Summary
DEAD-box proteins are ATPase enzymes that destabilize and unwind duplex RNA. Quantitative knowledge of the ATPase cycle parameters is critical for developing models of helicase activity. However, limited information regarding the rate and equilibrium constants defining the ATPase cycle of RNA helicases is available, including the distribution and flux of populated biochemical intermediates, the catalytic step(s) that limits the enzymatic reaction cycle, and how ATP utilization and RNA interactions are linked. We present a quantitative kinetic and equilibrium characterization of the rRNA-activated ATPase cycle mechanism of DbpA, a DEAD-box rRNA helicase implicated in ribosome biogenesis. rRNA activates the ATPase activity of DbpA by promoting a conformational change after ATP binding that is associated with hydrolysis. Chemical cleavage of bound ATP is reversible and occurs via a gamma phosphate attack mechanism. ADP-Pi and RNA binding display strong thermodynamic coupling, which causes DbpA-ADP-Pi to bind rRNA with > 10-fold higher affinity than with bound ATP, ADP or in the absence of nucleotide. The rRNA-activated steady-state ATPase cycle of DbpA is limited both by ATP hydrolysis and Pi release, which occur with comparable rates. Consequently, the predominantly populated biochemical states during steady-state cycling are the ATP- and ADP-Pi-bound intermediates. Thermodynamic linkage analysis of the ATPase cycle transitions favors models in which rRNA duplex destabilization is linked to strong rRNA and nucleotide binding. The presented analysis of the DbpA ATPase cycle reaction mechanism provides a rigorous kinetic and thermodynamic foundation for developing testable hypotheses regarding the functions and molecular mechanisms of DEAD-box helicases.
Keywords: DEAD-box protein, RNA, helicase, ATPase, kinetics, nucleotide
Introduction
RNA helicases are ATPase enzymes that destabilize and unwind double-stranded RNA structures, refold RNA and disrupt RNA-protein complexes1 - 4. RNA helicases that belong to the superfamily II of molecular motor helicase enzymes known as DEAD- or the related DExH/D-box proteins5, which are present in every organism from viruses to humans and play fundamental roles in viral transcription and replication, protein translation, proper pre-mRNA splicing and editing, ribosome biogenesis, RNA export to the cytoplasm, and RNA degradation6. Recent work has provided insight to the mechanism of DEAD-box helicase destabilization and unwinding7 - 9, but the RNA-activated ATPase cycle and its linkage to duplex destabilization and unwinding has for the most part remained uncharacterized.
Detailed mechanistic studies of DEAD-box RNA helicases have been hindered by a requirement for additional regulatory subunits, leaky ATPase activity in the absence of RNA, and low RNA substrate specificity10. DbpA is non-processive ATP-dependent 7 helicase that destabilizes and unwinds short rRNA duplexes 11. DbpA is unique among the E. coli DEAD-box family in that its ATPase activity is specifically activated by the peptidyl-transferase center (PTC) of the 23S ribosomal RNA12, thus it provides a unique model system in which to examine enzymatic activation by RNA. DbpA is not activated by 5S rRNA, 16S rRNA, tRNA, native 50S or native 70S ribosomes, but is activated by denatured 70S ribosomes13, favoring a physiological role in ribosome biogenesis and/or assembly.
It is well established that many classes of nucleic acid helicases couple the free energy from catalytic cycles of ATP binding, hydrolysis and product release to mechanical work in the form of nucleic acid duplex unwinding14; 15 and translocation16. However, limited information is available regarding the ATPase cycle of RNA helicases, including the rate and equilibrium constants defining the enzymatic reaction cycle, the distribution and flux of populated biochemical intermediates, what catalytic step(s) limits catalytic cycling, and how ATP utilization and RNA interactions are linked17; 18. In particular, it is unclear how RNA binding increases the DEAD-box protein ATPase activity, which can be achieved by accelerating ATP binding, hydrolysis, and/or product release.
We present in this study a quantitative kinetic and equilibrium characterization of the rRNA-activated ATPase cycle mechanism of DbpA. The experimental approach and analysis employed allows us to define in quantitative terms a) the ATPase cycle reaction pathway, b) the distribution of populated biochemical intermediates, c) the free energy changes associated with individual ATPase cycle transitions, d) the biochemical reaction(s) that limit ATP utilization, e) the nucleotide and RNA binding linkage (i.e. how binding of one affects the other), and f) how the catalytic ATPase activity of DbpA is activated by rRNA. Quantitative knowledge of the rRNA-dependent ATPase cycle of DbpA will serve as the basis for developing mechanistic models of how ATP utilization is coupled to RNA helicase activity. Given the high homology among all identified prokaryotic and eukaryotic DEAD-box helicases, the detailed analysis of the DbpA ATPase cycle reaction mechanism provides a rigorous kinetic and thermodynamic foundation from which testable hypotheses regarding the functions and molecular mechanisms of DEAD-box helicases can be developed.
Results
Steady-state ATPase activity of DbpA
The ATPase activity of DbpA is accelerated >500-fold from <0.01 s-1 to ∼5 s-1 by PTC-RNA under our experimental conditions (Figure 1). The Michaelis constant (Km) for ATP in the presence of saturating PTC-RNA is 65 μM and the apparent Michaelis constant for 153-mer PTC-RNA in the presence of saturating ATP is 1.0 nM (Table 1), comparable to published values measured under slightly different conditions12.
Figure 1. Steady state ATPase activity of DbpA.
A. PTC-RNA concentration-dependence of DbpA steady state turnover velocity at saturating (2 mM) ATP. B. ATP concentration-dependence of DbpA steady-state turnover velocity at saturating (120 nM) PTC-RNA. The solid lines through the data points are the best fits to Eq. 9. Uncertainty bars represent the standard errors from the fits. The steady-state ATPase rate is presented as the number of ATP molecules hydrolyzed per DbpA per second.
Table 1.
