Abstract
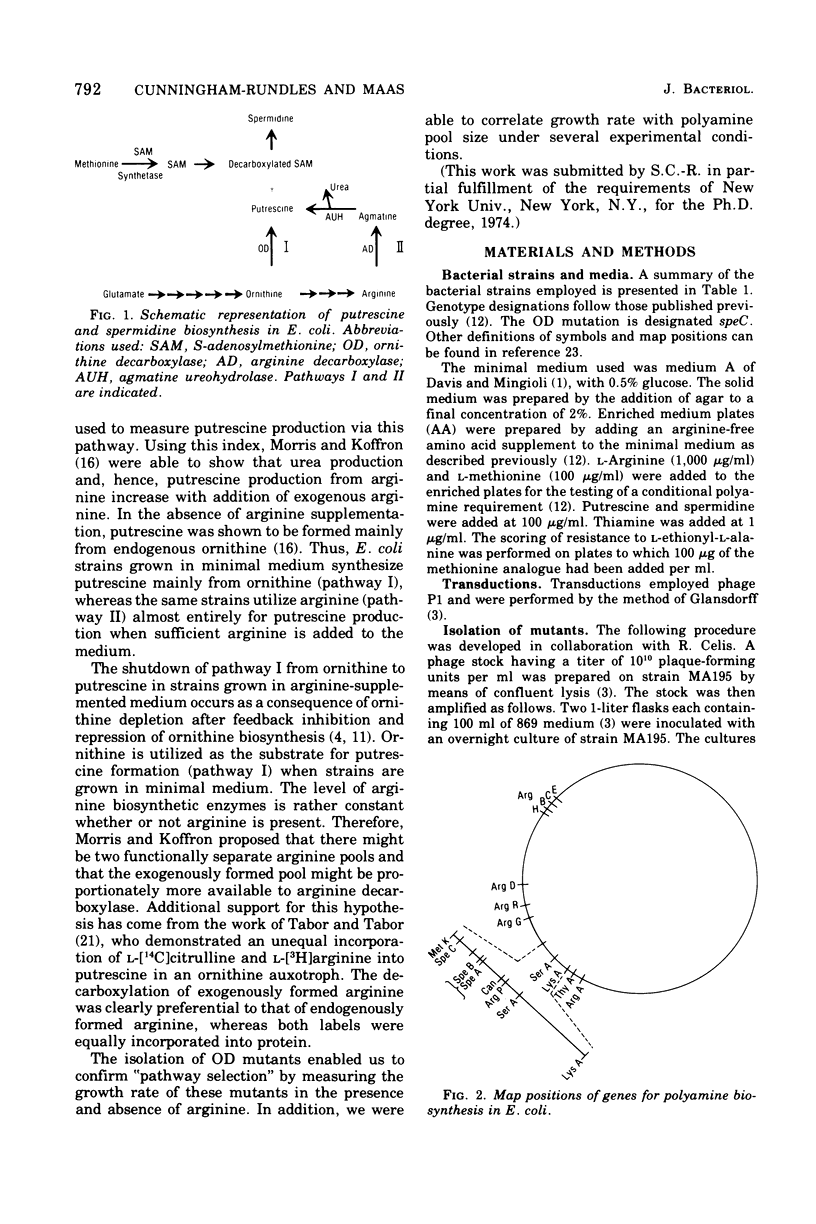
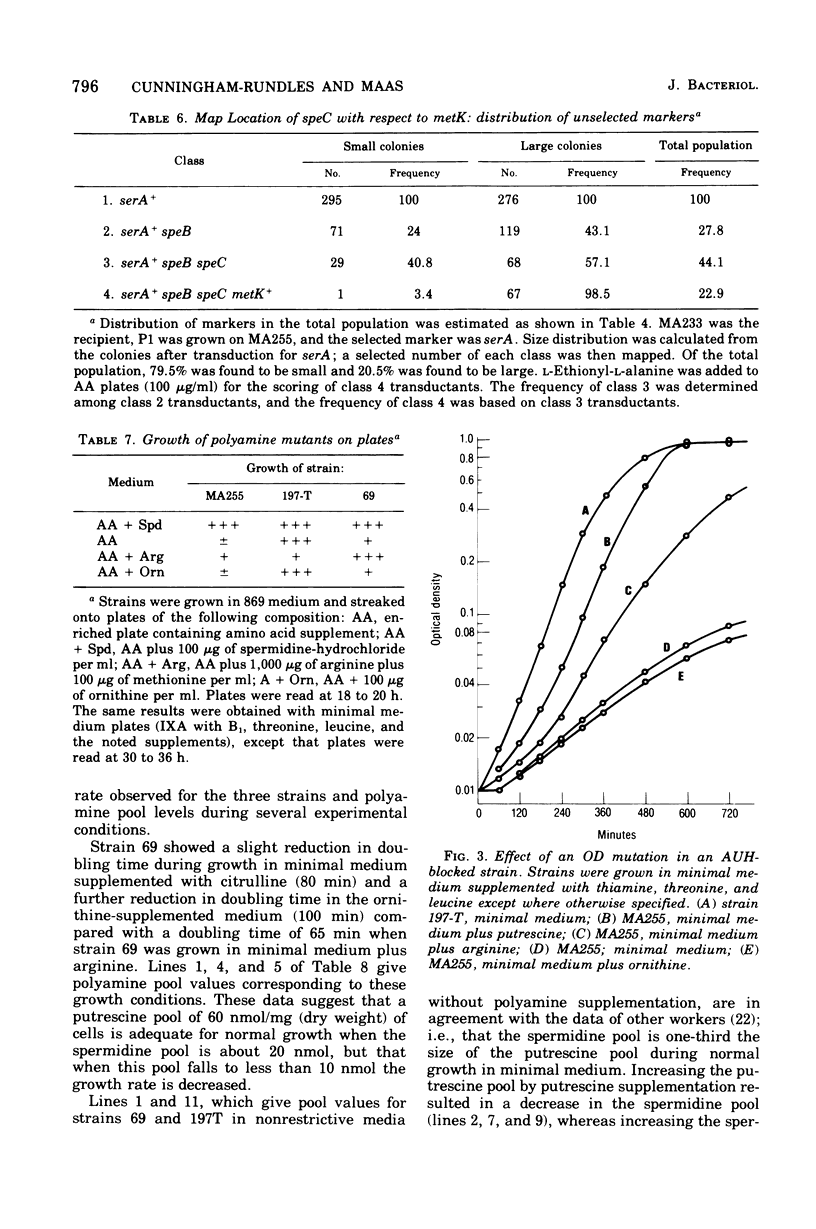
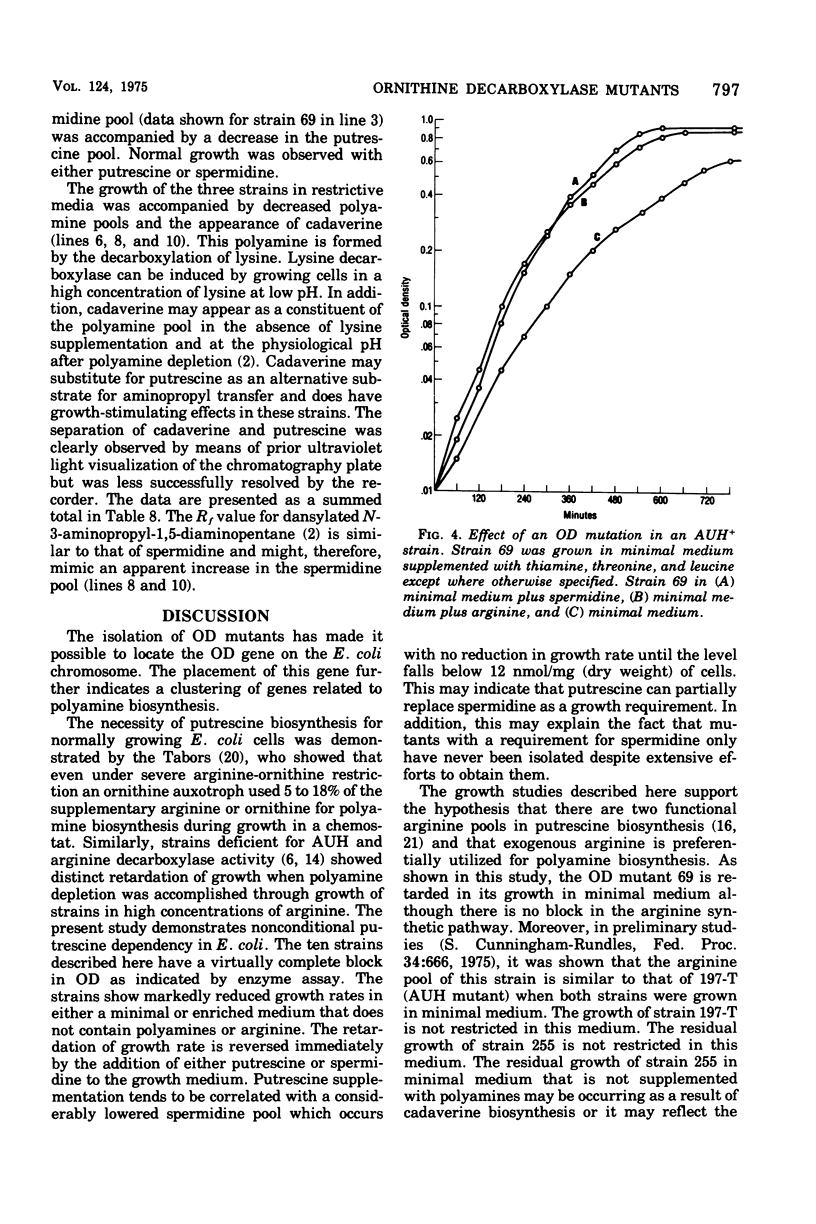
Several Escherichia coli K-12 mutants blocked in the synthesis of ornithine decarboxylase (OD) were isolated after transduction for serA+ in a strain (MA197) blocked in agmatine ureohydrolase (AUH) with a mutagenized phage lysate of P1. The new double-polyamine mutants were characterized by an unconditional polyamine dependence; either putrescine or spermidine was required for normal growth. The mutational block was varified by the demonstration of a virtual absence of OD activity in cellular extracts. The mutation, designated speC, was mapped by P1 transduction in several strains