an approximate population of 550 000 (Crosby 2003), which is about 74% of today's population. As judged from an analysis of the records of the San Francisco hospital (Hrenoff 1941), the 1918 pandemic affected all ages, sexes and races. Clinical symptoms included severe headache, prostration, muscle and joint pain, rapidly rising fever and chills, and general malaise. Other less characteristic manifestations of influenza included epistaxis, sore throat, cough, rhinitis, laryngitis, gastro-enteric upsets and leucopenia (Hrenoff 1941). When followed by pneumonia, influenza was potentially more lethal (Vaughn 1921). Generally, influenza spreads very quickly owing to the short incubation period and, consequently, the short serial interval (the sum of the mean latent period and the mean duration of infectiousness) of about 3–6 days (Khakpour et al. 1969; Kilbourne 1977).
Control measures implemented during the pandemic included education campaigns on prevention, isolation, face mask use and prohibition of public events, but there is no quantitative evidence on their effectiveness (Hrenoff 1941). For instance, mask use as a preventive measure was much criticized owing to the lack of general adoption (Capps 1918). The effectiveness of these campaigns was publicly debated at the time as, for example, 78% of the nurses at the San Francisco Hospital contracted influenza, although this facility was considered to have one of the best isolation services in this city. Consequently, public announcements were run in local newspapers calling for volunteers to help in overburdened hospitals (Hrenoff 1941), which may have increased transmission opportunities. Neither an influenza vaccine nor antiviral medications were available at the time.
2.2 Data sources
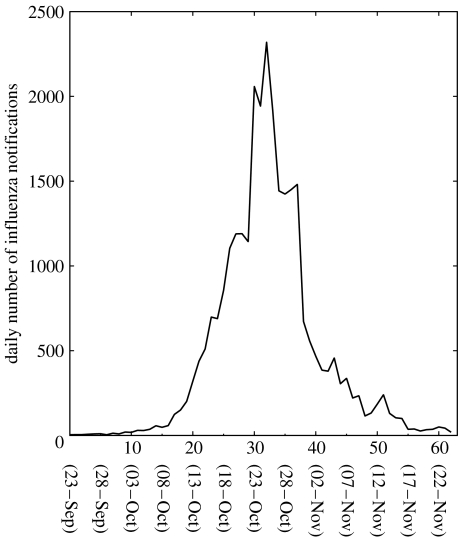
Daily epidemic data for the autumn influenza wave (September–November) in the city of San Francisco, California were obtained from public records as reported to the city health department (Department of Hygiene 1922; figure 1). Since the health department was aware of the rapidly spreading pandemic influenza in the United States before the autumn wave started in San Francisco, epidemic data were critically inspected and are believed to have been recorded rather precisely (Hrenoff 1941). Nevertheless, levels of underreporting (or overreporting once the epidemic was well publicized) are unknown quantitatively. We adopted the date of the first reported (index) case—23 September, 1918—as the starting date of the epidemic. See electronic supplementary material for the original data.
Figure 1.
Daily number of influenza notifications in San Francisco, California during the 1918–1919 influenza pandemic (Department of Hygiene 1922).
The total notified case fatality proportion (CFP) of the 1918 autumn pandemic wave in the city of San Francisco was 6.7% (Department of Hygiene 1922). The mortality from influenza in the San Francisco hospital (26%) was much greater than for the city as a whole owing to the large number of patients who were brought to the hospital in the final stages of disease progression, often with pneumonia as a complication (Hrenoff 1941).
2.3 Estimation of the reproduction number
(i) Method 1: estimating R from the intrinsic growth rate
The reproduction number is typically estimated from the early epidemic phase, during which the epidemic runs its free course in the absence of interventions and effects of susceptible depletion are small. To this end, it is common to assume an initial exponential-growth phase, which is characteristic of most human infectious diseases (Anderson & May 1991). Thus, one of the most common approaches to computing the reproduction number consists of estimating first the initial exponential-growth rate (r) for the cumulative number of cases by fitting a straight line b0+rt to the ‘best’ length of its exponential phase (in logarithmic scale), which can be determined by the Χ2 goodness-of-fit test statistic (Favier et al. 2006). The reproduction number is then computed by substituting the estimate for r into an expression derived from the linearization of the susceptible–exposed–infectious–removed (SEIR) deterministic epidemic model (Lipsitch 2003) and is given by
| (2.1) |
where V is the mean serial interval; and f is the ratio of the mean infectious period to the mean serial interval.
(ii) Method 2: estimating R from a simple susceptible–exposed–infectious–removed model
We use an epidemic model of SEIR-type that classifies individuals as susceptible (S), exposed (E), infectious (I), recovered (R) and dead (D) (Anderson & May 1991). Susceptible individuals in contact with the virus enter the exposed class at the rate βI(t)/N, where β is the transmission rate; I(t) is the number of infectious individuals at time t; and N(t)=S(t)+E(t)+I(t)+R(t) is the total population at time t. The entire population is assumed to be susceptible at the beginning of the epidemic. Latent individuals (E) progress to the infectious class at the rate k (1/k is the mean latent period). We assume homogeneous mixing between individuals and, therefore, the fraction I(t)/N(t) is the probability of a random contact with an infectious individual in a population of size N(t). Since we assume that the time-scale of the epidemic is much faster than characteristic times for demographic processes (natural birth and death), these effect are not included in the model. Infectious individuals either recover or die from influenza at the mean rates γ and δ, respectively. Recovered individuals are assumed protected for the duration of the outbreak. The mortality rate is given by δ=γ[CFP/(1−CFP)], where CFP is the mean case fatality proportion. The transmission process can be modelled using the following system of nonlinear differential equations:
| (2.2) |
where the dot denotes time derivatives, and C(t) is the cumulative number of case notifications.
