Abstract
The Saccharomyces cerevisiae genes PRP2, PRP16, and PRP22 encode pre-mRNA splicing factors that belong to the highly conserved “DEAH” family of putative RNA helicases. We previously identified two additional members of this family, JA1 and JA2. To investigate its biological function, we cloned the JA1 gene and generated alleles carrying mutations identical to those found in highly conserved regions of other members of the DEAH family. A ja1 allele carrying a mutation identical to that in the temperature-sensitive (ts) prp22–1 gene conferred ts phenotype when integrated into the genome of a wild-type strain by gene replacement. Northern analysis of RNA obtained from the ts strain shifted to a nonpermissive temperature revealed accumulation of unspliced pre-mRNAs and excised intron lariats. Furthermore, analysis of splicing complexes showed that intron lariats accumulated in spliceosomes. The results presented indicate that JA1 encodes a pre-mRNA processing factor (Prp) involved in disassembly of spliceosomes after the release of mature mRNA. We have therefore renamed this gene PRP43.
Keywords: JA1 gene, splicing
Numerous genes from Escherichia coli to humans encode proteins that belong to a highly conserved superfamily that comprises many well characterized DNA helicases and putative RNA helicases. Proteins encoded by these genes share extensive homology over a 300-amino acid domain containing seven highly conserved motifs (reviewed in refs. 1 and 2). These proteins can be grouped into families according to the sequences of the seven motifs. For example, the “DEAD-box” family is characterized by the amino acid sequence D-E-A-D in motif II, while the “DEAH-box” family, also named after its invariable sequence in motif II, has an additional 300-amino acid C-terminal domain present only in DEAH-box proteins.
Although specific functions of the putative RNA helicases in pre-mRNA splicing have not yet been elucidated at the molecular level, it is likely that these proteins modulate the network of RNA–RNA interactions that occur during spliceosome assembly and splicing of pre-mRNA (3, 4). For example, in the early stages of spliceosome assembly the U4 and U6 small nuclear RNAs (snRNAs) are extensively base paired to form a double-stranded structure that is completely dissociated before the first catalytic step of splicing (5–7). Consistent with the need of energy to unwind the double-stranded structure, the DEIH-box protein Slt22/Brr2 has been implicated in the dissociation of the U4/U6 small nuclear ribonucleoprotein particle (snRNP) (P. Raghunathan and C. Guthrie, personal communication) as well as in the regulation of U2, U6, and U5 snRNA interactions (8). The DEAH-box factor Prp16 has been proposed to mediate conformational changes in the spliceosome (9) and to play a proofreading role in selecting the proper splice sites and branch point (10). The DEAD-box protein Prp5 has been proposed to mediate an ATP-dependent conformational change in the U2 snRNP prior to binding to the branch site of pre-mRNA (3, 11, 12). Furthermore, an ATP-dependent reaction catalyzed by the DEAH-box protein Prp2 (13) is required for activation of the spliceosome (14, 15), while the DEAH-box protein Prp22 is required for release of the mRNA from the spliceosome and initiation of spliceosome disassembly (16). The previous studies have thus established that members of this family of spliceosomal ATPases play a central role in the spliceosome assembly cycle and splicing.
We have previously identified two members of the DEAH family of putative RNA helicases, JA1 and JA2 (16). In this work we report the molecular cloning of JA1 and present evidence indicating that JA1 is involved in pre-mRNA splicing and constitutes a pre-mRNA processing (Prp) factor that we have renamed Prp43.
MATERIALS AND METHODS
Strains and Extracts.
Wild-type Saccharomyces cerevisiae strains used were SS330 (MATa ade2–101 hisΔ200 tyr-1 ura3–52) and J1003.IE (Mata His3–111 Ade2–1 Ura3–1 Leu2–3,112 Can100). Temperature-sensitive (ts) strains prp2–1, prp16–2, and prp22–1 were previously described (17). Splicing extracts were prepared as described (14).
General Methods.
Yeast cultures, transformations, and other techniques were as described (18). Sequencing was performed with the United States Biochemical Sequenase kit used as directed.
