Abstract
Glutamate dehydrogenase (L-glutamate:NADP+ oxidoreductase [deaminating], EC 1.4.1.4) has been purified from Escherichia coli B/r. The purity of the enzyme preparation has been established by polyacrylamide gel electrophoresis, ultracentrifugation, and gel filtration. A molecular weight of 300,000 +/- 20,000 has been calculated for the enzyme from sedimentation equilibrium measurements. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate and sedimentation equilibrium measurements in guanidine hydrochloride have revealed that glutamate dehydrogenase consists of polypeptide chains with the identical molecular weight of 50,000 +/- 5,000. The results of molecular weight determination lead us to propose that glutamate dehydrogenase is a hexamer of subunits with identical molecular weight. We also have studied the stability and kinetics of purified glutamate dehydrogenase. The enzyme remains active when heat treated or when left at room temperature for several months but is inactivated by freezing. The Michaelis constants of glutamate dehydrogenase are 1,100,640, and 40 muM for ammonia, 2-oxoglutarate, and reduced nicotinamide adenine dinucleotide phosphate, respectively.
Full text
PDF

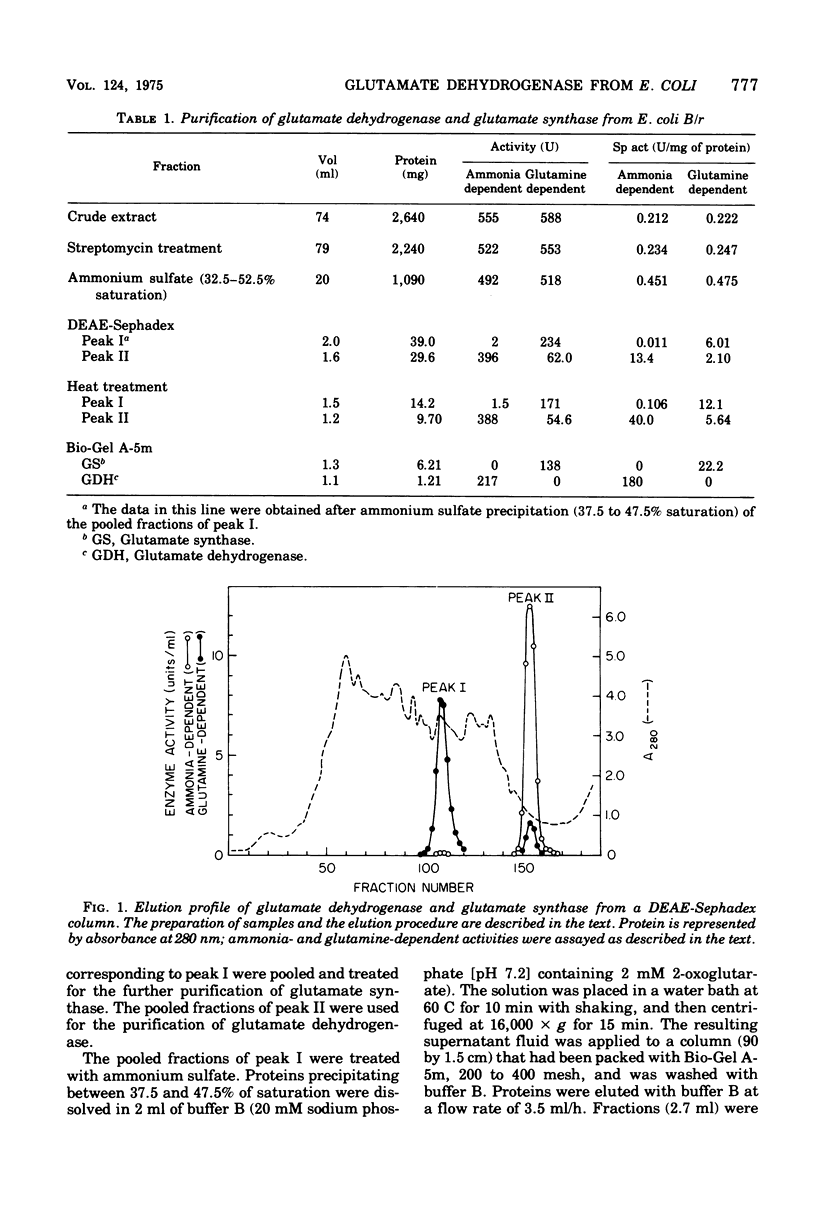

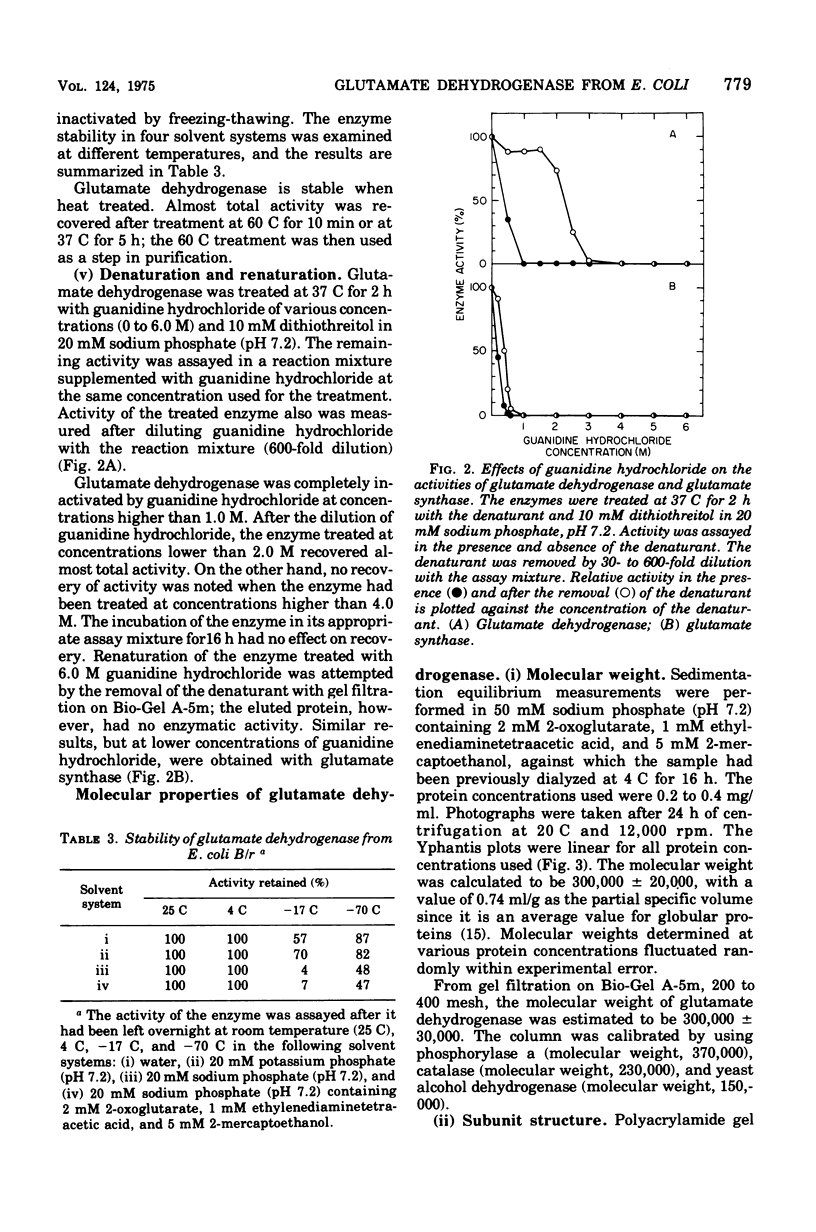
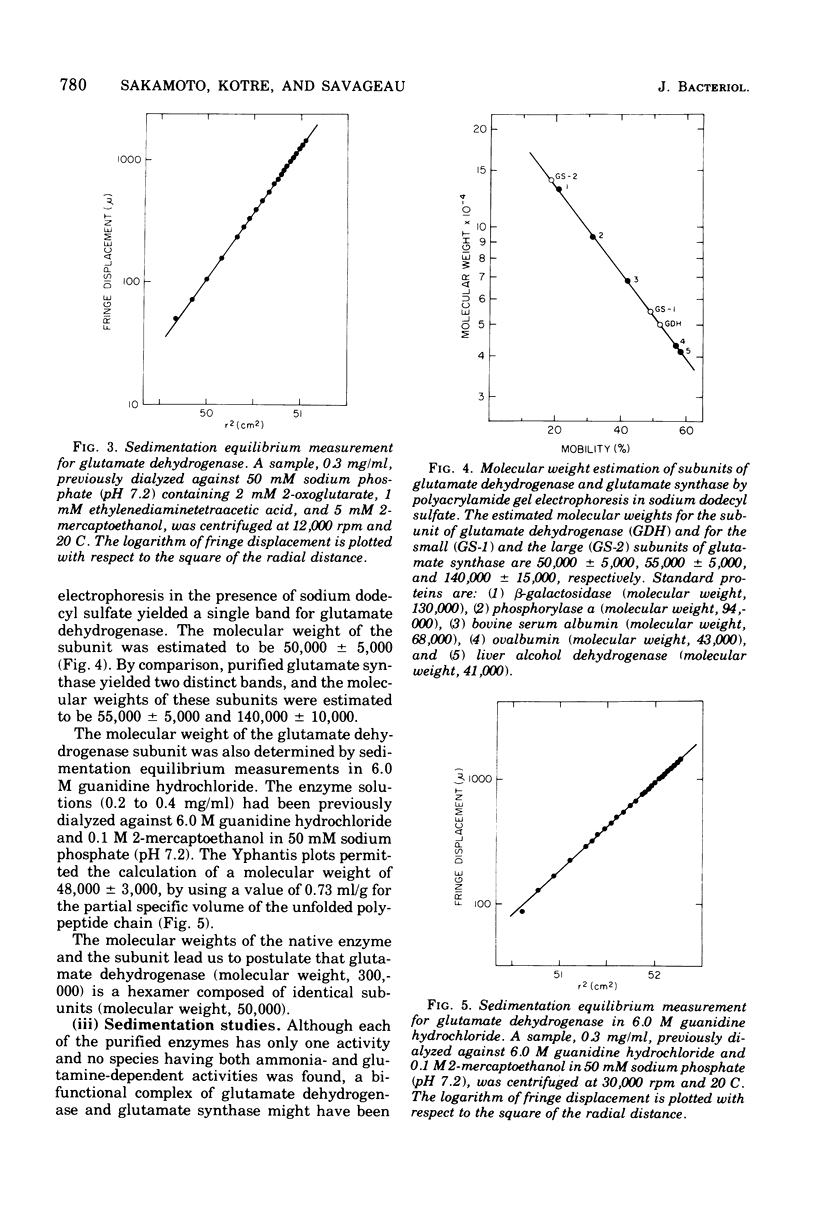
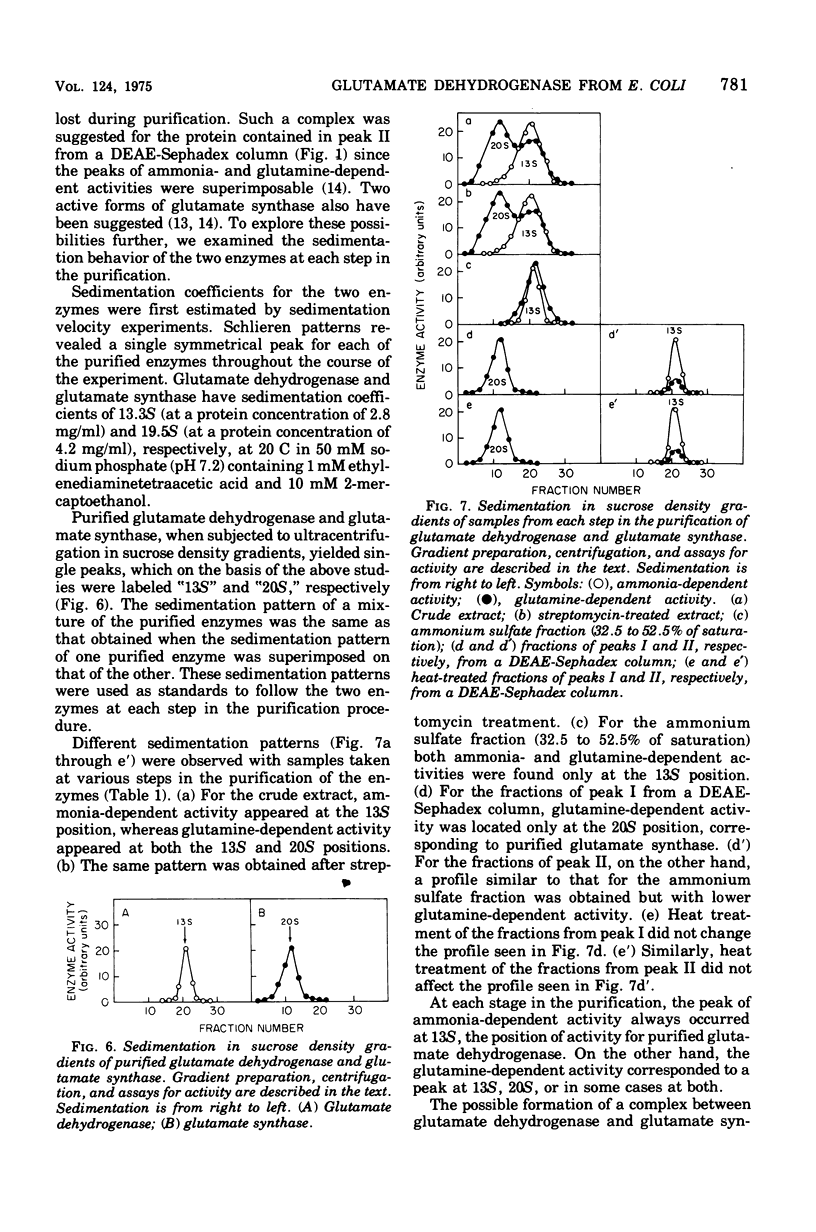


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J. 1965 Sep;96(3):595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal K. M., Smith E. L. Nicotinamide adenine dinucleotide phosphate-specific glutamate dehydrogenase of Neurospora. I. Isolation, subunits, amino acid composition, sulfhydryl groups, and identification of a lysine residue reactive with pyridoxal phosphate and N-ethylmaleimide. J Biol Chem. 1973 Sep 10;248(17):6002–6008. [PubMed] [Google Scholar]
- Coulton J. W., Kapoor M. Purification and some properties of the glutamate dehydrogenase of Salmonella typhimurium. Can J Microbiol. 1973 Apr;19(4):427–438. doi: 10.1139/m73-071. [DOI] [PubMed] [Google Scholar]
