Abstract
Yeast splicing factor Prp43, a DEAH box protein of the putative RNA helicase/RNA-dependent NTPase family, is a splicing factor that functions late in the pre-mRNA splicing pathway to facilitate spliceosome disassembly. In this paper we report cDNA cloning and characterization of mDEAH9, an apparent mammalian homologue of Prp43. Amino acid sequence comparison revealed that the two proteins are ≈65% identical over a 500-aa region spanning the central helicase domain and the C-terminal region. Expression of mDEAH9 in S. cerevisiae bearing a temperature-sensitive mutation in prp43 was sufficient to restore growth at the nonpermissive temperature. This functional complementation was specific, as mouse mDEAH9 failed to complement mutations in related splicing factor genes prp16 or prp22. Finally, double label immunofluorescence experiments performed with mammalian cells revealed colocalization of mDEAH9 and splicing factor SC35 in punctate nuclear speckles. Thus, the hypothesis that mDEAH9 represents the mammalian homologue of yeast Prp43 is supported by its high sequence homology, functional complementation, and colocalization with a known splicing factor in the nucleus. Our results provide additional support for the hypothesis that the spliceosomal machinery that mediates regulated, dynamic changes in conformation of pre-mRNA and snRNP RNAs has been highly conserved through evolution.
Pre-mRNA splicing in yeast and mammalian systems is a multistep process involving recognition and assembly of pre-mRNA splice sites into spliceosomal complexes, intron excision and exon ligation mediated by two transesterification reactions, and release of splice products concomitant with spliceosome disassembly (1–3). In contrast to self-splicing group I introns, pre-mRNA splicing requires ATP and numerous protein factors at multiple steps in the pathway. Among these factors are a group of genetically defined Saccharomyces cerevisiae genes (PRP2, -5, -16, -22, -28, -43) that are members of a family of “DEAD/DEAH box” proteins that contain conserved motifs characteristic of RNA-dependent NTPases and RNA helicases (4–6). More recently, an additional candidate pre-mRNA splicing factor (Slt22) with RNA-dependent ATPase activity has been identified in association with the U4/U6.U5 tri-snRNP in yeast (7, 8). The DEAD/DEAH class of splicing factor proteins may help catalyze the intricate series of highly ordered small nuclear RNA (snRNA)–pre-mRNA and snRNA–snRNA interactions required for successful splicing. Alternatively, such proteins may serve a kinetic proofreading function to enhance fidelity of splicing (9).
Extensive genetic and biochemical analysis has allowed temporal correlation of specific PRP gene functions with specific steps in spliceosome assembly, catalysis, and spliceosome disassembly. PRP2 and Slt22 are required early in assembly before the first catalytic step (8, 10–12); PRP16 activity is essential for the second catalytic step (13, 14); and PRP22 functions during spliceosome disassembly (15). Results in the accompanying paper (16) support the hypothesis that PRP43 acts at a late stage of the disassembly process: alteration of a conserved helicase motif in mutant prp43 allele yielded a phenotype characterized by excess accumulation of spliceosomal complexes retaining excised intron lariats. However, many mechanistic details of the reactions catalyzed by this intriguing class of enzymes, including the precise RNA substrate and the secondary structural change presumably effected by each, remain to be fully elucidated.
Thus far, few counterparts of these yeast DEAD/DEAH box splicing factors have been identified in Drosophila or mammalian systems. Evidence that such factors are expressed in mammalian cells was provided by the recent identification and characterization of HRH1, a mammalian homologue of yeast Prp22. Wild-type HRH1 protein exhibits extensive sequence homology with Prp22, functionally complements prp22 mutations in yeast, and is localized to nuclear splicing domains in cultured cells (17, 18). Moreover, dominant-negative mutants of HRH1 inhibit pre-mRNA splicing in vivo and inhibit release of spliced products from spliceosomal complexes in vitro (18). These results strongly support the hypothesis that HRH1 and Prp22 are functional homologues.
In this paper we identify a mammalian homologue of a second yeast DEAH box splicing factor, Prp43. The mouse homologue, designated mDEAH9, was cloned initially by reverse transcriptase (RT)/PCR using degenerate oligonucleotides to conserved helicase motifs. By the criteria of amino acid sequence homology to Prp43, functional complementation of prp43 mutants, and immunofluorescence localization of mDEAH9 in the nucleus, mDEAH9 appears to represent a mammalian homologue of yeast splicing factor Prp43.