| Steady-state parameters | ||
| kcat (-PTC) | < 0.01 s-1 | (NADH assay, Figure 1A) |
| kcat (+PTC) | 5.3 (± 0.2) s-1 | (NADH assay, Figure 1A) |
| 5.2 (± 0.3) s-1 | (NADH assay, Figure 1B) | |
| kcat (+PTC), predicted | 4 s-1 | (Equation 8) |
| Km, ATP (+PTC) | 65 (± 14) μM | (NADH assay, Figure 1B) |
| 82 (± 33) | (PiBiP, Figure 4) | |
| Km, ATP (+PTC) predicted | 23 μM | (Equation 8) |
| K m, PTC | 1.0 (± 0.3) nM | (NADH assay, Figure 1A) |
| Km, PTC, predicted | 5.2 nM | (Equation 8) |
| ATP binding | ||
| k +T | 1.4 (± 0.2) μM-1 s-1 | (mantATP; Figure 2C) |
| k -T | 101 (± 7) s-1 | (mantATP; Figure 2C) |
| K T | 72 (± 11) μM | (mantATP, ratio of rate constants) |
| k +RT | 1.2 (± 0.4) μM-1 s-1 | (mantATP; Figure 2C) |
| k -RT | 61 (± 19) s-1 | (mantATP; Figure 2C) |
| K RT | 51 (± 23) μM | (mantATP ratio of rate constants) |
| 86 (± 56) μM | (ATP, PiBiP, K0.5 of lag, Figure 4B) | |
| Hydrolysis & Pi release | ||
| K H | ≫ 1 | (PiBiP sequential mixing, quench flow) |
| k+RH + k-RH + k-RPi | 19.5 (± 4.1) s-1 | (PiBiP; Figure 4B) |
| 10.8 (± 3.6) s-1 | (mantATP slow rate maximum; Figure 2D) | |
| Pc = k-RH/(k-RH + k-RPi) | 0.2 | (18O Exchange) |
| k +RH | 11.9 s-1 | (18O Exchange & Pi release) |
| k -RH | 1.5 s-1 | (18O Exchange & Pi release) |
| K RH | 0.13 | (18O Exchange & Pi release) |
| k -RPi | 6.1 s-1 | (PiBiP &18O Exchange; Figure 4B) |
| k +RPi | <0.06 mM-1 s-1 | (calculated from KRPi and k-RPi) |
| K RPi | > 100 mM | (effect of Pi on mantADP release) |
| K Pi | > 100 mM | (effect of Pi on mantADP release) |
| ADP binding | ||
| k +1D | 3.3 (± 0.3) μM-1 s-1 | (mantADP; Figure 4B) |
| k -1D | 170 (± 10) s-1 | (mantADP; Figure 4B) |
| 130 (± 2) s-1 | (mantADP dissociation time course; Figure 4B) | |
| K 1D | 51 (± 6) μM | (mantADP ratio of rate constants Figure 4B) |
| K 2D | ≫1 | (no slow phase in mantADP binding) |
| k +1RD | 4.1 (± 0.2) μM-1 s-1 | (mantADP; Figure 4B) |
| k -1RD | 137 (± 20) s-1 | (mantADP; Figure4B) |
| K 1RD | 33 (± 5) μM | (mantADP ratio of rate constants; Figure 4B) |
| k +2RD | 32 (± 18) s-1 | (mantADP; Figure 4C) |
| k -2RD | 77 (± 10) s-1 | (mantADP; Figure 4C) |
| K 2RD | 2.4 (± 1.3) | (mantADP; Figure 4C) |
| K RD, overall | 23 (± 4) μM | (mantADP; Equation 3) |
| RNA bindingc | ||
| K R, overall | 16 (± 2) nM | (EMSA, data not shown) |
| K TR, overall d | 11 nM | (calculated from detailed balance) |
| K PiR, overall e | ≪ 1.5 nM | (estimated from detailed balance) |
| K 2DR, overall f | ≪25 nM | (estimated from detailed balance) |
| K 1DR, overall g | 10 nM | (calculated from detailed balance) |
Ki defined as k-i/k+i
Conditions: 75 mM KCl, 20 mM K-HEPES (pH 7.5), 5 mM MgCl2, 1 mM DTT, 25 °C.
Ki represent dissociation equilibrium constants for overall PTC-RNA binding
Calculated from KTR = KRT KR/KT
Calculated from KPiR = KTR KRH/KH
Calculated from K2DR = K1DR K2RD/K2D
Calculated from K1DR = K1RD KR/K1D
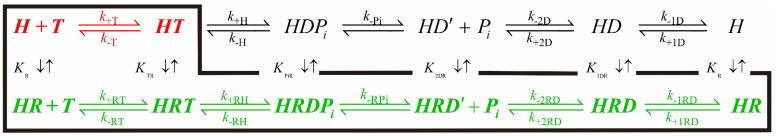
DbpA exists as a non-associating monomer in the absence19 and in the presence of PTC-RNA, during both equilibrium conditions19 and steady state ATP cycling12. We present evidence below that two distinct DbpA-ADP states exist when bound to PTC-RNA. Therefore, the minimum reaction scheme that accounts for ATPase activity (ATP binding, hydrolysis and product release) free in solution and when bound to RNA, assuming that product release is sequential and Pi dissociation precedes ADP release, is defined by ten biochemical states, fifteen equilibrium, and thirty rate constants (Scheme 1). We use this reaction scheme as a framework for interpreting the transient kinetic experiments described in the following sections. We have omitted the non-specific collision complexes20, which almost certainly exist, because they were not characterized in this study. We provide in the Appendix the explicit solution of the differential equations describing the formation and loss of the biochemical intermediates depicted in Scheme 1, and the equations used to analyze the kinetics of ATP binding, hydrolysis and product release measured in this study.
Scheme 1. The rRNA-activated DbpA ATPase cycle.
The predominant cycling pathway at saturating rRNA is highlighted in green. The intermediates populated in the absence of RNA are colored red.
Nucleotide binding
The fluorescent mant-analogs were used to measure ATP and ADP binding to DbpA and DbpA-PTC complex. Previous measurements demonstrate that even though the mant fluorophore slightly alters the kinetics and affinity of unlabelled nucleotides ∼2-fold21, mant-nucleotides can be used to reliably monitor DbpA-nucleotide binding.
Time courses of fluorescence change after mixing DbpA with mantATP (Figure 2B) or mantADP21 follow single exponentials with observed rate constants that depend linearly on the [mant-nucleotide] (Figure 2C and 3B), consistent with simple one-step binding mechanisms21, yielding association rate constants from the slopes (k+T = 1.4 μM-1 s-1 and k+D = 3.3 μM-1 s-1), dissociation rate constants from the intercepts (k-T = 101 s-1; k-D = 170 s-1) and overall equilibrium binding affinities from the ratio of rate constants (KT = 72 μM and KD = 51 μM; Table 1). The mantADP affinity (KD) is comparable to published values measured with mantADP21 or radiolabelled ADP22. The DbpA-mantADP dissociation rate constant (k-D) measured directly by competition with unlabeled ADP is 130 s-1 (data not shown and21), comparable to the value obtained from the intercept of Figure 3B. The mantADP dissociation rate constant was unaffected by 100 mM Pi (data not shown), suggesting that the affinity of Pi for DbpA-ADP is >100 mM.
Figure 2. Kinetics of mantATP binding to DbpA and DbpA·PTC-RNA.
A. Time courses of mantATP binding assayed by resonance energy transfer from DbpA tryptophans to bound mantATP. The smooth lines through the data represent the best fits to double exponentials. Final concentrations: 1 μM DbpA-PTC-RNA and (lower to upper) 30 μM, 40 μM, 60 μM and 100 μM mantATP. B. Time course of 50 μM mantATP binding to 1 μM DbpA (green) or 1 μM DbpA·PTC-RNA (blue). The solid lines are the best fits to single (DbpA) or double (DbpA-PTC) exponentials. C. [mantATP]-dependence of the fast observed rate constant of binding to DbpA ( ) or DbpA·PTC (
) or DbpA·PTC ( ). Solid lines are the fits to Eq. 33 of Appendix.. D. [mantATP]-dependence of the slow observed rate constant for binding to DbpA·PTC-RNA (
). Solid lines are the fits to Eq. 33 of Appendix.. D. [mantATP]-dependence of the slow observed rate constant for binding to DbpA·PTC-RNA ( ). The solid line represents the best fit to Eq. 34 of Supplementary Material with the apparent Kd constrained to 51 μM as predicted from the ratio of the rate constants determined from data in Panel C. Uncertainty bars represent standard errors in the fits.