We use least-square fitting to look for the model trajectory that best matches the epidemic time series. Specifically, we fit the cumulative number of cases given by equation C(t) to the cumulative number of case notifications. We implemented a least-square fitting procedure in Matlab (The Mathworks Inc.) using the built-in routine lsqcurvefit in the optimization toolbox. The latent period was fixed to 1/k=1.9 days and the recovery period was set to 4.1 days, as in previous studies (Mills et al. 2004). We then estimate the transmission rate β and the initial number of exposed and infectious individuals, assuming E(0)=I(0). The basic reproduction number is given by the product of the mean transmission rate and the mean infectious period, .
(iii) Method 3: estimating R using a complex susceptible–exposed–infectious–removed model
We apply this method to estimate the reproduction numbers from two different sets of data: (i) exponential-growth phase (i.e. as in Methods 1 and 2); and (ii) model fit to the entire epidemic curve.
Our complex SEIR model was developed originally for studying the transmissibility and the effect of hypothetical interventions for the 1918 influenza pandemic in Geneva, Switzerland (Chowell et al. 2006). In this model, individuals are classified as susceptible (S), exposed (E), clinically ill and infectious (I), asymptomatic and partially infectious (A), diagnosed and reported (J), recovered (R) and dead (D). The birth and natural death rates are assumed to have a common rate μ (60-year life expectancy as in Chowell et al. 2006). The entire population is assumed susceptible at the beginning of the pandemic wave. Susceptible individuals in contact with the virus progress to the latent class at the rate β(I(t)+J(t)+qA(t)/N(t)), where β is the transmission rate, and 0<q<1 is a reduction factor in the transmissibility of the asymptomatic class (A). Since there is no evidence for the effectiveness of interventions, and a high burden was placed upon the sanitary and medical sectors, diagnosed/hospitalized individuals (J) are assumed equally infectious. Although it is difficult to explicitly evaluate the difference in infectiousness between the general community and hospital, we roughly made this assumption as 78% of the nurses of the San Francisco Hospital contracted influenza (Hrenoff 1941). A more rigorous assumption requires either statistical analysis of more detailed time-series data (Cooper & Lipsitch 2004) or an epidemiological comparison of specific groups by contact frequency (Nishiura et al. 2005). The total population size at time t is given by N(t)=S(t)+E(t)+I(t)+A(t)+J(t)+R(t). We assumed homogeneous mixing of the population and, therefore, the fraction (I(t)+J(t)+qA(t))/N(t) is the probability of a random contact with an infectious individual. A proportion 0<ρ<1 of latent individuals progress to the clinically infectious class (I) at the rate k, while the remaining (1−ρ) progress to the asymptomatic partially infectious class (A) at the same rate k (fixed to 1 per 1.9 days Mills et al. 2004). Asymptomatic cases progress to the recovered class at the rate γ1. Clinically infectious individuals (class I) are diagnosed (reported) at the rate α or recover without being diagnosed (e.g. mild infections, hospital refusals) at the rate γ1. Diagnosed individuals recover at the rate γ2=1/(1/γ1−1/α) or die at rate δ. The mortality rates were adjusted according to the CFP, such that δ=[CFP/(1−CFP)](μ+γ2).
The transmission process can be modelled using the following system of nonlinear differential equations:
| (2.3) |
The cumulative number of influenza notifications, our observed epidemic data, is given by C(t). Seven model parameters (β, γ1, α, q, ρ, E(0) and I(0)) are estimated from the epidemic curve using least-square fitting (as in Method 2). The reproduction number for model (2.3) is given by (Chowell et al. 2006)
| (2.4) |
and the clinical reporting proportion is given by
| (2.5) |
(iv) Method 4: estimating using Bayesian inference of stochastic SIR
As a final method, we use a stochastic version of a standard SIR model. This method estimates the effective reproduction number, , defined as the actual average number of secondary cases per primary case at time t (for t>0) (Haydon et al. 2003; Wallinga & Teunis 2004; Nishiura et al. 2006) and is typically less than R0. Precise estimates of are of importance for outbreak evaluation and management; shows time-dependent variation with the decline in susceptible individuals (intrinsic factors) and with the implementation of control measures (extrinsic factors). It may also increase over time owing to changes in population structure or pathogen evolution.
Such formulation, as we show briefly below (see also Bettencourt & Ribeiro 2006), takes into account the probabilistic nature of contagion processes and allows direct estimation of the probability distribution of the effective reproduction number , from real-time data, without the need for parameter search and optimization as in Methods 1–3. In this sense, the four methods address the problem of modelling and estimation in complementary ways. To see this, consider a standard SIR model (a version of an SEIR model can be formulated, but is more complex), such that on average
| (2.6) |
and R and D classes receive progressed infections in the same manner as the simple SEIR described above and were thus omitted here for simplicity. The stochastic version of the model is formulated as usual by taking the rates on the right-hand side of the population equations to determine the mean change λ over the time τ of the several population classes, which is in practice extracted from a probability distribution P[λ] with average λ. In the estimation procedure described below, P is taken to be a Poisson distribution, which is the maximal entropy distribution for a discrete process for which only the average is known. If information is also available about the statistics of fluctuations, a more general distribution, such as a Negative Binomial, can be employed instead.