MATERIALS AND METHODS
Strategy for Cloning mDEAH9.
Degenerate oligonucleotide primers were designed to hybridize with two conserved sequences encoding the “GKT” and “DEAH” amino acid motifs. The primers used were as follows. Sense strand primer 1, 5′-CGGAATTCACIGGITCIGGIAA(G/A)AC-3′; antisense strand primer 2, 5′-CGGAATTC(T/C)(T/C)(T/G/C)(A/G)T(C/G)IGC(T/C)TC(A/G)-3′. Underlined bases represent EcoRI sites added to facilitate subcloning. Mouse erythroleukemia (MEL) cell RNA was amplified by RT/PCR, yielding a mixture of products of ≈300–310 nucleotides. A subcloned fragment with high homology to PRP43 was used as a probe for isolation of multiple overlapping cDNAs from two different MEL cell cDNA libraries. One library, constructed in the plasmid vector pXM, was a gift of Alan D’Andrea (Dana–Farber Cancer Institute, Harvard Medical School, Boston). The other was purchased from CLONTECH (Palo Alto, CA) and was constructed in λgt10. Both libraries yielded clones with identical sequence in extensive regions of overlap.
Complementation of Temperature-Sensitive prp43.
Yeast expression vector pRS426pG1 (a gift of T.-H. Chang, Ohio State University) was modified by insertion of an EcoRI site at the BamHI cloning site, and mDEAH9 cDNA was inserted into the EcoRI site. Plasmid DNAs were introduced into S. cerevisiae by the LiCl technique and transformants were selected by growth on uracil− agar plates at the permissive temperature of ≈23°C.
Immunofluorescence.
WI38 cells, diploid human fetal lung fibroblasts, were grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) containing 10% fetal bovine serum, 4 mM glutamine, and antibiotics (50 units/ml penicillin and 50 μg/ml streptomycin). Cells grown on coverslips for 1–2 days were rinsed twice in phosphate-buffered saline (PBS) at pH 7.4, fixed for 10 min at 4°C in PBS containing 2% formaldehyde/0.2% Triton X-100, further permeabilized with 1% Triton X-100 for 10 min at room temperature, immersed in cold acetone for 5 min, and rinsed twice in PBS. All subsequent incubations were for 1 hr at room temperature in PBS containing 10 mg/ml bovine serum albumin and additions as indicated. After incubation in 10% goat serum to block nonspecific protein binding, fixed cells were incubated with the two primary antibodies: affinity-purified rabbit IgG against mDEAH9, and mouse monoclonal antibody to SC-35 (19) (hybridoma cell supernatant, a gift of X.-D. Fu, Univ. of California, San Diego). After washing, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Molecular Probes) and tetramethylrhodamine-conjugated goat anti-mouse IgG (Sigma). Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) to confirm nuclear localization of immunofluorescent signals and washed, and the coverslips were mounted on slides by using Vectashield (Vector Laboratories). Alternate fixation methods (omitting the final acetone step or substituting ethanol for acetone) gave essentially the same results, although the nuclear signals were somewhat more diffuse.
Confocal Microscopy.
A Bio-Rad confocal laser scanning microscope (MRC 1000) equipped with a Nikon Diaphot 200 with a fluor 60× 1.4 numerical aperature Planapo oil-immersion lens was used to obtain optical sections of WI38 cells. An argon laser (λ = 488 nm) was used to excite fluorescein and a helium/neon laser (λ = 543 nm), to excite rhodamine. Images spanning the entire nucleus were constructed by means of Bio-Rad software. The limit of resolution in this system was 0.2 μm.
RESULTS
To clone candidate splicing factor cDNAs of the DEAD/H box family of putative RNA helicases, we amplified MEL cell RNA with oligonucleotide primers corresponding to two conserved helicase motifs (GKT and DEAH) (Fig. 1A). A diffuse band of ≈300–310 nt was obtained, which upon subcloning and sequencing was found to include fragments of at least three distinct cDNAs with homology to the DEAH box family. One of these (mDEAH6) exhibited high homology to the yeast splicing factor PRP22 (15) and near identity with its human homologue HRH1 (data not shown). A second cDNA fragment, mDEAH9, was highly homologous to the newly described yeast splicing factor PRP43 (16) and the partial cDNA encoding human HRH2 (17). The mDEAH9 cDNA was utilized as a probe for screening traditional cDNA libraries (from MEL cells), as well as GenBank expressed sequence tag (EST) libraries, allowing assembly of an ≈2.7-kb cDNA predicted to encode a protein of 758 amino acids (Fig. 1B).