Electrophoresis of Total RNA.
For Northern blot analysis of total yeast RNA, 10 μl of total RNA (15 μg) was treated with 16 μl of dimethyl sulfoxide, 5.3 μl of glyoxal, and 1 μl of 320 mM sodium phosphate, pH 7.0. The mixture was heated at 50°C for 60 min, and the sample was supplemented with bromophenol blue (BPB) and electrophoresed (7 hr, 90 mA) in a 25-cm-long horizontal 1.2% agarose gel in 10 mM sodium phosphate, pH 7.0, with constant buffer recirculation. In this system the excised intron comigrated with BPB. This system provides good resolution of the RNA species produced during in vivo splicing of actin pre-mRNA and of snRNAs.
Plasmids and Constructs.
pRS303EV− was generated by deletion of the HincII–EcoRV fragment from the integrative plasmid pRS303 (19). pRSEVJA1sn was obtained by cloning the EcoRI–SacII fragment of the PRP43 gene (lacking its promotor and first 48 codons) into the same sites of pRS303EV−. pRSJA1 carries the complete PRP43 gene, including the HindIII and SacII sites upstream and downstream of the PRP43 ORF, respectively, inserted into the same sites of pRS316 (19). pRSJA1sn contains the truncated EcoRI–ScaII PRP43 fragment inserted into pRS316. pYCP.JA1 was isolated from a genomic library and contains an ≈15-kb genomic fragment inserted into pYCP50.
RESULTS AND DISCUSSION
Molecular Cloning.
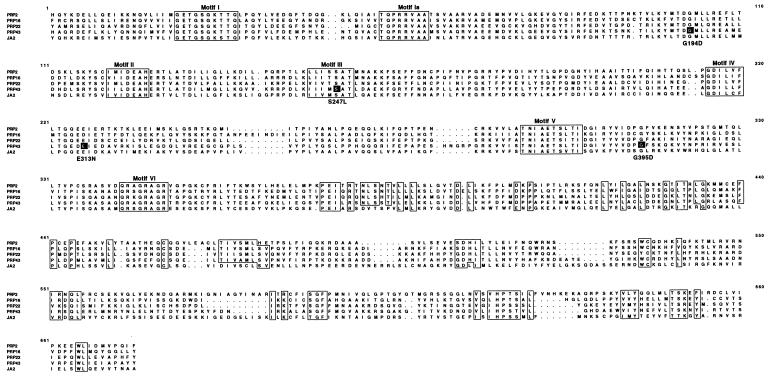
A fragment of the PRP43 gene (JA1), was isolated from PCR products obtained with degenerate oligonucleotides designed to amplify members of the DEAH family of genes (16), and this gene fragment was used as a probe for colony hybridization to obtain the complete gene from a yeast genomic library. DNA sequencing revealed a 2,301-nt ORF that included the 600-bp PCR probe. The PRP43 ORF predicts a 767-amino acid, 87.5-kDa polypeptide, Prp43, that can be divided into three domains. The 100-amino acid N-terminal domain is unique to Prp43 and shows no homology to other known proteins; the central domain, residues 100–440, contains a highly conserved DEAH-box putative RNA-helicase domain homologous to splicing factors Prp2, Prp16, and Prp22; and the 300-residue C-terminal domain of unknown function is uniquely conserved in DEAH-box proteins (16). No sequence homology has been found to date between the C-terminal domain of DEAH proteins and any other proteins (Fig. 1).
Figure 1.
Alignment of DEAH family proteins, including the helicase and C-terminal domains. The alignment includes residues 96–736 of Prp43. The complete sequences of Prp43 and JA2 have been deposited in GenBank, accession numbers U41851 and AF005090, respectively. The seven highly conserved motifs (I–VI) of the putative helicase domain are boxed. The positions of single amino acid changes introduced by mutagenesis are indicated with black boxes. Highly conserved motifs of the C-terminal domain are also boxed.
Generation of Conditional Mutants.