Epidemiological reports are given usually, not in terms of infectious individuals but rather as a tally of cases, which at the time of reporting may have progressed. Thus, it is advantageous to write our estimation procedure in terms of the change in the cumulative number of cases C(t). New cases at time t are given in terms of the increase in cumulative case numbers as ΔC(t)=C(t)−C(t−τ), where τ denotes the time-interval between successive reports and may vary over time. In our dataset, τ=1 day. Note that C(t)=I(t)+R(t)+D(t) and, consequently, equation (2.6) implies . It follows from this relation and from integrating the dynamical equation for I(t) in (2.6) that the relation between the average change in case numbers between two consecutive periods is
| (2.7) |
where we used and , and for the SIR model. The approximate equality here assumes that S(t)/N(t) remains approximately constant over the period τ, but may vary across successive periods. Given that τ=1 day and that the susceptible population is much larger than the number of infected per day, especially in the beginning of the outbreak, this is usually an excellent approximation. Note that these relations imply in turn that .
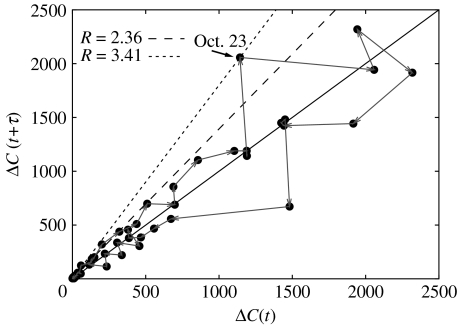
Now, recall that relation (2.7) holds only on the average. However, if fluctuations are small compared with the mean, then the effective reproduction number can be estimated directly from a new case time-delay diagram (i.e. a plot of ΔC(t+τ) versus ΔC(t)), without any more complex estimation, as shown in figure 2. Specifically, relation (2.7) implies that is the slope of the tangent at the origin in this case time-delay diagram trajectory (grey line in figure 2). This trajectory eventually crosses the line with slope unity as susceptibles are depleted and becomes less than one. Such plots also help to provide an intuition about the magnitude of case fluctuations, and identify time periods when cases may have jumped, signalling changes in the population structure, effects of control interventions, pathogen characteristics or, more probably, artefacts in the reporting. We will return to these points in §4.
Figure 2.
The course of the outbreak can be visualized in an epidemic time-delay diagram of new cases ΔC at consecutive times (black dots). For data that are not too stochastic, this provides a very simple method to estimate R, via the tangent at the origin (dashed lines) of the initial growth trajectory (grey arrows). Jumps in case numbers (indicated for 22–23 Oct) lead to greater uncertainty in the estimation of the reproduction number.
In general, the probabilistic formulation of the model implies that, given (and other parameters such as γ) and ΔC(t), we can predict the distribution of future case numbers as
| (2.8) |
The probabilistic formulation for future cases is equivalent, via Bayes' theorem, to the estimation of the probability distribution for , viz.
| (2.9) |
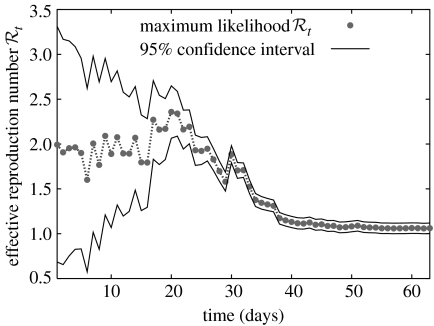
where is the prior, which reflects any a priori knowledge of the distribution of (or can be given by a uniform distribution otherwise); and the denominator is a normalization factor. Thus, knowledge of two or more new case reports, and the adoption of a probabilistic contagion model, results, via Bayes' theorem, in the estimation of the probability distribution function for , as the posterior. This estimation scheme is then iterated, whereby the probability distribution for from previous reports, the posterior at time t, is used as the prior for new cases, at t+τ. From these successive distributions, maximum-likelihood (the value corresponding to the probability maximum) estimates or averages are read out, as well as bounds corresponding to desired levels of confidence. Since successive case reports improve the estimation in this iterative Bayesian scheme by reducing uncertainty and simultaneously tends to decrease owing to depletion of susceptibles, we associate the maximum of with the best estimator for R (figure 6).
Figure 6.
Sequential Bayesian estimation of the full distribution of leads to the estimation of its maximum-likelihood value (grey dots) and 95% CIs (solid black lines). Uncertainty, measured by the width of the CI, decreases with more case observations. The estimates eventually lead to smaller owing to depletion of susceptibles. At late times, as a result of averaging periods in which the epidemic grows and declines.
This class of method becomes particularly useful for estimation of when the data are very stochastic, such as for emerging infectious diseases, and for sequential estimation in real time, as data stream in. As a disadvantage, it does not estimate R directly but rather its effective value resulting from the convolution of R with the population fraction of susceptibles, which varies over time. Other applications of this method to time-series for H5N1 avian influenza in humans, and to other seasonal and pandemic datasets, are given by Bettencourt & Ribeiro (2006).
2.4 Quantifying parameter uncertainty
Confidence intervals for R estimates were constructed for Methods 2 and 3 by generating sets of realizations of the best-fit curve C(t) using parametric bootstrap (Efron & Tibshirani 1986). Each realization of the cumulative number of case notifications Ci(t) (i=1, 2, …, m) is generated as follows: for each observation C(t) for t=2, 3, …, n days generate a new observation for t≥2 () that is sampled from a Poisson distribution with mean C(t)−C(t−1) (the daily increment in C(t) from day t−1 to day t). The corresponding realization of the cumulative number of influenza notifications is given by , where t=1, 2, 3, …, n. The reproduction number was then estimated from each of 1000 simulated epidemic curves. The distribution of estimated reproduction numbers can be used to construct 95% CIs. For Method 3, fitting a complex model (with seven parameters in this case) comes at the cost of increased potential variation for these estimates. Difficulties with the fitting procedure occur if the model cannot be uniquely determined from the data leading to unbounded variances for the estimated parameters. This simulation study allowed us to explore the identifiability of model parameters. Lack of identifiability can be recognized when large perturbations in the model parameters generate small changes in the model output (Pillonetto et al. 2003). Our results indicate that our parameter estimates are stable to perturbations around the model output.