Figure 1.
(A) Domain structure of mouse mDEAH9 and yeast Prp43 proteins. Numbers represent percent identity of amino acid sequence in each domain. Highest homology (≈72% identity) is observed in the central domain, which contains motifs shared with the DEAD box family of putative RNA helicases. Homology is much weaker (<30%) in the N-terminal region. Arrows indicate position of primers used to amplify a region of mDEAH9 in the early cloning experiments. (B) Amino acid sequence comparison of mDEAH9, Prp43, and a related Caenorhabditis elegans protein. The N-terminal residues 1–30 of mDEAH9 were deduced by sequence analysis of a mouse embryo expressed sequence tag (EST) clone (GenBank accession no. W84193) and the remainder from MEL cDNAs. The source of other sequences is as follows: human HRH2, ref. 17; C. elegans, accession no. U13644.
The deduced amino acid sequence of mDEAH9 revealed a protein with the typical tripartite structure found in other members of the DEAH splicing factor proteins: a unique N-terminal domain, a central helicase domain containing conserved motifs shared by both DEAD and DEAH-type members of the RNA helicase family, and a C-terminal domain characteristic of the DEAH subclass. Homology between mouse mDEAH9 and S. cerevisiae Prp43 is very high in the helicase domain (72% identity) and in the C-terminal domain (64% identity), but much lower in the N-terminal region. For comparison, mDEAH9 and HRH2 are 98% identical in the helicase domain. Interestingly, the primary sequence of the N-terminal domain exhibits two repeating sequences of unknown importance: a stretch of predominantly charged amino acids with alternating acidic and basic residues, and another region with seven repeats of the tripeptide (A/T)-H-S (Fig. 1B).
Pairwise comparison of the full-length mDEAH9 sequence to other known DEAH proteins supports the hypothesis that it represents the mammalian homologue of Prp43. Over the ≈500-aa region spanning the helicase and C-terminal domains, mDEAH9 is more closely related to Prp43 than it is to other putative helicases from mammalian sources (HRH1, RNA helicase A), to the Drosophila mle protein, or to yeast splicing factors Prp2, Prp16, and Prp22. Indeed, mDEAH9 was found to be more closely homologous to Prp43 than to any other predicted open reading frame in the recently completed S. cerevisiae sequence (data not shown). Conversely, Prp43 is more similar to mouse mDEAH9 than it is to other yeast members of this family, including Prp2, Prp16, and Prp22.
Homology searches of the GenBank databases for genes similar to PRP43/mDEAH9 revealed close homologues in plants (Arabidopsis, not shown) and nematodes (C. elegans). Alignment of these sequences demonstrated a consistently high degree of sequence identity from the GKT nucleotide binding motif almost to the C terminus, a span of ≈500 aa (Fig. 1B). In contrast, the length and primary sequence of the N-terminal domains is more variable between species. Of note, however, this domain in all of the higher eukaryotic homologues is rich in charged residues, with multiple simple repeats of basic and acidic residues, suggesting that these conserved elements may be functionally important.
To explore whether the extensive structural homology between mouse mDEAH9 and yeast Prp43 proteins is indicative of a conserved physiological function, we tested whether mDEAH9 expression in vivo could functionally complement prp43 mutations. For this study we employed a yeast strain bearing a temperature-sensitive mutation (Gly-395 → Asp) in the helicase domain of Prp43, which can grow normally at 23°C but not 35°C (16). Mutant yeast cells transformed with the yeast expression vector pRS426pG1 containing mDEAH9 cDNA were tested for growth at the nonpermissive temperature. As shown in Fig. 2, mDEAH9 cDNA rescued the growth defect at 35°C, allowing growth of single colonies within 4–6 days. Cells transformed in parallel with the expression vector alone did not yield single colonies at 35°C even after 14 days. Control experiments revealed that this functional complementation was specific—i.e., mDEAH9 did not rescue growth of temperature-sensitive alleles of prp22 or prp16 (not shown). Positive control plasmid p316JA1, encoding the wild-type Prp43 protein, promoted growth at 35°C after only 2–3 days. Thus, mDEAH9 complements the prp43 mutation specifically, but less efficiently than the native yeast protein.