- Coulton J. W., Kapoor M. Studies on the kinetics and regulation of glutamate dehydrogenase of Salmonella typhimurium. Can J Microbiol. 1973 Apr;19(4):439–450. doi: 10.1139/m73-072. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Davies G. E., Stark G. R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc Natl Acad Sci U S A. 1970 Jul;66(3):651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. M., Westlake D. W. Purification and characterization of glutamic acid dehydrogenase and -ketoglutaric acid reductase from Peptococcus aerogenes. Can J Microbiol. 1972 Jun;18(6):881–892. doi: 10.1139/m72-136. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Miller R. E., Stadtman E. R. Glutamate synthase from Escherichia coli. An iron-sulfide flavoprotein. J Biol Chem. 1972 Nov 25;247(22):7407–7419. [PubMed] [Google Scholar]
- Savageau M. A., Kotre A. M., Sakamoto N. A possible role in the regulation of primary animation for a complex of glutamine: -ketoglutarate amidotransferase and glutamate dehydrogenase in Escherichia coli. Biochem Biophys Res Commun. 1972 Jul 11;48(1):41–47. doi: 10.1016/0006-291x(72)90341-5. [DOI] [PubMed] [Google Scholar]
- Veronese F. M., Nyc J. F., Degani Y., Brown D. M., Smith E. L. Nicotinamide adenine dinucleotide-specific glutamate dehydrogenase of Neurospora. I. Purification and molecular properties. J Biol Chem. 1974 Dec 25;249(24):7922–7928. [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Winnacker E. L., Barker H. A. Purification and properties of a NAD-dependent glutamate dehydrogenase from Clostridium SB4. Biochim Biophys Acta. 1970 Aug 15;212(2):225–242. doi: 10.1016/0005-2744(70)90203-2. [DOI] [PubMed] [Google Scholar]
- YPHANTIS D. A. EQUILIBRIUM ULTRACENTRIFUGATION OF DILUTE SOLUTIONS. Biochemistry. 1964 Mar;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]
- Yarrison G., Young D. W., Choules G. L. Glutamate dehydrogenase from Mycoplasma laidlawii. J Bacteriol. 1972 May;110(2):494–503. doi: 10.1128/jb.110.2.494-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]