For the case of Method 4, uncertainty bounds for the effective reproduction number are obtained directly from the probabilistic nature of the model for future cases, which is transformed, given a case time series, via Bayes' theorem, into the probability distribution of . Average and maximum-likelihood values for are extracted from such distributions, as well as bounds on at 95% confidence intervals. In the results shown in figure 6 and table 1, we started the estimation at the initial time, with a Gaussian prior for with average and variance , which is fairly unbiased in the expected range for R and is characterized by a 95% CI of [0, 4]. As indicated above, the distribution for at subsequent times uses the posterior at the previous time as prior, thus incorporating the time-series up to that time in the estimation.
Table 1.
Estimates of the reproduction number for the autumn wave of the Spanish flu pandemic in San Francisco, California. n.a., not applicable. The number of data points is smaller than the number of parameters being estimated (seven parameters for the complex SEIR model). Note that the stochastic SIR method provides the effective reproduction number at time t, while the other methods estimate the reproduction number by fitting the models to a specified number of epidemic days of data. The number of degrees of freedom (d.f.) is different by method. Initial growth rate, simple SEIR and complex SEIR estimate 1, 2 and 7 parameters, respectively. d.f. was determined by the difference between the observed number of epidemic days, n, and the number of parameters to be estimated (e.g. for the complex SEIR, d.f. at 17 days was n−7=10).
| initial growth rate | simple SEIR | complex SEIR | stochastic SIR | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| epidemic days | R | R 95% CI | R | R 95% CI | R | R 95% CI | 95% CI | |
| 5 | 5.78 | (3.80, 8.15) | 3.72 | (2.01, 5.44) | n.a. | n.a. | 1.96 | (0.83, 3.09) |
| 17 | 2.98 | (2.73, 3.25) | 2.38 | (2.16, 2.60) | 2.20 | (1.55, 2.84) | 2.10 | (1.21, 2.95) |
3. Results
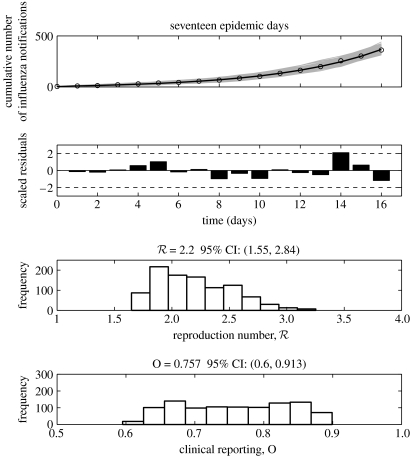
We estimated the reproduction number for the autumn wave of the Spanish flu pandemic in San Francisco, California from daily case reports using four different methods. While Methods 2 (simple SEIR) and 3 (complex SEIR) suggested a 17-day duration as the best length of the initial exponential-growth phase (figure 3), Method 1 (a pure exponential-growth approximation) indicated a 5-day duration as the best length of exponential growth based on the goodness-of-fit. The estimates of the reproduction number obtained from the four methods were found to be consistent with each other (in the range , with overlapping CIs) when using an initial epidemic phase comprising 17 days (table 1 and figures 4–6). Although we also explored the goodness-of-fit statistic for the remaining epidemic days, there were no other clear candidates for the cut-off (e.g. there was no interval which suggests a second minimum of the goodness-of-fit statistic). However, Method 1 (with a 5-day exponential phase) yielded an estimate of the reproduction number significantly larger than those obtained from the other methods (table 1), and associated with very large uncertainty. Method 4 estimated the effective reproduction number to be 2.10 (95% CI: 1.21, 2.95) by using the first 17 epidemic days and 2.36 (2.07, 2.65) including the entire fall wave (maximum effective in figure 6).
Figure 3.
The Χ2 density provided by three different models (Methods 1–3) to the initial epidemic growth phase of the cumulative number of influenza notifications as a function of the length of the initial epidemic phase. Using the goodness-of-fit statistic, the initial growth phase is predicted to be 5 days by Method 1 and 17 days by Methods 2 and 3.
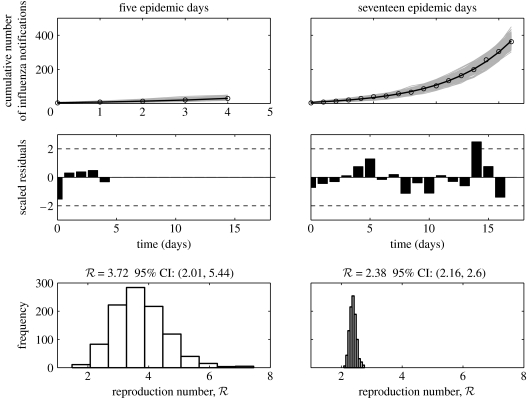
Figure 4.
Model fits, residuals plots and the resulting distributions of the reproduction number obtained after fitting the simple SEIR epidemic model (Method 2) to the initial phase of the autumn influenza wave using 5 and 17 epidemic days of the Spanish flu pandemic in San Francisco, California. In the top panel, the epidemic data of the cumulative number of reported influenza cases are the circles, the solid line is the model best-fit and the solid grey lines are 1000 realizations of the model fit to the data obtained through parametric bootstrapping as explained in the text.