Figure 2.
Functional complementation of prp43 mutant by mDEAH9 cDNA. S. cerevisiae transformed with the indicated plasmids was streaked out on uracil− agar plates and incubated at nonpermissive (35°C) temperatures for 7 days. Expression of mDEAH9, but not the vector alone, rescued growth at nonpermissive temperatures.
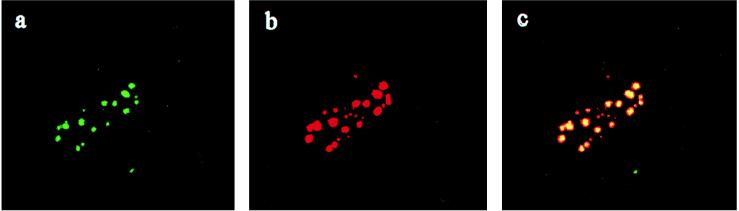
The ability of mouse mDEAH9 to rescue the growth defect in prp43 temperature-sensitive mutant yeast suggests that mDEAH9 is a functional homologue of PRP43. If indeed mDEAH9 functions in the mammalian pre-mRNA splicing pathway, this hypothesis predicts that mDEAH9 should be a nuclear protein, and it might colocalize with known splicing factors in the nucleus. To examine the subcellular localization of mDEAH9, we performed a series of immunofluorescence experiments, using (i) affinity-purified rabbit polyclonal antibodies directed against a synthetic peptide in the unique N terminus of mDEAH9, and (ii) a well characterized mouse monoclonal antibody against splicing factor SC35 (19). The human fibroblast cell line WI38 stained with anti-mDEAH9 plus FITC-conjugated anti-rabbit IgG (Fig. 3A) and anti-SC35 plus rhodamine-conjugated anti-mouse IgG (Fig. 3B), yielded punctate “speckled” patterns of nuclear fluorescence. Most importantly, these patterns were largely coincident, as indicated by the yellow generated when the two patterns were superimposed (Fig. 3C). 4′,6-Diamidino-2-phenylindole staining confirmed that the speckled patterns were indeed nuclear; little or no cytoplasmic staining was observed (not shown). These immunofluorescence studies demonstrate that mDEAH9 and SC35 colocalize in nuclear speckles believed to represent storage sites for pre-mRNA splicing factors (20), consistent with a role for mDEAH9 in pre-mRNA splicing.
Figure 3.
Compartmentalization of mDEAH9 in nuclear speckles. Images show a human WI38 fibroblast processed for double-label immunofluorescence with antibodies against both mDEAH9 and SC35. (×1,200.) (A) mDEAH9 staining visualized with an FITC-conjugated anti-rabbit IgG (green). (B) Anti-SC35 staining of the same cells visualized with rhodamine-conjugated anti-mouse IgG (red). (C) Superimposition of the anti-mDEAH9 plus anti-SC35 immunofluorescence signals, showing colocalization of the two antigens (yellow). Individual focal planes containing both fluorochromes also show overlap, although the areas stained by SC-35 (red) are slightly larger than the mDEAH9-stained speckles.
To characterize the expression of mDEAH9 in various mouse tissues, Northern blot analysis was performed (Fig. 4). All tissues tested, including heart, brain, spleen, lung, liver, skeletal muscle, kidney, testis, and MEL cells (not shown) expressed an mRNA of ≈3.4 kb (Fig. 4 Upper). Differences in signal intensity were largely attributable to variations in RNA loading, as indicated by the intensity of hybridization with an actin cDNA probe (Fig. 4 Lower). This apparently ubiquitous expression of mDEAH9 is consistent with a role as an essential splicing factor.
Figure 4.
Expression of mDEAH9 mRNA in mouse tissues. A multiple tissue Northern blot (CLONTECH), containing 2 μg of total RNA from the indicated mouse tissues, was hybridized with a 32P-labeled mDEAH9 cDNA probe (Upper) or an actin cDNA probe (Lower) under stringent conditions. Lanes: 1, heart; 2, brain; 3, spleen; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; and 8, testis.