). The solid line represents the best fit to Eq. 34 of Supplementary Material with the apparent Kd constrained to 51 μM as predicted from the ratio of the rate constants determined from data in Panel C. Uncertainty bars represent standard errors in the fits.
Figure 3. Kinetics of mantADP binding to DbpA-PTC-RNA.
A. Time courses of mantADP binding assayed by resonance energy transfer from DbpA tryptophans to bound mantADP. The smooth lines through the data represent the best fits to double exponentials. Final concentrations: 1 μM DbpA·PTC-RNA and (lower to upper) 10 μM, 20 μM, 30 μM, 40 μM and 60 μM mantADP. B. [mantATP]-dependence of the observed rate constant binding to DbpA and DbpA·PTC (fast phase ( ) and slow phase (
) and slow phase ( ). Both solid lines are the fits to Eq. 2. C. The [mantADP]-dependence of the slow observed rate constant of binding mantADP to DbpA-PTC-RNA. Uncertainty bars represent standard errors in the fits. The solid line is the best fit to Eq. 2. D. Time course of mantADP dissociation from DbpA-PTC-RNA after mixing an equilibrated sample of 1 μM DbpA-PTC-RNA and 125 μM mantADP with 2 mM MgADP. The smooth lines are the best fits of the data to double exponentials with a fast observed rate constant of 169.8 (± 4.5) s-1 comprising 82% of the amplitude and a slow observed rate constant of 39.7 (± 2.7) s-1.
). Both solid lines are the fits to Eq. 2. C. The [mantADP]-dependence of the slow observed rate constant of binding mantADP to DbpA-PTC-RNA. Uncertainty bars represent standard errors in the fits. The solid line is the best fit to Eq. 2. D. Time course of mantADP dissociation from DbpA-PTC-RNA after mixing an equilibrated sample of 1 μM DbpA-PTC-RNA and 125 μM mantADP with 2 mM MgADP. The smooth lines are the best fits of the data to double exponentials with a fast observed rate constant of 169.8 (± 4.5) s-1 comprising 82% of the amplitude and a slow observed rate constant of 39.7 (± 2.7) s-1.
Time courses of fluorescence change after mixing DbpA-PTC with mantATP or mantADP are biphasic and follow double exponentials (Figure 2A, 2B, 3A), indicating that binding is monitoring at least two biochemical transitions. We favor a sequential, two-step binding mechanism (Scheme 1) over a parallel reaction pathway with binding to a mixed population because the relative amplitudes of the two phases measured in association (Figure 3A) and dissociation (Figure 3D; described below) reactions differ23. Accordingly, the fast observed rate constants (kfast) are dominated by nucleotide binding and the slow observed rate constants (kslow) arise from an isomerization(s) subsequent to nucleotide binding. We assume that the two high fluorescence states have similar intensities, so the observed fluorescence reflects the sum of the concentrations of these two species. The experimentally observed rate constants are independent of the relative intensities of these states, so the outcomes of the analysis presented below are unaffected if this assumption does not apply. The 2′- and 3′-mant nucleotide isomers exist as an equilibrium mixture (60% 3′ isomer) that interconvert slowly with a half life of ∼10 minutes at pH 7.5, 25 °C24. The two isomers may bind DbpA with different kinetics, yielding the biphasic time courses, so measurements were also done with 2′-deoxy isomers labeled at the 3′ hydroxyl. The fluorescence changes associated with 2′-deoxymantATP binding are considerably smaller than with mixed 2′- and 3′- labeled isomers (data not shown), but the time courses remain biphasic, require a sum of two exponentials to describe the data, and are qualitatively similar, as reported for Rep DNA helicase24.
The fast observed rate constant (kfast) values depend linearly on the [mant-nucleotide] (Figure 2C and 3B). The fast phase is much more rapid than the slow phase in the case of mantATP binding (i.e. kfast ≫ kslow; Figure 2). Under these conditions, kfast = k+RT[mantATP] + k-RT (Eq. 33 of Appendix), permitting the mantATP association rate constant (k+RT = 1.2 μM-1 s-1) and the dissociation rate constant (k-RT = 61 s-1) to be determined from the slope and intercept of the [mantATP]-dependence of kfast, respectively (Figure 2C), and the mantATP binding affinity (KRT = 51 μM) from the ratio of the rate constants (Table 1).
The slow phase observed with mantATP represents the approach to steady state following initial binding and has contributions from ATP hydrolysis (k+RH), or more likely a conformational change preceding rapid ATP hydrolysis (these are kinetically equivalent), ATP resynthesis (k-RH), and Pi release (k-RPi). ADP release does not contribute the slow phase of ATP binding (Appendix) because it is more rapid than the preceding steps (discussed below). The limited [mantATP] range examined precludes reliable determination of the [mantATP]-dependence of the observed slow phase rate constant (kslow). However, kslow is predicted to depend hyperbolically on the [mantATP] (Eq. 34 of Appendix, discussed below) with a maximum observed rate constant at saturating [mantATP] that is equal to the sum of the ATP hydrolysis, ATP resynthesis and Pi release rate constants (k+RH + k-RH + k-RPi ∼11 s-1, Fig. 2D). Measurements presented below with unmodified ATP (Figure 4B) confirm this prediction.
Figure 4. Transient kinetics of Pi release from DbpA-PTC-RNA.
A. Time courses of transient Pi release after mixing 1 μM DbpA-PTC-RNA with excess ATP in the presence of 5 μM (final) MDCC-PiBiP. Solid lines are the best fits to Eq. 38 in Appendix. B. [ATP]-dependence of the observed rate constant that describe the lag phase component of the time courses. The solid line is the fit using Eq. 6.
The kfast ≫ kslow condition is not fulfilled with mantADP binding to DbpA-RNA (Figure 3B). Consequently, the square root approximation25 cannot be applied, so unconstrained global fitting of the [mantADP]-dependence of kfast and kslow (Figure 3B; Table 1) to the following quadratic rate constant equation(s)23:
(Eq. 1).
was used to determine the mantADP binding (k+1RD = 4.1 μM-1 s-1), dissociation (k-1RD = 137 s-1), and isomerization (k+2RD = 32 s-1, k-2RD = 77 s-1) rate constants (Table 1). The corresponding mantADP binding (K1RD) and isomerization (K2RD) equilibrium constants calculated from the ratios of the rate constants are 33 μM and 2.4, respectively (Table 1).
The rate and equilibrium constants determined from the association time courses predict that the time course of irreversible mantADP release from an equilibrated DbpA-RNA-mantADP complex will be biphasic with a fast observed rate constant (koff-fast) of ∼190 s-1 and a slow observed rate constant (koff-slow) of 55 s-1 according to the following equation(s)23:
| (Eq.2) |
Experimental time courses of irreversible mantADP release are biphasic with observed rate constants of 170 s-1 and 40 s-1 (Figure 3D), consistent with two high fluorescence DbpA-RNA-mantADP states (HRD and HRD’) that exist in a reversible equilibrium (Scheme 1) defined by the rate constants determined from the association kinetics (Table 1). Inclusion of up to 100 mM Pi does not affect dissociation mantADP from DbpA-RNA (data not shown), suggesting that the affinity of Pi binding to DbpA-ADP-RNA is > 100 mM.