Figure 5.
Model fits, residuals plots and the resulting distributions of the reproduction number and the proportion of the clinical reporting obtained after fitting the complex SEIR epidemic model (Method 3) to the initial phase of the autumn influenza wave using 17 epidemic days of the Spanish flu pandemic in San Francisco, California. In the top panel, the epidemic data of the cumulative number of reported influenza cases are the circles, the solid line is the model best-fit and the solid grey lines are 1000 realizations of the model fit to the data obtained through parametric bootstrapping as explained in the text.
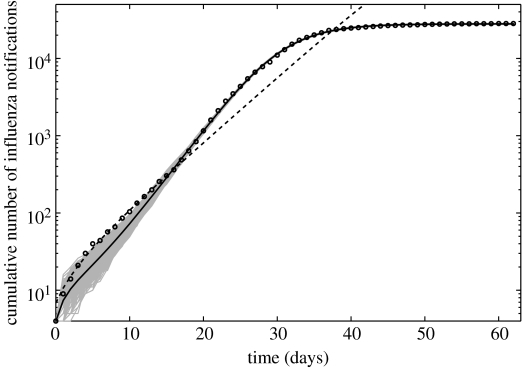
While the simple SEIR model was unable to describe the entire epidemic course, the complex SEIR fitted reasonably well the entire pandemic curve (63 epidemic days) with a clinical reporting percentage of 55.5% (95% CI: 52.1–58.8) and a reproduction number (95% CI: 3.45–3.62), which is higher than that obtained using 17 epidemic days (2.20 (95% CI: 1.55–2.84)). However, a closer look at the complex SEIR model fit to the entire pandemic wave reveals a systematic deviation from case numbers for the initial epidemic phase (figure 7). This effect is owing to features of the data, which show in later periods two 1-day large increments in case numbers, which lead to larger estimate of R, as also suggested by Method 4. Accommodating these features together with the initial growth phase, in a model with fixed parameters in time, leads to the higher expected value of the reproduction number. Nevertheless, we note that the CI for the estimate in this period obtained via Method 4 overlap with the estimate obtained for the early period, primarily owing to the larger uncertainty associated with the R estimate obtained at day 17 (table 1). These points are further discussed in §4.
Figure 7.
The complex SEIR model fit (solid line) to the entire epidemic curve (circles) and the simple SEIR model fit (dashed line) calibrated using the initial exponential phase (17 days) are shown for comparison (cumulative cases are shown in logarithmic scale). Solid grey lines are 1000 realizations of the complex SEIR model fit to the data obtained through parametric bootstrapping as explained in the text.
4. Discussion
We used four distinct approaches to model the progression of pandemic influenza in the city of San Francisco, California, in 1918–1919, measured by daily case reports, and estimate the corresponding reproduction number. The first three methods were used to obtain R estimates by fitting the model solutions to an early exponential-growth phase. The complex SEIR (Method 3) and stochastic SIR (Method 4) models were also used to obtain an estimate of the reproduction number from the entire epidemic curve. The fourth method assumes an underlying probabilistic epidemic model (while the former three are purely deterministic) and estimates the effective reproduction number via a Bayesian data assimilation scheme of the case time-series. This approach leads to the estimation of the probability distribution of , which is successively improved (in the sense of uncertainty reduction) as each new report streams in, potentially in real time. Nevertheless, the omission of a short latency period into the SIR framework could potentially slightly underestimate the reproduction number (Wearing et al. 2005). The four methods presented here provided estimates in the range R=2–3 that are in good agreement with each other for data from the initial epidemic phase, which was explicitly determined by using the goodness-of-fit test statistic (Favier et al. 2006). There are several important messages arising from our analysis.
First, the mean R estimate derived from the initial intrinsic growth rate (Method 1) using the first 17 epidemic days was found to be slightly higher (i.e. approx. ) than mean estimates derived from all other methods ( and 2.2 from the simple and the complex SEIR, respectively, and from the stochastic SIR method). This discrepancy may be partly attributable to the assumption incorporating the depletion of susceptible individuals in Methods 2–4, which decreases the estimate of R. Indeed, the goodness-of-fit obtained using equation (2.1) with two fitting parameters was always worse than that obtained from Methods 2 (two fitting parameters) and 3 (seven fitting parameters). R estimates obtained using 17 epidemic days appeared to be robust to parameter uncertainties (figures 4 and 5) and to slightly different assumptions and initial conditions (e.g. estimation of three parameters: β, E(0) and I(0); details not shown). However, when we took 5 days as the length of the exponential phase (as predicted by Method 1), our R estimates differed substantially from one another. This may imply that 17 days was a more appropriate cut-off point for the exponential phase, although it was not possible to explicitly identify a unique length of the initial epidemic phase from either of these two possibilities. Since assuming a simple exponential-growth phase at the initial epidemic phase (Method 1) relies on a theoretical approximation, it is difficult for this simple method to always be excellent (Heffernan et al. 2005). Moreover, a weakness of the assumption on the exponential growth of cases was criticized during the epidemic of severe acute respiratory syndrome (SARS) (Razum et al. 2003). The clinical features of influenza further complicate the interpretation of case notifications owing to potential substantial underreporting and large numbers of asymptomatic infections (Cauchemez et al. 2006; Glass et al. in press). As a general recommendation, our study suggests that Method 1, assuming the theoretical exponential-growth approximation, should be used only with careful consideration of the data and firm understanding of the underlying assumptions.