Clues to the biological function of a newly identified gene can be obtained by studying ts and dominant-negative (dn) alleles. Many ts and dn alleles of the DEAD and DEAH families have point mutations within the highly conserved motifs of this protein family. We reasoned that introduction of some of these point mutations at homologous positions in Prp43 was likely to yield prp43 alleles exhibiting similar phenotypes. We performed oligo-directed site mutagenesis on a truncated copy of PRP43, (i.e., JA1sn, lacking a promoter and an N-terminal domain), to generate prp43 alleles carrying the mutations G194D, E315N, G395E, and S247L, which are homologous to mutations found in prp2–1, prp16–2, prp22–1 and prp2-dn1, respectively, all DEAH-box genes. (Fig. 1). The mutagenized truncated gene was inserted into a yeast integration plasmid (pRS303EV−) carrying the HIS3 marker. The resulting construct was linearized at the unique EcoRV site within the PRP43 fragment and was used to transform a wild-type haploid strain. The mutant sequence integrated into the genome by homologous recombination, disrupting the wild-type sequence and conferring a histidine-independent (HIS+) phenotype.
Screening of a small number of HIS+ transformants revealed several colonies with an apparent ts phenotype at 37°C: G194D, 9/34; E315N, 7/62; and G395E, 25/35 (numbers indicate ts colonies/total colonies). Due to the generation of duplicated sequences by the integration event, simple recombination events are expected to occur at a frequency of 10−4 (20), causing the appearance of revertants. Therefore, colonies with a ts phenotype were streaked and replated several times to isolate strains with a stable phenotype. Stable ts HIS+ G395E transformants were obtained; however, no ts G194D or E315N transformants were isolated, suggesting that strains carrying these alleles may be unstable. The same results were obtained with two different wild-type strains as recipient. In each case, G194D transformants grew very slowly and colonies remained very small, indicating that this mutation has an unconditional deleterious effect. Repeated attempts to obtain stable ts E315N transformants were unsuccessful.
G395E Transformants Exhibit a ts Phenotype.
The presence of the G395E mutation in a stable transformant (prp43–1) was confirmed by sequencing PCR products from genomic DNA (data not shown). We found that prp43–1 cells grew normally at 23°C but could not grow on yeast extract/peptone/dextrose (YPD) agar at 37°C, thus showing a ts phenotype. However, in liquid YPD medium, prp43–1 cells cultured at 23°C and shifted to 37°C, began to grow slowly after a prolonged lag phase lasting up to 24 hr and reached a wild-type growth rate after several generations. Interestingly, prp43–1 cells actively growing at 37°C in liquid YPD were still unable to grow at 37°C when transferred to YPD agar, indicating that growth in liquid medium at nonpermissive temperature is not due to the generation of revertants. Furthermore, after plates were kept for 4 days at 37°C and shifted back to 23°C, growth resumed, indicating that the effects of the mutation were not lethal at 37°C.
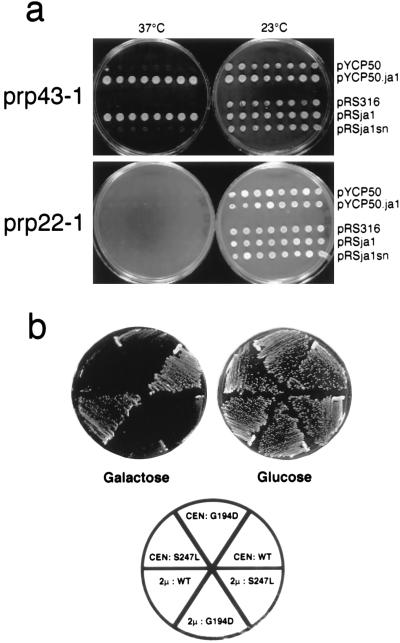
We confirmed that the ts phenotype was due to the prp43–1 allele by complementation experiments using a pRS316 plasmid carrying a wild-type copy of PRP43 (pRSJA1), a truncated PRP43 gene (pRSJA1sn), or a pYCP50 genomic clone carrying a 15-kb fragment containing PRP43. Only the plasmids carrying the complete PRP43 gene restored growth at 37°C (Fig. 2a). These results demonstrate that the ts phenotype of the prp43–1 strain was indeed caused by the mutant gene. In contrast, none of these plasmids could complement the ts phenotype of prp22–1 cells carrying an identical mutation in the prp22 gene, indicating that the two genes are functionally distinct.