The overall affinity for ADP binding to DbpA-PTC-RNA (KRD, overall), accounting for both DbpA-RNA-ADP states, calculated using the following expression 23:
| (Eq.3) |
is 23 μM. The overall affinity estimated from the [ADP]-dependence of the total transient kinetic time courses amplitudes (Figure 3A) yields a comparable value of 38 μM (data not shown).
ATP hydrolysis
Time courses of ATP hydrolysis by DbpA and DbpA-PTC-RNA were measured by chemical quench flow with 32P-ATP and by HPLC with ATP. There is no detectable hydrolysis of ATP (up to 1 mM) by DbpA in the absence of PTC-RNA. In contrast, ATP is hydrolyzed in the presence of PTC-RNA, although no significant burst phase could be resolved (data not shown). The lack of product formation with DbpA supports the interpretation that hydrolysis of bound ATP does not occur in the absence of PTC-RNA and that the slow phase of mantATP binding, present with DbpA-PTC-RNA but absent from DbpA alone (Figure 2), arises from ATP hydrolysis (or more likely an isomerization preceding rapid hydrolysis). Sequential mixing experiments of transient Pi release presented below further support this interpretation.
Pi release
Time courses of Pi release after mixing DbpA-PTC-RNA with ATP (Figure 4A) display a distinct lag phase that is well described by an exponential with negative amplitude 20 followed by a linear, steady-state phase (Eq. 38 in Appendix). The observed lag phase represents the formation of (at least one) intermediate preceding Pi release on the approach to steady state turnover with a rate constant that is comparable or slower than Pi release, consistent with the lack of a burst phase in the time course of ATP hydrolysis measured by chemical quench-flow.
The steady state rate determined from the slope of the linear phase depends hyperbolically on the [ATP] and yields a Km for ATP of 82 μM (Table 1; data not shown). The observed rate constant of the lag phase (klag) also depends hyperbolically on the [ATP] (Figure 4B). The hyperbolic [substrate]-dependence of klag eliminates mechanism in which the observed lag phase arises from an initial slow isomerization of the enzyme that limits productive ATP binding, consistent with the nucleotide binding measurements (Figure 2). The hyperbolic [ATP]-dependence of klag is approximated by (Eq. 34 in Appendix):
| (Eq.4) |
which simplifies to the sum of the ATP hydrolysis (k+RH), ATP resynthesis (k-RH) and Pi release (k-RPi) rate constants at saturating [ATP]:
| (Eq.5) |
Pi release is irreversible in the presence of phosphate binding protein, so Pi rebinding does not contribute to the observed relaxations. The intercept (i.e. limit as [ATP] approaches 0, klag,0) is proportional to:
| (Eq. 6) |
The best fit of the [ATP]-dependence of klag to a hyperbola yields a maximum observed rate constant (klag,∞) of 19.5 s-1 at saturating [ATP] and an intercept (klag,0) of 8.8 s-1. The midpoint of the hyperbola (i.e. apparent Kd, Figure 4B) is equal to the ATP affinity (KRT, Eq. 37 in Appendix) and yields a value of 86 μM, within a factor of 2 of that determined from the kinetics of mantATP binding (51 μM, Figure 2).
Time courses of Pi release after mixing saturating PTC-RNA (1 μM) with DbpA (0.8 μM) that had been mixed with saturating ATP (2 mM) and aged for various times (18 ms - 25 s) are superimposable (data not shown), independent of the ageing time, and identical to time courses of Pi release measured by mixing DbpA·PTC-RNA complex with saturating ATP (2 mM). That is, equilibrating DbpA with ATP up to 25 s does not bypass the observed lag phase in Pi release that arises from slow ATP hydrolysis (or a conformational change limiting rapid ATP hydrolysis) preceding steady-state ATP turnover. These data indicate that DbpA does not hydrolyze bound ATP during the ageing time, either because it is very slow and/or the equilibrium constant for ATP hydrolysis favors ATP synthesis (KRH = k-RH/k+RH ≫ 1), consistent with the chemical quench flow measurements, and the conclusion that RNA activates the DbpA ATPase cycle by promoting an isomerization that allows chemical cleavage of bound ATP.
Isotope Exchange
We used oxygen isotopic exchange26; 27 to determine the reversibility of the ATP hydrolysis reaction (k-RH) in the presence of PTC-RNA. If hydrolysis occurs without reversal (k-RH ≪ k-RPi) then one and only one water-derived oxygen will be incorporated into the released Pi. Reversible resynthesis of ATP from ADP and Pi before release of Pi, however, can result in the incorporation of additional oxygen atoms from water into Pi. The partition coefficient
| (Eq. 7) |
for the fraction of DbpA-RNA-ADP-Pi that partitions back to DbpA-RNA-ATP can be calculated from the extent of incorporation of these extra water oxygens into the Pi28. For the ∼50% enrichment of the water in 18O as used here, hydrolysis without reversal will incorporate one water-derived oxygen per Pi and will thus yield equal amounts of the species with 0 and 1 18O per Pi and none of the species with 2, 3 or 4 oxygens per Pi (as indicated by the fit in Figure 5 at Pc = 0). The observation of a significant amount of the species with 2 18O per Pi with DbpA (Figure 5) indicates that reversal of hydrolysis does occur. A Pc value of 0.2 was obtained as the best fit to a random mode28, which corresponds to a Pi release rate constant (k-RPi) that is ∼4 times faster than the ATP resynthesis rate constant (k-RH).
Figure 5. Distribution of 18O-labeled Pi species after hydrolysis of ATP by DbpA-PTC-RNA in 49% 18O-water.
Inset-Distribution of 18O-2 and 18O-3 species. Note that only a trace of 18O-3 species due to natural abundance will be detected if there was no exchange.
Knowledge of the Pc value, klag,∞ and klag,0 allow us to determine the values of the fundamental rate constants defining ATP hydrolysis, ATP resynthesis and Pi release using Eqs. 6-8. ATP hydrolysis (k+RH) occurs at 11.9 s-1, ATP resynthesis (k-RH) occurs at 1.5 s-1 and Pi release (k-RPi) occurs at 6.1 s-1. The equilibrium constant for ATP hydrolysis determined from the ratio of the rate constants (KRH = k-RH/k+RH) is 0.13 (Table 1).
PTC-RNA binding
The affinity of nucleotide-free DbpA for 153mer PTC-RNA (KR in Scheme, 1) determined by electrophoretic mobility shift assay under our conditions is 16 nM (Table 1, data not shown), comparable to published values measured under slightly different buffer conditions 22. The value of Km,RNA measured from [RNA]-dependent ATPase is determined by the Kd’s for RNA binding to DbpA intermediate states populated during ATP cycling (Appendix). Therefore, comparison of the Km,RNA value with the affinity of RNA in the absence of nucleotide (KR) provides information regarding the RNA affinity of populated cycling intermediates
If Km,RNA< KR, at least one nucleotide-bound intermediate that binds RNA more strongly than nucleotide-free DbpA must be populated during ATPase cycling. If Km,RNA> KR, at least one populated intermediate binds RNA more weakly than nucleotide-free DbpA. The PTC-RNA binding affinity in the absence of nucleotide (16 nM) is larger than the Km,RNA value measured during steady-state cycling (1 nM), indicating that at least one nucleotide-bound intermediate state(s) populated during steady-state cycling binds PTC-RNA with a higher affinity (< 16 nM) than nucleotide-free DbpA.