Second, we found some qualitative differences associated with the intrinsic and extrinsic dynamics in the simple and the complex SEIR models (Methods 2 and 3). While the R estimates from the initial epidemic phase were similar for the two models, the simple SEIR model was unable to describe the course of the entire autumn pandemic wave (using the R estimate based on the exponential-growth fit). This inability may be attributable to both (i) intrinsic dynamical factors linked to the epidemiology of influenza (e.g. asymptomatic infection, mortality rate), and (ii) its extrinsic dynamics which are the result of human intervention (e.g. diagnostic rate, isolation of infectious individuals in hospital settings and behaviour changes among susceptible individuals to avoid potential contacts). On the other hand, the complex SEIR model, even using the obtained R estimate from the exponential phase, reasonably realized the observed shape and scale of the entire epidemic curve. This might be also problematic from a modelling perspective, in particular, for a model based on an autonomous system (i.e. the system without time-varying assumptions). Time-varying extrinsic dynamics, which cannot be discarded during the Spanish flu, were not explicitly incorporated into the complex SEIR model. For instance, implicit time-varying parameters were the base of several models for SARS (Chowell et al. 2003; Massad et al. 2005; Hsieh & Cheng 2006). Moreover, it should be remembered that the intrinsic parameters are likely to vary during the course of an epidemic (e.g. the serial interval was shortened with time during the SARS epidemic (Lipsitch 2003)). Systematic consideration of the processes that may lead to time-varying parameters remains an open question in studies of pandemic influenza, which we reserve for future research.
Third, estimates of R obtained from the complex SEIR model were found to be sensitive to the number of epidemic days adopted in the estimation. Specifically, the complex SEIR model when fitted to the entire pandemic wave (as in Chowell et al. 2006) using the Spanish flu pandemic in Geneva) yielded a higher R than that obtained when the same model was fitted to the exponential phase only. This difference in the R estimates can be explained by examining the residual plot obtained from the fit of the complex SEIR model to the entire epidemic curve. Specifically, the goodness-of-fit of the model to the initial exponential phase worsened compared with the goodness-of-fit obtained when the same model was fitted to the initial exponential phase only (figure 7).
Fourth, the type of data employed in this study is likely to become available when the next pandemic arrives. Thus, it is worth pointing out the lessons learned from these data analyses. First, we note that the data of the pandemic in San Francisco used here are based on the daily case notification, which is different from other modelling studies of pandemic influenza (Mills et al. 2004; Gani et al. 2005), where data were aggregated over longer time periods (e.g. a week). Daily reporting data are characterized by smaller numbers and are thus generally more sensitive, in relative terms, to changes in reporting rates and population behaviour. For instance, a dramatic increase in incidence from 1143 to 2058 occurred from 22 to 23 October, which has a direct effect on the uncertainty of the reproduction number estimates. This jump in incidence may have resulted from reaction to official announcements before and during the preceding weekend, possibly leading to an increase in the reporting rate in the beginning of the week, which most probably coincided with peak of the growth of cases. In fact, during 22–23 October, alarm may have influenced population behaviour (On 18 October, the Board of Health declared the situation as ‘grave’ leading to closures of public places including schools and churches) (Crosby 2003). Moreover, on 22 October, a full-page ad appeared in the Chronicle in which the Mayor, Board of Health, Red Cross, Postal Department, Chamber of Commerce, Labour Council and other organizations proclaimed, ‘wear a mask and save your life!’ ‘A gauze mask is ninety nine percent proof against influenza’ (Crosby 2003). This jump in incidence over 1 day is a major source of uncertainty in estimating R, which can be readily visualized from a time-delay diagram of new cases at consecutive days (figure 2). To illustrate this point quantitatively, consider that Method 4 provides a maximum-likelihood estimator for , given any two consecutive case reports, as
| (4.1) |
which between 22 and 23 of October gives a mean effective reproduction number of 3.41. In estimating R from cumulative data, or indeed via a Bayesian method without a narrow prior, the effect of this jump in case reports leads to a substantial increase in the estimates, explaining why fits to the entire curve, via Methods 3 and 4, result in larger values for the reproduction number.
Fifth, an important challenge in epidemic modelling lies in the realistic representation of features of disease spread. One of the most important features of the transmission dynamics of influenza might be asymptomatic infection and underreporting. (Thus, the complex SEIR model originally assumed an elaborate structure to comply with these characteristics.) However, when dealing with data characterized by random missing observations, statistical approaches with an explicit assumption of missing data may more accurately estimate the parameters of interest (Cauchemez et al. 2006; Glass et al. in press). Thus, a combination of deterministic models and statistical methods is desirable to model real-time noisy data and should be required in future studies. Further, it should be noted that the interpretation of the estimates of the reproduction number using classical epidemic models that assume homogeneous mixing is probably one of the most delicate tasks. For example, it is worth noting that even Method 4 required a random-mixing assumption. Whereas this might be a disadvantage of this method, compared with the use of the serial interval distribution (Wallinga & Teunis 2004) which assumed independence of transmission events, the serial interval distribution of pandemic influenza is unavailable today. (Instead, Method 4 yields an explicit distribution of by using Bayesian estimation.) Recent studies have explored the role of heterogeneous contact networks (Meyers et al. 2006), and some researchers suggest that an appropriate estimate of the reproduction number is not feasible without explicit information about the structure of contacts (Breban et al. 2005). However, modellers have so far not succeeded in estimating the transmission potential of droplet infections with explicit contact structures, because the contact is obviously very difficult to measure and quantify. In particular, when we deal with the issue of Spanish influenza, the estimation must be performed based on very limited information, which was originally collected without consideration for their utility for quantitative estimation.