Figure 2.
(a) Complementation of the prp43–1 ts phenotype. The prp43–1 and prp22–1 strains were transformed with the following plasmids: pYCP50, control vector; pYCP.ja1, genomic pYCP50 clone containing an ≈15-kb fragment including PRP43; pRS316, control vector; pRSJA1, pRS316 carrying PRP43; and pRSJA1sn, pRS316 carrying a truncated PRP43 gene. From each transformation, eight colonies were randomly selected and arranged in rows for ts testing. The upper panel shows the growth of prp43–1 transformants. The lower panel shows growth of prp22–1 transformants at the indicated temperatures. (b) Constructs containing the PRP43 gene under control of the GAL1 promotor were subcloned into the low-copy-number CEN plasmid pRS416 and the high-copy-number 2μ plasmid pRS426. Identical clones were also prepared for PRP43 alleles carrying the mutations G194D and S247L. All constructs were used to transform wild-type yeast cells. Uracil-independent (URA+) transformants were isolated and tested for growth on glucose and galactose. The plasmid and PRP43 allele harbored by the transformants is shown in the schematic below.
G194D and S247L Mutations Confer a dn Phenotype.
The mutagenesis approach described above was successful for the generation of a new ts strain, prp43–1, with the G395E mutation. Among other mutant constructs tested we observed that G194D transformants grew very slowly at 23°C, producing very small colonies that reverted to wild-type growth after replica plating. No ts colonies were isolated, suggesting that this mutation has an unconditional deleterious phenotype. We then tested whether the prp43 alleles carrying the mutations G194D or S247L confer a dn phenotype. The same amino acid changes in Prp2 confer ts and dn phenotypes, respectively. In addition, the same Ser to Leu change in the conserved SAT motif of a human Prp22 homologue (HRH1) also results in a dn phenotype (21). We constructed CEN and 2μ plasmid derivatives (pRS416 and pRS426, respectively) containing the wild-type, G194D, or S247L prp43 alleles under control of the GAL1 promotor (p416G.ja1, p416Gja1.194, p416Gja1LAT; and p426G.ja1, p426Gja1.194, p426Gja1LAT, respectively). These constructs were used to transform wild-type yeast cells, and transformants were tested for growth on synthetic minimal medium (18) containing glucose (SM-Glc) or galactose (SM-Gal) agar plates at 23°C. The GAL1 promoter is off in the presence of glucose and is turned on in the presence of galactose. All transformants could grow well on SM-Glc plates. However, those carrying G194D or S247L alleles failed to grow on SM-Gal plates, demonstrating that these mutations have a dn phenotype (Fig. 2b). Therefore we assigned the names prp43-dn1 and prp43-dn2 to prp43 alleles carrying the G194D and S247L mutations, respectively.
Interestingly, after initial growth in liquid culture in SM-Glc, prp43-dn2 transformants continued to grow well after the pelleted cells were washed and resuspended in SM-Gal liquid medium. However, prp43-dn2 cells growing in liquid SM-Gal were unable to grow when transferred to SM-Gal solid medium. This behavior is similar to that observed with the ts phenotype of prp43–1 (discussed above), which after growing in liquid medium at 37°C, failed to grow when transferred to solid medium. This puzzling observation indicates that the requirement for Prp43 function is greater for cells growing on solid media. Consistent with this interpretation, tetrad dissection of a diploid carrying a single copy PRP43::HIS null allele produced only 2 viable (HIS−) spores, indicating that PRP43 is essential for growth in solid media. The reason for the different media-specific requirements remains to be understood.
Prp43 Is Involved in mRNA Splicing.