Knowledge of the affinities of nucleotide-free DbpA for PTC-RNA (KR) and nucleotide for DbpA (KT and KD) and DbpA-PTC-RNA (KRT and KRD) provides the information required to explain in thermodynamic terms how nucleotide bound to DbpA modulates the RNA binding affinity and vice-versa (i.e. how the two binding sites are linked/coupled). Detailed balance allows the PTC-RNA affinity of DbpA with bound ATP to be calculated from KTR = KR KRT/KT (Scheme 1, Table 1). The affinities of DbpA with bound ADP (K1DR and K2DR) or ADP-Pi (KPiR) for PTC-RNA can be determined in a similar manner (Table 1).
Initial ATP or ADP binding weakly affects the affinity of DbpA for PTC-RNA and vice versa (KT ∼ KRT, KD ∼ KRD, and KR ∼ KTR ∼ K1RD), indicating that RNA and ATP or ADP binding are weakly coupled in DbpA. DbpA with bound ADP-Pi binds PTC-RNA with a ≫10-fold higher affinity than nucleotide free DbpA or DbpA with bound ATP or ADP, revealing that strong allosteric RNA and nucleotide binding linkage exists when ADP-Pi is bound to DbpA. This observation suggests that DbpA-RNA with bound ADP-Pi is significantly populated during steady-state ATP cycling and contributes to the low Km,RNA value. We hypothesize that formation of an “activated”, hydrolysis-competent, strong ATP binding state (omitted from Scheme 1 for simplicity) precedes and limits rapid chemical cleavage of the ATP γ-phosphate bond 29; 30; 31 and that this state resembles and behaves similar to DbpA-ADP-Pi with respect to the strong RNA binding affinity. The activated, ATP hydrolysis competent and post-hydrolysis ADP-Pi states of DbpA may be mimicked to some extent by AMPpNp binding, which increases the PTC-RNA binding affinity of DbpA22, though more work needs to be done to confirm that DbpA-PTC with bound AMPpNp reliably mimics an on-pathway reaction intermediate.
Distribution of Biochemical States
Knowledge of the ATPase cycle rate and equilibrium constants permits determination of the steady-state distribution of the biochemical intermediates populated during ATP cycling (Figure 6). The predominantly populated intermediates under our experimental conditions are the ADP-Pi-bound (∼53%) and the ATP-bound (∼33%) states. The significant populated mole fraction of the ADP-Pi-bound state and it’s high affinity for PTC-RNA explains why the Km,RNA value measured from steady-state ATPase cycling activity is smaller than the binding affinity in the absence of nucleotide (KR). However, the distribution of biochemical states populated under in vivo nucleotide concentrations (ATP ∼3 mM, ADP ∼0.25 mM and Pi ∼5 mM)32 shifts to the ADP-bound state (∼ 60%), with ADP-Pi- and ATP-bound states comprising a smaller fraction (∼25% and ∼15%, respectively) than under our experimental conditions, indicating that the weak RNA binding conformations of DbpA are predominantly (∼75%) populated in vivo.
Figure 6. Distribution of the DbpA-RNA biochemical states populated during steady-state ATP cycling.
HR represents DbpA- RNA complex, HRT represents DbpA- RNA-ATP, HRDPi represents DbpA-RNA-ADP-Pi, and HRDT represents the sum of both DbpA-RNA-ADP states depicted in Scheme 1.
Discussion
DbpA ATPase activity and activation by PTC-RNA
PTC-RNA accelerates the maximum DbpA ATPase activity (kcat) ∼500-fold from <0.01 s-1 to ∼ 5 s-1 (Table 1). Nucleotide (ATP and ADP) binding and dissociation from DbpA are very rapid (Table 1), indicating that ATP turnover by DbpA in the absence of PTC-RNA is limited by some other step(s) in the cycle (Scheme 1, top pathway) such as ATP hydrolysis and/or Pi release. Time courses of mantATP and mantADP binding to DbpA with bound PTC-RNA are biphasic, in contrast to only one phase in the absence of PTC-RNA. The observed fast phases reflect nucleotide binding and dissociation and are similar in the presence and absence of PTC-RNA (Table 1), indicating that PTC-RNA has minimal effects on nucleotide binding to DbpA; rather, PTC-RNA binding promotes an isomerization after ATP or ADP binding. The significance of the additional ADP binding state is unclear and because it is weakly populated (<10% mole fraction) during steady-state ATP cycling, we will not consider it further in this study. However, the slow phase observed with mantATP reflects ATP hydrolysis, ATP resynthesis, and Pi release, indicating that PTC-RNA activates the DbpA ATPase activity by promoting any or all of these transitions. The high DbpA-PTC-RNA binding affinities22 (Table 1) and the rapid rates of rebinding (data not shown) implies that dissociation of the complex is negligible22 during steady state cycling and that cycling exclusively along the lower pathway depicted in Scheme 1 when PTC-RNA is present.
PTC-RNA activates the steady-state ATPase activity of DbpA by shunting DbpA along a different ATP hydrolysis pathway (lower path in Scheme 1). In the absence of PTC-RNA, DbpA does not hydrolyze bound ATP to an appreciable extent, suggesting a slow, rate-limiting ATP hydrolysis that is accelerated by PTC-RNA. Alternatively, ATP hydrolysis by DbpA could be rapid, but the equilibrium largely favor the ATP state (i.e. ATP resynthesis is more rapid than hydrolysis and neither are rate limiting). Our data cannot distinguish between these two possibilities. Nonetheless, the outcome is the same: DbpA without bound PTC-RNA binds but does not hydrolyze ATP. Because both DbpA and DbpA-PTC-RNA bind ATP, but no hydrolysis occurs without RNA, PTC-RNA binding must induce conformational rearrangement of DbpA that allows ATP hydrolysis to occur. PTC-RNA may also modulate the rate of Pi release from DbpA. However, PTC-RNA activation of DbpA ATPase activity is achieved by promoting ATP hydrolysis, which does not occur significantly in its absence.
The PTC-RNA activated steady-state ATPase cycle of DbpA is limited both by a step after ATP binding (interpreted as a step preceding rapid ATP hydrolysis) and Pi release, which occur with comparable rates. ATP hydrolysis is readily reversible, but Pi release (∼6 s-1) occurs ∼4-fold more quickly than ATP resynthesis (∼1.5 s-1). Pi release can be considered to be essentially irreversible since rebinding is slow and overall binding weak (Table 1). Slow ATP hydrolysis and Pi release causes the ATP- and ADP-Pi-bound intermediates to be predominantly populated during steady-state ATP cycling under our conditions (2 mM ATP, <10 μM ADP). However, the distribution favors population of the ADP-bound states under physiological nucleotide concentrations (∼3 mM ATP, ∼250 μM ADP).