In conclusion, we produced estimates of the reproduction number for pandemic influenza using four different methods and analysed their advantages and disadvantages, given daily reporting data for the city of San Francisco. The exponential-growth assumption (Method 1) may be reasonable and simple, but we have to keep in mind that the assumption tends to be statistically flawed. Whereas further methodological improvements and empirical information are needed to further clarify the reproduction number for Spanish influenza, our analysis indicates that its reproduction number, aggregated at the level of San Francisco, lies in the range of 2.0–3.0. While our estimates are broadly consistent with previous values derived by fitting epidemic models to mortality and morbidity time-series data of the 1918 flu pandemic (Mills et al. 2004; Gani et al. 2005; Chowell et al. 2006), values of the reproduction number for seasonal influenza derived from indirect estimates are, in some cases, one order of magnitude higher (Gog et al. 2003; Dushoff et al. 2004). Our estimates of the reproduction number for pandemic influenza strongly suggest a tighter range of uncertainty than has previously been assumed, as well as targets for public health interventions in the case of future similar pandemics that, while very challenging, may not be impossible to tackle.
Acknowledgments
G.C. was supported by a Director's Postdoctoral Fellowship from Los Alamos National Laboratory. H.N. received financial support from the Banyu Life Science Foundation International. L.M.A.B. was supported by the Laboratory Directed Research and Development Program at LANL, and thanks R. Ribeiro for discussions and collaboration on Method 4.
Supplementary Material
References
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans. [Google Scholar]
- Antonovics J, Hood M.E, Howell Baker C. Molecular virology: was the 1918 flu avian in origin? Nature. 2006;440:E9. doi: 10.1038/nature04824. [DOI] [PubMed] [Google Scholar]
- Arino J, Brauer F, van den Driessche P, Watmough J, Wu J. Simple models for containment of a pandemic. J. R. Soc. Interface. 2006;3:453–457. doi: 10.1098/rsif.2006.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt, L. M. A. & Ribeiro, R. M. Submitted. Detecting early human transmission of H5N1 avian influenza. Proc. Natl Acad. Sci. USA.
- Breban R, Vardavas R, Blower S. Linking population-level models with growing networks: a class of epidemic models. Phys. Rev. E. 2005;72:046110. doi: 10.1103/PhysRevE.72.046110. [DOI] [PubMed] [Google Scholar]
- Capps J.A. Measures for the prevention and control of respiratory infections in military camps. JAMA. 1918;71:448–450. [Google Scholar]
- Cauchemez S, Carrat F, Viboud C, Valleron A.J, Boelle P.Y. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat. Med. 2004;23:3469–3487. doi: 10.1002/sim.1912. [DOI] [PubMed] [Google Scholar]
- Cauchemez S, Boelle P.Y, Donnelly C.A, Ferguson N.M, Thomas G, Leung G.M, Hedley A.J, Anderson R.M, Valleron A.J. Real-time estimates in early detection of SARS. Emerg. Infect. Dis. 2006;12:110–113. doi: 10.3201/eid1201.050593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell G, Fenimore P.W, Castillo-Garsow M.A, Castillo-Chavez C. SARS Outbreaks in Ontario, Hong Kong and Singapore: the role of diagnosis and isolation as a control mechanism. J. Theor. Biol. 2003;24:1–8. doi: 10.1016/S0022-5193(03)00228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell G, Ammon C.E, Hengartner N.W, Hyman J.M. Transmission dynamics of the great influenza pandemic of 1918 in Geneva, Switzerland: assessing the effects of hypothetical interventions. J. Theor. Biol. 2006;241:193–204. doi: 10.1016/j.jtbi.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Cooper B, Lipsitch M. The analysis of hospital infection data using hidden Markov models. Biostatistics. 2004;5:223–237. doi: 10.1093/biostatistics/5.2.223. [DOI] [PubMed] [Google Scholar]
- Crosby A.W. America's Forgotten Pandemic. The Influenza of 1918. 2nd edn. Cambridge University Press; Cambridge, UK: 2003. The second and third wave. Part III. Chapter 7. Flu in San Francisco; pp. 91–120. [Google Scholar]
- Department of Hygiene, Japanese Ministry of Interior. In: Influenza (Ryukousei Kanbou) Ministry of Interior; Tokyo, Japan: 1922. Epidemic records and preventive methods of influenza in the United States of America; pp. 431–484. Chapter 7, Section 2. [Google Scholar]
- Diekmann O, Heesterbeek J. Wiley; New York, NY: 2000. Mathematical epidemiology of infectious diseases: model building, analysis and interpretation. [Google Scholar]
- Dushoff J, Plotkin J.B, Levin S.A, Earn D.J. Dynamical resonance can account for seasonality of influenza epidemics. Proc. Natl Acad. Sci. USA. 2004;101:16 915–16 916. doi: 10.1073/pnas.0407293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1986;1:54–75. [Google Scholar]
- Favier C, et al. Early determination of the reproduction number for vector-borne diseases: the case of dengue in Brazil. Trop. Med. Int. Health. 2006;11:332–340. doi: 10.1111/j.1365-3156.2006.01560.x. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Cummings D.A, Fraser C, Cajka J.C, Cooley P.C, Burke D.S. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flahault A, et al. Modelling the 1985 influenza epidemic in France. Stat. Med. 1988;7:1147–1155. doi: 10.1002/sim.4780071107. [DOI] [PubMed] [Google Scholar]
- Gani R, Hughes H, Fleming D, Griffin T, Medlock J, Leach S. Potential impact of antiviral drug use during influenza pandemic. Emerg. Infect. Dis. 2005;11:1355–1362. doi: 10.3201/eid1109.041344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M.J, Gibbs A.J. Molecular virology: Was the 1918 pandemic caused by a bird flu? Nature. 2006;440:E8. doi: 10.1038/nature04823. [DOI] [PubMed] [Google Scholar]