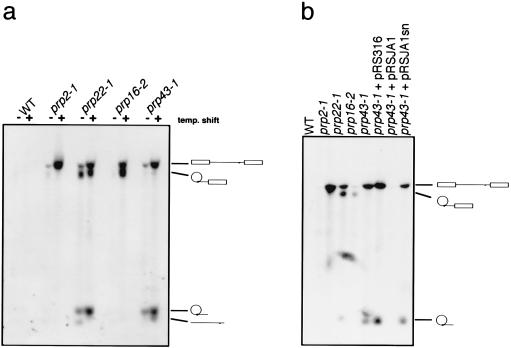
To investigate if Prp43 plays a role in RNA processing, we performed a Northern analysis of RNA prepared from prp43–1 cells after incubation at 37°C for 2 hr in liquid medium. Using a probe complementary to the intron of the actin pre-mRNA, we found that prp43–1 cells incubated at the nonpermissive temperature showed higher levels of pre-mRNA and excised intron lariats relative to wild-type cells. This phenotype differs from that of prp22–1 cells, which we here show to accumulate lariat intermediate as well as pre-mRNA and intron, from prp16–2 cells, which are blocked at the second step splicing, and from prp2–1 cells, which are blocked before the first step of splicing (Fig. 3a). The ts splicing phenotype of prp43–1 was complemented by the wild-type PRP43 gene but not by a truncated PRP43 gene (Fig. 3b), demonstrating that the ts splicing phenotype is indeed due to the prp43–1 gene. This suggests that Prp43 is involved in nuclear pre-mRNA splicing. Analysis of U1, U2, U4, U5, and U6 snRNAs on the same Northern blot showed no fluctuations in the RNA levels, thus ruling out the possibility that the ts phenotype of prp43–1 is caused by destabilization of these snRNAs (not shown).
Figure 3.
(a) Northern blot analysis of total RNA prepared from yeast strains grown at room temperature (−) and after shift to 37°C for 2 hr (+). WT, wild type. The blot was probed with a radiolabeled DNA fragment complementary to sequences of the intron of the actin gene. (b) The indicated strains and prp43–1 transformed with various complementing plasmids were grown at room temperature and shifted to 37°C. Total RNA was prepared after temperature shift and analyzed by Northern blotting as in a. A schematic representation of each splicing product is shown to the right.
Intron Lariats Accumulate in Spliceosomes.
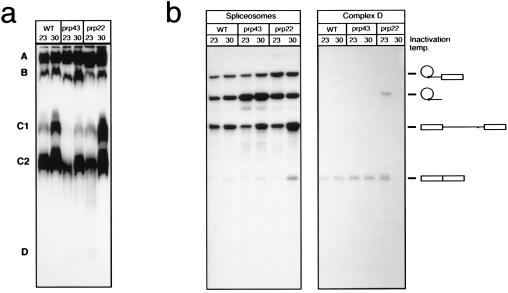
The splicing activity of yeast extracts prepared from ts prp mutant strains is often inactivated by brief heating at 30–37°C. We attempted to inactivate prp43–1 extracts under various conditions without success. However, when these extracts were compared with wild-type extracts, it was evident that prp43–1 extracts accumulated intron lariats independently of heat treatment (data not shown). To investigate possible effects of prp43–1 on spliceosome assembly and disassembly we analyzed the distribution of splicing products among the various splicing complexes formed in vitro. Extracts from wild-type, prp43–1, and prp22–1 strains were heated to 30°C for 1 hr. This treatment corresponds to the standard inactivation of prp22–1 extracts (16). After heat treatment, splicing of radiolabeled actin pre-mRNA was carried out at 15°C and the spliceosomal complexes formed were separated on a nondenaturing gel as previously described (22). The splicing complexes formed are shown in Fig. 4a. Complex A consists of at least three different spliceosome stages containing pre-mRNA, intermediates, or the excised intron lariats. Complex B is the prespliceosome and contains pre-mRNA only. C1 and C2 are a heterogeneous group of nonspecific complexes containing mostly pre-mRNA. Finally, complex D, which is often very diffuse and low in abundance, corresponds to a group of complexes containing mRNA and excised introns that result from spliceosome disassembly. A hallmark of the prp22–1 phenotype after heat inactivation is the complete lack of complex D formation due to failure to disassemble the spliceosome, resulting in accumulation of spliceosomes containing mature mRNA and intron lariats (16). The formation of splicing complexes in prp43–1 extracts was not affected by the heat treatment. However, elution of RNA from the complexes shown in Fig. 4a and analysis in denaturing 8 M urea gels showed that, in prp43–1 extracts, spliceosomes containing intron lariats (complex A) appear to be as much as 10-fold more abundant than spliceosomes containing pre-mRNA (note that because of the difference in length and base composition the radiolabel specific activity of introns is approximately half that of pre-mRNA), while spliceosomes containing intron appear to be only 2-fold higher than pre-mRNA in wild-type extracts. Furthermore, mRNA could be found at wild-type levels in complex D, indicating that the release of mRNA from spliceosomes was not affected by the prp43–1 mutation (Fig. 4b). The quantity and identity of the RNAs in complexes B, C1, and C2 from prp43–1 extracts were identical to the wild-type complexes (not shown). These observations suggest that the observed in vivo stabilization of excised introns in prp43–1 cells shifted to 37°C is due to retention of introns in spliceosomes that failed to disassemble.