The steady-state ATPase parameters (kcat, Km,ATP, and Km,RNA) are defined in terms of individual ATPase cycle rate constants according to the following expressions (Eqs. 25, 26 and 55 of Appendix):
| (Eq. 8) |
The values of these parameters predicted from the experimentally determined rate and equilibrium constants are comparable to the experimentally determined values (kcat predicted ∼4 s-1, observed ∼5 s-1; Km,ATP predicted ∼23 μM, observed ∼65 μM; and Km,RNA predicted ∼5 nM, observed ∼1 nM; Table 1), indicating that the applied model and analysis are valid, and that the experimentally determined rate and equilibrium constants are consistent with the overall steady-state ATPase cycling behavior of DbpA. When we account for the ADP (7 μM) present under our experimental conditions (Eq. 42 of Appendix), the predicted Km,ATP value increases slightly (∼27 μM; Table 1), while the kcat and Km,RNA are unaffected since they are independent of [ADP]. Under physiological nucleotide conditions (3 mM ATP, 250 μM ADP; Figure 6), the apparent Km,ATP increases dramatically to 190 μM due to competition with ADP, accounting for the shift in the distribution of populated ATP- and ADP-bound states (Figure 6).
Communication between RNA and nucleotide binding sites
PTC-RNA does not affect the affinity of DbpA for ATP or ADP, and as predicted from thermodynamic linkage, neither ATP nor ADP binding significantly affects the affinity of DbpA for PTC-RNA. That is, nucleotide (ATP and ADP) and PTC-RNA binding are weakly coupled. This does not mean that there are no nucleotide- or RNA-dependent conformational changes in DbpA. However, if significant conformational rearrangement of DbpA is coupled to nucleotide or PTC-RNA binding alone, these do not affect binding of the other ligand. The structures of the DEAD-box protein, UAP56, with and without bound ADP33 are consistent with small structural perturbations being coupled to nucleotide binding. There is no high resolution structure available of a DEAD-box protein with bound RNA and no bound adenine nucleotide. The weak coupling predicts that if the DbpA nucleotide binding site were perturbed by PTC-RNA binding, it would be in a way that does not affect initial ATP or ADP binding.
In contrast, strong thermodynamic coupling is displayed with ADP-Pi and RNA binding. DbpA with bound ADP-Pi binds PTC-RNA with > 10-fold higher affinity than in the absence of nucleotide, suggesting that DbpA undergoes significant conformational change when both RNA and ADP-Pi are bound. The high-resolution crystal structure of the Vasa DEAD-box protein with bound RNA and AMPpNp shows a closure of the nucleotide-binding cleft34. Binding of AMPpNp may mimic an intermediate populated after initial ATP binding and preceding Pi release, though the possibility that it represents an off-pathway intermediate cannot be eliminated. Therefore, it is likely that the conformational change that occurs after initial ATP binding to DbpA is a closure of the nucleotide-binding cleft separating the two RecA domains that comprise the DbpA helicase core domain19; 35. Cleft closure would promote non-covalent interactions between bound Mg2+ and ATP with the conserved helicase motifs across the cleft of the DbpA core domain34, which would favor strong Mg2+ and ATP binding and formation of the ATP hydrolysis competent, strong PTC-RNA binding conformation (not shown in Scheme 1, discussed above). The hydrolysis-competent ATP state likely binds PTC-RNA and nucleotide with high affinity as does the post-hydrolysis, ADP-Pi conformation (Scheme 1).
Our analysis confirms previous quantitative equilibrium binding measurements revealing the weak coupling between ADP and PTC-RNA binding22. Strong coupling between ATP and PTC- RNA binding to DbpA was concluded from the same study22. However, as clarified by the authors, this conclusion was based on measurements done with AMPpNp, which binds differently than ATP22. DbpA with bound AMPpNp may better represent a chemical state populated after initial ATP binding (an activated ATP state omitted from Scheme 1 for simplicity, discussed below) and preceding product release.
ATPase cycle and free energy coupling
Unwinding of duplex RNA by DbpA is coupled to ATP utilization2;19. The ATP-dependence of efficient rRNA duplex unwinding by DbpA favors models in which destabilization and unwinding are coupled to one or more reaction steps along the DbpA ATPase cycle7; 11. Since nucleotide binding has minimal effects on PTC-RNA binding affinity (i.e. ATP and ADP binding processes are weakly coupled to PTC-RNA binding), it is very unlikely that large conformational rearrangement or unwinding of RNA occurs during initial ATP and ADP binding. On the other hand, the transiently populated DbpA-ADP-Pi state (and/or the strong ATP binding, hydrolysis competent state discussed above) binds rRNA ≫15-fold more strongly, indicating nucleotide-linked conformational rearrangement of the DbpA-PTC-RNA complex.
The free energy change associated with hydrolysis of ATP (ΔGo’ hydrolysis) in solution and at standard state is ∼ -40 kJ mol-1 under our conditions36. In contrast, the total free energy change (ΔGo’ tot) associated with one catalytic turnover of ATP by DbpA-PTC-RNA, which reflects the sum of the free energy change for ATP hydrolysis (∼ -40 kJ mol-1) and the free energy change associated with work production37, is < -0.5 kJ mol-1 (ΔGo’tot = ΔGo’RT + ΔGo’RH - ΔGo’RPi -ΔGo’2RD - ΔGo 1RD; Table 2), considerably less than the value of ΔGo’hydrolysis. The difference between ΔGo’hydrolysis and ΔGo’tot indicates that the hydrolysis free energy is transferred to and stored in the DbpA-PTC-RNA complex, suggesting that the final product is a high energy, DbpA-PTC-RNA complex in which the RNA is destabilized, unwound or unfolded and/or the DbpA is in a strained conformation. If the coupling efficiency is high and used to disrupt RNA secondary structure, a single consumed ATP molecule could destabilize a maximum of 4-5 duplex RNA base pairs38. The reported 10-mer rRNA duplex unwinding rate constant of DbpA is much slower39 (< 0.01 s-1) than ATP turnover. The large difference suggests that futile (i.e. nonproductive) ATPase cycling occurs, presumably due to competitive reannealing of the partially unwound >4-5 base pair long segment.
Table 2.