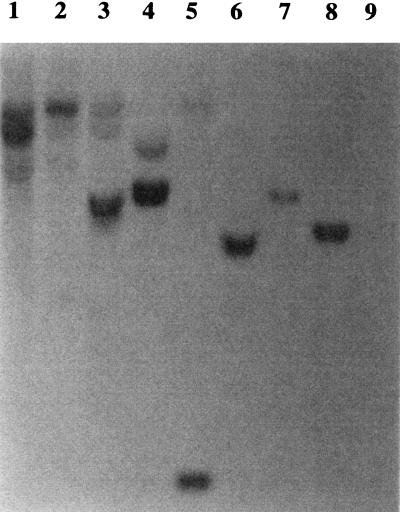
Finally, phylogenetic conservation of mDEAH9 gene sequences was examined by Southern blot hybridization to genomic DNA from several species on a Zoo-Blot (Fig. 5). Strong cross-hybridization of mouse mDEAH9 cDNA was observed to DNA from human, monkey, rat, dog, cow, rabbit, and chicken origin. However, despite the considerable amino acid sequence homology between mDEAH9 and Prp43 proteins, little or no cross-hybridization was seen between mDEAH9 cDNA and S. cerevisiae genomic DNA (Fig. 5, lane 9). This is due to considerable sequence divergence at the wobble base of many codons, which lowers the overall DNA sequence homology. As a control for specificity, the Zoo-Blot was reprobed with a partial cDNA representing the mouse homologue of HRH1, designated mDEAH6. A distinct pattern of hybridizing genomic fragments was observed, confirming that individual DEAH hybridization probes do not cross-react with related family members under the conditions employed (results not shown).
Figure 5.
Conservation of mDEAH9 gene sequences among vertebrate species. A Zoo-Blot (CLONTECH) containing 8 μg of EcoRI-digested genomic DNA was hybridized with 32P-labeled mDEAH9 cDNA probe under stringent conditions. Lanes: 1, human; 2, monkey; 3, rat; 4, mouse; 5, dog; 6, cow; 7, rabbit; 8, chicken; and 9, S. cerevisiae.
DISCUSSION
In this paper we characterize a mouse cDNA, designated mDEAH9, that encodes a novel member of the DEAH box family of putative RNA helicases and RNA-dependent NTPases. Several lines of evidence support the hypothesis that mDEAH9 represents a mammalian homologue of the yeast splicing factor Prp43 (16). First, the degree of phylogenetic sequence conservation is remarkable. The mouse mDEAH9 and S. cerevisiae Prp43 proteins are >65% identical over 500 amino acids, a level of homology comparable to that seen between yeast (Tif) and mouse (eIF-4A) translation initiation factors. Another manifestation of its evolutionary conservation, at the nucleotide level, is the specific cross-hybridization of mDEAH9 cDNA with genomic DNA from various species (Fig. 5). Second, mDEAH9 was able to complement a temperature-sensitive prp43 mutant so as to facilitate growth at otherwise nonpermissive temperatures, demonstrating conservation of function between mDEAH9 and Prp43. Third, immunofluorescence experiments show that mDEAH9 colocalizes with splicing factor SC35 in nuclear speckles, the presumptive storage site for pre-mRNA splicing factors (21). The latter experiment demonstrates that mDEAH9 is localized exactly as might be predicted for a candidate splicing factor. Finally, mDEAH9 appears by Northern blot analysis to be widely expressed among mouse tissues, as would be expected for a constitutive component of the cellular splicing machinery.
The homology of mDEAH9 to DEAD/H box proteins in general, and to Prp43 in particular (16), provides clues as to its possible biochemical function in pre-mRNA splicing. mDEAH9, like the related splicing factors Prp2, Prp16, and Prp22, is likely to possess RNA-dependent NTPase activity and may account in part for the ATP requirement during pre-mRNA splicing (9). Moreover, since some DEAD/H box proteins have ATP-dependent RNA unwinding activity, mDEAH9 is a candidate RNA helicase as well. Experiments with a prp43 temperature-sensitive mutant are consistent with a role late in the splicing process, during spliceosome disassembly (16); mDEAH9 may thus function after the biochemical steps of splicing to promote release of splicing products and recycling of components for another round of splicing. It is interesting to speculate that mDEAH9 may bind to one or more of the splicing product RNAs and utilize an RNA-dependent NTPase activity to promote a critical conformational change in either the RNA itself (e.g., by helicase action) or in an interacting spliceosomal protein; either of these may be required for disassembly and recycling of spliceosomal components.