Figure 4.
(a) Splicing extracts from wild type (WT), prp43–1, and prp22–1 were heated for 60 min at 23°C or 30°C as indicated. Standard splicing reactions were then set with radiolabeled actin pre-mRNA and the previously heated extracts. Splicing complexes formed were resolved by nondenaturing PAGE. See text for description of complexes. (b) Splicing complexes were sliced off the gel in a and their eluted RNA contents were resolved by denaturing PAGE.
Prp43 and Pre-mRNA Splicing.
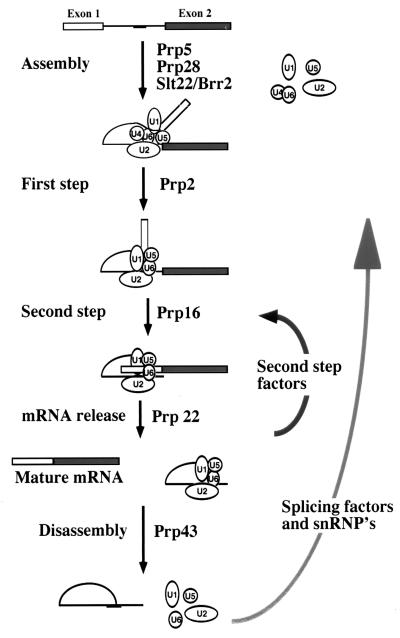
We have shown that the prp43–1 mutation induces accumulation of pre-mRNA and excised intron lariats in vivo, suggesting a role in pre-mRNA splicing. prp43–1 splicing extracts yielded increased levels of intron lariats; however, they were as active as wild-type extracts and produced wild-type levels of all other splicing products, indicating that spliceosome assembly and the two catalytic steps of the splicing reaction were not affected by the prp43–1 mutation. In this respect, it is interesting to compare the phenotypes of prp22–1 and prp43–1. In vivo, prp43–1 cells accumulate pre-mRNA and intron lariats but no intermediate products. In contrast, our Northern analyses show that prp22–1 cells not only accumulate introns and pre-mRNA as previously described (17) but also accumulate intermediate lariats. This observation suggests that limiting splicing factor(s) required for the second step of splicing are released from the spliceosome concomitant with or after the release of the mRNA but before the release of the intron lariat. Thus, when prp43–1 cells are switched to nonpermissive temperature, all spliceosomes containing pre-mRNA are able to complete the first and second steps of splicing and release the mRNA. However as intron-containing spliceosomes accumulate, various factors and snRNPs required for spliceosome assembly and the first step of splicing are depleted, causing accumulation of pre-mRNA. A possible explanation for our observations is that Prp43 may play a role in accelerating spliceosome disassembly after the release of mRNA, thus ensuring efficient recycling of splicing factors. An interesting hypothesis is that Prp43 mediates spliceosome dissasembly by inducing an ATP-dependent comformational change in one or more snRNPs. As illustrated in Fig. 5, multiple putative RNA helicases are required during spliceosome assembly and the catalytic steps of splicing. Extensive RNA conformational changes occur during spliceosome assembly that may be mediated by RNA-dependent ATPases. Additional energy-dependent conformational changes may also be required to induce spliceosome disassembly and to return snRNPs to their initial state. The requirement of RNA-dependent ATPases for the release of splicing products and spliceosome disassembly suggests that this is a highly ordered process that regulates the recycling of splicing factors.