Equilibrium constants and standard Gibbs free energy changes associated with DbpA, PTC-RNA (153 mer) and nucleotide association at 25 °C
| K d | ΔGo’assoc (kJ mol-1) | |
|---|---|---|
| K T | 72 μM | -23.6 |
| K RT | 51 μM | -24.5 |
| K H | ≫ 1 | ≫ 0 |
| K RH | 0.13 | -5.1 |
| K Pi | > 100 mM | < 5.7 |
| K RPi | > 100 mM | < 5.7 |
| K 2D | ≫ 1 | ≫ 0 |
| K 2RD | 2.4 | 2.2 |
| K 1D | 51 μM | -24.5 |
| K 1RD | 33 μM | -25.6 |
| K R | 16 nM | -44.5 |
| K TR | 11 nM | -45.3 |
| K PiR | ≪ 1.5 nM | ≪ -50.4 |
| K 1DR | 10 nM | -45.6 |
| K 2DR | ≪ 25 nM | ≪ -43.4 |
K1DR = K1RD KR/K1D
KTR = KRT KR/KT
KPiR = KTR KRH/KH
K2DR = K1DR K2RD/K2D
We propose that the strong PTC-RNA binding transition is linked to rRNA duplex destabilization. The slow hydrolysis rate (i.e. large activation energy) and large PTC-RNA binding free energy change (Table 2) of this transition is consistent with significant conformational rearrangement of the DbpA-PTC-RNA complex. Consistent with the proposed destabilization mechanism, the high-resolution structure of the DEAD-box RNA helicase, Vasa34, with bound ssRNA and AMPpNp undergoes a significant conformational change that favors closing of the nucleotide binding cleft and bending of ssRNA that precludes formation of Watson-Crick base pairing with a complementary strand. The observation that DbpA with bound AMPpNp strongly binds but does not unwind PTC-RNA duplexes7 seems at variance with the proposed pathway. However, this observation can be accounted for if DbpA-AMPpNp populated a strong RNA binding state that destabilized short rRNA duplexes but did not dissociate the destabilized strands.
Scheme 1 and the experimental rate and equilibrium constants (Table 1) are consistent with strong PTC-RNA binding coinciding with an isomerization after ATP binding, interpreted as being a conformational change that precedes and limits ATP cleavage. Subsequent product release favors formation of the weak RNA binding conformation (Tables 1 and 2). Weak RNA binding enables DbpA to transfer the unwound rRNA product to other factors (e.g. the ribosomal proteins associated with the 50S subunit of the ribosome) and regenerate free DbpA capable of initiating another round of catalysis.
Materials and Methods
Reagents and Proteins
All chemicals and reagents were the highest purity commercially available. Millipore MlliQ® dispensed water that had been treated with DEPC for 8 hours and autoclaved was used in all procedures. RNAse activity was undetectable in all reagents, buffers and protein preparations. ATP (99+% purity as assayed by HPLC, data not shown) was purchased from Roche Molecular Biochemicals (Indianapolis, IN) and ADP (Sigma A-5285, 99+% purity by HPLC, data not shown) was purchased from Sigma (St. Louis, MO). Nucleotide concentrations were determined by absorbance using ε 259 of 15,400 M-1 cm-1. The N-methylanthraniloyl (mant) derivatives of ADP, 2′-deoxyADP, ATP, and 2′-deoxyATP were synthesized23 and concentrations determined using ε255 of 23,300 M-1 cm-1. A molar equivalent of MgCl2 was added to nucleotides immediately before use.
DbpA was overexpressed and purified as described 2; 21. The [DbpA] was determined by extinction coefficient method21; 40. MDCC (7-diethylamino-3-((((2- maleimidyl)ethyl)amino)carbonyl)coumarin)-labeled phosphate binding protein (PiBiP) was expressed, purified and labeled as described41.
RNA synthesis
A DNA fragment of 153 bp containing a sequence from the E. coli 23S rRNA (2454-2606 nt) was cloned into an EcoRI and BamHI restriction site of (pTZ-18R). PTC-RNA was prepared by in-vitro transcription of BamHI digested pTZ-18R-PTC with T7 RNA polymerase. Transcripts were treated with DNase I, phenol extracted (pH 4.7), passed through a G-50 mini-spun column (Roche Molecular Biochemical) and precipitated in ethanol. [RNA] were calculated using ε 260 = 8600 M-1 cm-1 nucleotide-1. PTC-RNA was refolded12 in 50 mM K-HEPES (pH 7.5), 50 mM KCl, 5 mM MgCl2.
Steady state ATPase activity
The steady-state ATPase activity of DbpA (2-20 nM) was measured by absorbance (λ = 340 nm) using the ATP regenerating, NADH coupled assay 12; 42at 25 (±0.1) °C in KMg75 buffer (75 mM KCl, 20 mM K-HEPES (pH 7.5), 5 mM MgCl2, 1 mM DTT) supplemented with saturating (2 mM) ATP while varying the [PTC-RNA], or with saturating (120 nM) PTC-RNA and varying the [ATP]. The [RNA] or [ATP]-dependence of the steady state ATPase rate (V in units of ATP sec-1 DbpA-1) was fitted to the quadratic form of the Briggs-Haldane equation:
| (Eq. 9) |
where ko is the ATPase rate of DbpA alone, kcat is the turnover rate at saturating [S], Km is the apparent Michaelis constant for substrate activation, [H]tot is the total [DbpA], and and [S]tot is the total [PTC-RNA] or [ATP]. This equation assumes a 1:1 binding stoichiometry, which has been confirmed for PTC-RNA-activated ATPase activity of DbpA12. The [ADP] under our conditions of 2 mM ATP is ∼7 μM 42.
Pre-steady state kinetic analysis
All transient kinetic fluorescence measurements were made in KMg75 buffer using an Applied Photophysics SX.18MV-R stopped-flow apparatus thermostatted at 25 ± 0.1 °C. The concentrations stated are final concentrations after mixing. Time courses were fitted to a sum of exponentials by nonlinear least squares fitting using Pro-K software provided with the instrument. Uncertainties are reported as standard errors in the fits unless stated otherwise.
Mant-nucleotide binding kinetics were measured by energy transfer from DbpA tryptophans (λex = 280 nm) to bound mant-nucleotide (fluorescence monitored at 90° through a 400-nm long-pass colored glass filter) under pseudo-first order conditions with [nucleotide] ≫ [DbpA] or [DbpA-RNA]21. Transient Pi release was measured using the fluorescently labeled mutant43 of PiBiP. Background Pi was removed from all solutions, syringes and the instrument with 7-methylguanosine (0.5-1 mM) and purine nucleoside phosphorylase (0.1 units mL-1). There was ∼5-fold enhancement in the fluorescence of MDCC-PiBiP with Pi binding (λex = 436 nm, 460 nm long pass emission filter). The rate and equilibrium constants of Pi binding to MDCC-PiBiP under experimental conditions comparable to those used in this study are k+ = 117 μM-1 s-1, k- = 24 s-1, and Kd = 0.20 μM41.
Time courses of [α-32P]ATP hydrolysis were made at 25 °C in KMg75 buffer with a KINTEK (Austin, Texas) RQF-3 quench flow apparatus31. Experiments were done by mixing 23 μM DbpA or with 23 μM DbpA-PTC RNA 51mer with 20 μM ATP, ageing for various times then quenching with 5 M formic acid. Samples were spotted (2 μL) on a Cellulose F TLC plates (Analtech, Newark, DE) and resolved in 0.6 M KH2PO4 pH 3.4 for 45 min. Plates were exposed to phosphor screen, read using Storm PhosphorImager®, Molecular Dynamic (GE Healthcare), and quantitated using ImageQuant software, Molecular Dynamic (GE Healthcare). Formation of ADP from the hydrolysis of ATP (1 mM) by DbpA was also measured by HPLC44.
Measurement of Oxygen Isotopic Exchange during ATP Hydrolysis
Hydrolysis of ATP was performed in KMg75 buffer containing 49% [18O]-water supplemented with 2 mM ATP, 4 mM phosphoenolpyruvate, and 100 units mL-1 pyruvate kinase to regenerate ATP. The reactions were quenched with acid and the Pi was isolated and analyzed for 18O content27.