- Glass, K., Becker, N. & Clements, M. In press. Predicting case numbers during infectious disease outbreaks when some cases are undiagnosed. Stat. Med. ( 10.1002/sim.2523) [DOI] [PubMed]
- Gog J.R, Rimmelzwaan G.F, Osterhaus A.D, Grenfell B.T. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc. Natl Acad. Sci. USA. 2003;100:11 143–11 147. doi: 10.1073/pnas.1830296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon D.T, Chase-Topping M, Shaw D.J, Matthews L, Friar J.K, Wilesmith J, Woolhouse M.E. The construction and analysis of epidemic trees with reference to the 2001 UK foot-and-mouth outbreak. Proc. R. Soc. B. 2003;270:121–127. doi: 10.1098/rspb.2002.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan J.M, Smith R.J, Wahl L.M. Perspectives on the basic reproductive ratio. J. R. Soc. Interface. 2005;2:281–293. doi: 10.1098/rsif.2005.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrenoff A.K. The influenza epidemic of 1918–1919 in San Francisco. The military surgeon. 1941;89:805–811. [Google Scholar]
- Hsieh Y.H, Cheng Y.S. Real-time forecast of multiphase outbreak. Emerg. Infect. Dis. 2006;122:122–127. doi: 10.3201/eid1201.050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N.P, Mueller J. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- Khakpour M, Saidi A, Naficy K. Proved viraemia in Asian influenza (Hong Kong variant) during incubation period. Br. Med. J. 1969;4:208–209. doi: 10.1136/bmj.4.5677.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne E. Influenza pandemics in perspective. JAMA. 1977;237:1225–1228. doi: 10.1001/jama.237.12.1225. [DOI] [PubMed] [Google Scholar]
- Koopman J. Modeling infection transmission. Annu. Rev. Public Health. 2004;25:303–326. doi: 10.1146/annurev.publhealth.25.102802.124353. [DOI] [PubMed] [Google Scholar]
- Lipsitch M, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A.L. The dependence of viral parameter estimates on the assumed viral life cycle: limitations of studies of viral load data. Proc. R. Soc. B. 2001;268:847–854. doi: 10.1098/rspb.2000.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longini I.M, Jr, Halloran M.E. Strategy for distribution of influenza vaccine to high-risk groups and children. Am. J. Epidemiol. 2005;161:303–306. doi: 10.1093/aje/kwi053. [DOI] [PubMed] [Google Scholar]
- Longini I.M, Jr, Koopman J.S, Monto A.S, Fox J.P. Estimating household and community transmission parameters for influenza. Am. J. Epidemiol. 1982;115:736–751. doi: 10.1093/oxfordjournals.aje.a113356. [DOI] [PubMed] [Google Scholar]
- Longini I.M, Jr, Seaholm S.K, Ackerman E, Koopman J.S, Monto A.S. Simulation studies of influenza epidemics: assessment of parameter estimation and sensitivity. Int. J. Epidemiol. 1984;13:496–501. doi: 10.1093/ije/13.4.496. [DOI] [PubMed] [Google Scholar]
- Longini I.M, et al. Containing pandemic influenza at the source. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- Massad E, Burattini M.N, Lopez L.F, Coutinho F.A. Forecasting versus projection models in epidemiology: the case of the SARS epidemics. Med. Hypothesis. 2005;65:17–22. doi: 10.1016/j.mehy.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers L.A, Newman M.E.J, Pourbohloul B. Predicting epidemics on directed contact networks. J. Theor. Biol. 2006;240:400–418. doi: 10.1016/j.jtbi.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Mills C.E, Robins J.M, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H, et al. Rapid awareness and transmission of severe acute respiratory syndrome in Hanoi French Hospital, Vietnam. Am. J. Trop. Med. Hyg. 2005;73:17–25. [PubMed] [Google Scholar]
- Nishiura H, Schwehm M, Kakehashi M, Eichner M. Transmission potential of primary pneumonic plague: time inhomogeneous evaluation based on historical documents of the transmission network. J. Epidemiol. Commun. Health. 2006;60:640–645. doi: 10.1136/jech.2005.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A, et al. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J. Virol. 1997;71:7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillonetto G, Sparacino G, Cobelli C. Numerical non-identifiability regions of the minimal model of glucose kinetics: superiority of Bayesian estimation. Math. Biosci. 2003;184:53–67. doi: 10.1016/S0025-5564(03)00044-0. [DOI] [PubMed] [Google Scholar]
- Razum O, Becher H, Kapaun A, Junghanss T. SARS, lay epidemiology, and fear. Lancet. 2003;361:1739–1740. doi: 10.1016/S0140-6736(03)13335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley S, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- Taubenberger J.K, Reid A.H, Lourens R.M, Wang R, Jin G, Fanning T.G. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- The World Health Organization (WHO). Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2006_07_04/en/index.html [04 July 2006].
- Ungchusak K, et al. Probable person-to-person transmission of avian influenza A (H5N1) New Eng. J. Med. 2005;352:333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- Vaughn W.T. Influenza: an epidemiological study. Am. J. Hyg. 1921 Monograph No. 1. [Google Scholar]
- Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004;160:509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearing H.J, Rohani P, Keeling M.J. Appropriate models for the management of infectious diseases. PLoS Med. 2005;2:e174. doi: 10.1371/journal.pmed.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G, Bean W.J, Gorman O.T, Chambers T.M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.