Figure 5.
Seven putative RNA helicases required during spliceosome assembly, catalytic steps, and spliceosome disassembly are shown. While some limiting second-step splicing factors may leave the spliceosome at the time of mRNA release, many splicing factors, including snRNPs, are not recycled until the excised intron lariat is released.
The study of yeast splicing factors has been instrumental for the identification and cloning of mammalian homologues, including a human homologue of PRP22, HRH1, for which a role in mRNA release has been demonstrated (21, 23). In the accompanying paper (24), Gee and colleagues describe the isolation of a mouse homologue of PRP43, mDEAH9, which can complement the ts growth defect of prp43–1.
Searches of the yeast genome database revealed only five ORFs encoding DEAH-box proteins. Four of them correspond to the splicing factors Prp2, Prp16, Prp22, and Prp43. The biological function of the fifth DEAH-box gene (JA2) has not yet been elucidated; however, it is tempting to speculate that it also may be involved in pre-mRNA splicing. The approach taken here to generate ts and dn alleles should be instrumental to answer this question and to further investigate the role of Prp43 and other DEAD/DEAH-box proteins in splicing.
Acknowledgments
We thank P. Raghunathan and C. Guthrie for sharing results prior to publication, Yan Wang for review of the manuscript, and J. Messenger for sequencing of the JA2 gene. This work was supported by National Institutes of Health Grant GM32637.
ABBREVIATIONS
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein
- ts
temperature-sensitive phenotype
- dn
dominant-negative phenotype
Footnotes
References
- 1.Fuller-Pace F V, Lane D P. In: Nucleic Acids and Molecular Biology. Eckstein F, Lilley D M J, editors. Vol. 6. Berlin: Springer; 1992. pp. 159–173. [Google Scholar]
- 2.Schmid S R, Linder P. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 3.Lamond A I. BioEssays. 1993;15:595–603. doi: 10.1002/bies.950150905. [DOI] [PubMed] [Google Scholar]
- 4.Kramer A. In: Pre-mRNA Processing. Lamond I L, editor. Heidelberg: Springer; 1995. pp. 35–64. [Google Scholar]
- 5.Madhani H D, Guthrie C. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen T W. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- 7.Wassarman D A, Steitz J A. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 8.Xu D, Nouraini S, Field D, Tang S J, Friesen J D. Nature (London) 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 9.Schwer B, Guthrie C. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess S M, Guthrie C. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 11.Dalbadie-McFarland G, Abelson J. Proc Natl Acad Sci USA. 1990;87:4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Day C L, Dalbadie-McFarland G, Abelson J. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 13.Chen J H, Lin R J. Nucleic Acids Res. 1990;18:6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S H, Lin R J. Mol Cell Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S H, Lin R J. Proc Natl Acad Sci USA. 1993;90:888–892. doi: 10.1073/pnas.90.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Company M, Arenas J E, Abelson J N. Nature (London) 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 17.Vijayraghavan U, Company M, Abelson J. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 18.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alani E, Cao L, Kleckner N. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno M, Shimura Y. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 22.Arenas J E, Abelson J N. Proc Natl Acad Sci USA. 1993;90:6771–6775. doi: 10.1073/pnas.90.14.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono Y, Ohno M, Shimura Y. Mol Cell Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gee S, Krauss S W, Miller E, Aoyagi K, Arenas J, Conboy J G. Proc Natl Acad Sci USA. 1997;94:11803–11807. doi: 10.1073/pnas.94.22.11803. [DOI] [PMC free article] [PubMed] [Google Scholar]