Kinetic modeling
Simulations of reaction time courses and equilibrium distribution of biochemical states were performed with Tenua (provided by Dr. D. Wachsstock, available free at http://www.geocities.com/tenua4java/), which is based on the kinetic simulation program KINSIM developed by Carl Frieden and colleagues45.
Supplementary Material
Acknowledgement
We thank Dr. Eckhard Jankowsky (Case Western Reserve) for comments on the manuscript. This work is supported by grants from the National Science Foundation (MCB-0546353), National Institutes of Health (GM071688), and American Heart Association (0655849T) to E.M.D.L.C. W.C. is supported by American Heart Association postdoctoral fellowship award 0625997T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Henn A, Medalia O, Shi SP, Steinberg M, Franceschi F, Sagi I. Visualization of unwinding activity of duplex RNA by DbpA, a DEAD box helicase, at single-molecule resolution by atomic force microscopy. Proc Natl Acad Sci U S A. 2001;98:5007–5012. doi: 10.1073/pnas.071372498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:3569–74. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–4. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 5.Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. Birth of the D-E-A-D box. Nature. 1989;337:121–2. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 6.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–41. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 7.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. Embo J. 2001;20:5503–12. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–6. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 9.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linder P. Dead-box proteins: a family affair-active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–80. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is a 3′ -> 5′ RNA helicase. Biochemistry. 2005;44:7903–11. doi: 10.1021/bi050033x. [DOI] [PubMed] [Google Scholar]
- 12.Tsu CA, Uhlenbeck OC. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–96. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 13.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23S rRNA. Embo J. 1993;12:3619–26. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–90. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 15.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 16.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–47. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1 A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–93. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 18.von Hippel PH, Delagoutte E. Macromolecular complexes that unwind nucleic acids. Bioessays. 2003;25:1168–77. doi: 10.1002/bies.10369. [DOI] [PubMed] [Google Scholar]
- 19.Talavera MA, Matthews EE, Eliason WK, Sagi I, Wang J, Henn A, De La Cruz EM. Hydrodynamic characterization of the DEAD-box RNA helicase DbpA. J Mol Biol. 2006;355:697–707. doi: 10.1016/j.jmb.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 20.Robblee JP, Cao W, Henn A, Hannemann DE, De La Cruz EM. Thermodynamics of nucleotide binding to actomyosin V and VI: a positive heat capacity change accompanies strong ADP binding. Biochemistry. 2005;44:10238–49. doi: 10.1021/bi050232g. [DOI] [PubMed] [Google Scholar]
- 21.Talavera MA, De La Cruz EM. Equilibrium and kinetic analysis of nucleotide binding to the DEAD-box RNA helicase DbpA. Biochemistry. 2005;44:959–70. doi: 10.1021/bi048253i. [DOI] [PubMed] [Google Scholar]
- 22.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 23.Hannemann DE, Cao W, Olivares AO, Robblee JP, De La Cruz EM. Magnesium, ADP, and actin binding linkage of myosin V: evidence for multiple myosin V-ADP and actomyosin V-ADP states. Biochemistry. 2005;44:8826–40. doi: 10.1021/bi0473509. [DOI] [PubMed] [Google Scholar]
- 24.Moore KJ, Lohman TM. Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 1 Use of fluorescent nucleotide analogues. Biochemistry. 1994;33:14550–64. doi: 10.1021/bi00252a023. [DOI] [PubMed] [Google Scholar]
- 25.Johnson KA. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- 26.Hackney DD, Rosen G, Boyer PD. Subunit interaction during catalysis: alternating site cooperativity in photophosphorylation shown by substrate modulation of [18O]ATP species formation. Proc Natl Acad Sci U S A. 1979;76:3646–50. doi: 10.1073/pnas.76.8.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olivares AO, Chang W, Mooseker MS, Hackney DD, De La Cruz EM. The tail domain of myosin Va modulates actin binding to one head. J Biol Chem. 2006;281:31326–36. doi: 10.1074/jbc.M603898200. [DOI] [PubMed] [Google Scholar]
- 28.Hackney DD, Stempel KE, Boyer PD. Oxygen-18 probes of enzymic reactions of phosphate compounds. Methods Enzymol. 1980;64:60–83. doi: 10.1016/s0076-6879(80)64005-1. [DOI] [PubMed] [Google Scholar]
- 29.Wong I, Lohman TM. A two-site mechanism for ATP hydrolysis by the asymmetric Rep dimer P2S as revealed by site-specific inhibition with ADP-A1F4. Biochemistry. 1997;36:3115–25. doi: 10.1021/bi9621977. [DOI] [PubMed] [Google Scholar]
- 30.Yengo CM, De la Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of myosin V. Biochemistry. 2002;41:8508–17. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]
- 31.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci U S A. 1999;96:13726–31. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.John L, Ingraham FCN. Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology. ASM Press; Washington, D.C.: 1987. [Google Scholar]
- 33.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci U S A. 2004;101:17628–33. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 35.Henn A, Shi SP, Zarivach R, Ben-Zeev E, Sagi I. The RNA Helicase DbpA Exhibits a Markedly Different Conformation in the ADP-bound State When Compared with the ATP- or RNA-Bound States. J Biol Chem. 2002;277:46559–46565. doi: 10.1074/jbc.M207438200. [DOI] [PubMed] [Google Scholar]
- 36.Phillips RC, George P, Rutman RJ. Thermodynamic data for the hydrolysis of adenosine triphosphate as a function of pH, Mg2+ ion concentration, and ionic strength. J Biol Chem. 1969;244:3330–42. [PubMed] [Google Scholar]
- 37.Jencks WP. The utilization of binding energy in coupled vectorial processes. Adv Enzymol Relat Areas Mol Biol. 1980;51:75–106. doi: 10.1002/9780470122969.ch2. [DOI] [PubMed] [Google Scholar]
- 38.Xia T, SantaLucia J, Jr., Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–35. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 39.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–9. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 41.Henn A, De La Cruz EM. Vertebrate myosin VIIb is a high duty ratio motor adapted for generating and maintaining tension. J Biol Chem. 2005;280:39665–76. doi: 10.1074/jbc.M507667200. [DOI] [PubMed] [Google Scholar]
- 42.De La Cruz EM, Sweeney HL, Ostap EM. ADP inhibition of myosin V ATPase activity. Biophys J. 2000;79:1524–9. doi: 10.1016/S0006-3495(00)76403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–71. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 44.Samizo K, Ishikawa R, Nakamura A, Kohama K. A highly sensitive method for measurement of myosin ATPase activity by reversed-phase high-performance liquid chromatography. Anal Biochem. 2001;293:212–5. doi: 10.1006/abio.2001.5146. [DOI] [PubMed] [Google Scholar]
- 45.Barshop BA, Wrenn RF, Frieden C. Analysis of numerical methods for computer simulation of kinetic processes: development of KINSIM--a flexible, portable system. Anal Biochem. 1983;130:134–45. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- 46.Gutfreund H. Kinetics for the Life Sciences: Receptors,transmitters and catalysts. Cambridge University Press; Cambridge: 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.