One intriguing question about RNA processing concerns the extent to which the nuclear splicing machinery in higher eukaryotes resembles that in yeast, despite the vastly different magnitude and complexity of the splicing problem in the two types of organism. How similar might the splicing machinery be in S. cerevisiae—where most genes are intronless or contain a single intron with well conserved splice sites—versus higher eukaryotes—where most genes possess multiple introns with less well defined splice sites and often complicated alternative splicing patterns? It has been well established that the major snRNA components of the small nuclear ribonucleoprotein (snRNP) particles exhibit considerable conservation of secondary structure and function (e.g., see ref. 22). More recently, a number of core snRNP proteins with identifiable homologues in both yeast and higher eukaryotic systems have been reported, including components of the U1 snRNP (23, 24); the U2 snRNP (25–28); and the U5 snRNP (29, 30). Relatively little is known about conservation of non-snRNP splicing factors, but here too progress is being made. The recently described human HRH1 protein, a DEAD/H box protein and apparent homologue of S. cerevisiae Prp22, is one example. The structural and functional similarity between mDEAH9 and Prp43 represents a second example of evolutionary conservation of non-snRNP splicing factors. Such findings suggest that yeast and mammalian splicing machineries probably utilize similar strategies and protein components to orchestrate the dynamic base pairing interactions that occur during pre-mRNA splicing (31). Future biochemical experiments, testing the effects of quantitative and qualitative variations in mDEAH9 protein on the efficiency of pre-mRNA splicing, will be required to define more precisely the role of mDEAH9.
Acknowledgments
This work was supported by National Institutes of Health Grant HL45182 (J.G.C.).
ABBREVIATIONS
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein
- MEL
mouse erythroleukemia
- FITC
fluorescein isothiocyanate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF017153).
References
- 1.Moore M J, Query C C, Sharp P A. In: The RNA World. Gesteland R, Atkins J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–357. [Google Scholar]
- 2.Newman A J. Curr Opin Genet Dev. 1994;4:298–304. doi: 10.1016/s0959-437x(05)80057-7. [DOI] [PubMed] [Google Scholar]
- 3.Ares M J, Weiser B. Prog Nucleic Acids Res Mol Biol. 1995;50:131–159. doi: 10.1016/s0079-6603(08)60813-2. [DOI] [PubMed] [Google Scholar]
- 4.Wassarman D A, Steitz J A. Nature (London) 1991;349:463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- 5.Schmid S R, Linder P. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 6.Fuller-Pace F V. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 7.Laggerbauer B, Lauber J, Luhrmann R. Nucleic Acids Res. 1996;24:868–875. doi: 10.1093/nar/24.5.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu D, Nouraini S, Field D, Tang S J, Friesen J D. Nature (London) 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 9.Burgess S M, Guthrie C. Trends Biochem Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- 10.King D S, Beggs J D. Nucleic Acids Res. 1990;18:6559–6564. doi: 10.1093/nar/18.22.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S H, Lin R J. Proc Natl Acad Sci USA. 1993;90:888–892. doi: 10.1073/pnas.90.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plumpton M, McGarvey M, Beggs J D. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwer B, Guthrie C. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umen J G, Guthrie C. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 15.Company M, Arenas J, Abelson J. Nature (London) 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 16.Arenas J E, Abelson J N. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono Y, Ohno M, Shimura Y. Mol Cell Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohno M, Shimura Y. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 19.Fu X D, Maniatis T. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 20.Spector D L. Curr Opin Cell Biol. 1993;5:442–448. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- 21.Spector D L. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 22.Shuster E O, Guthrie C. Nature (London) 1990;345:270–273. doi: 10.1038/345270a0. [DOI] [PubMed] [Google Scholar]
- 23.Smith V, Barrell B G. EMBO J. 1991;10:2627–2634. doi: 10.1002/j.1460-2075.1991.tb07805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao H Y, Siliciano P G. Nucleic Acids Res. 1992;20:4009–4013. doi: 10.1093/nar/20.15.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brosi R, Groning K, Behrens S-E, Luhrmann R, Kramer A. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- 26.Bennett M, Reed R. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- 27.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 28.Gozani O, Feld R, Reed R. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 29.Pinto A L, Steitz J A. Proc Natl Acad Sci USA. 1989;86:8742–8746. doi: 10.1073/pnas.86.22.8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia B M, Anderson G J, Beggs J, Sharp P A. Proc Natl Acad Sci USA. 1990;87:3082–3086. doi: 10.1073/pnas.87.8.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsen T W. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]