and was shown to have a cotransduction frequency of 17.2% with serA. Map order was established as serA speB speC metK. A derivative of one of the OD mutants having wild-type levels of AUH and blocked in OD was utilized along with an OD AUH mutant and an OD+ AUH strain to explore the phenomenon of "pathway selection" using growth rate as a parameter. Polyamine pool studies were carried out simultaneously. The results presented here support the hypothesis of pathway selection, implying a preferential utilization of exogenous arginine rather than endogenously produced arginine in polyamine biosynthesis.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion A. S., Cohen S. S. Polyamine stimulation of nucleic acid synthesis in an uninfected and phage-infected polyamine auxotroph of Escherichia coli K12 (arginine-agmatine ureohydrolase-putrescine-spermidine-lysine-cadaverine). Proc Natl Acad Sci U S A. 1972 Jan;69(1):213–217. doi: 10.1073/pnas.69.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLANSDORFF N. TOPOGRAPHY OF COTRANSDUCIBLE ARGININE MUTATIONS IN ESCHERICHIA COLI K-12. Genetics. 1965 Feb;51:167–179. doi: 10.1093/genetics/51.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORINI L., GUNDERSEN W., BURGER M. Genetics of regulation of enzyme synthesis in the arginine biosynthetic pathway of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1961;26:173–182. doi: 10.1101/sqb.1961.026.01.022. [DOI] [PubMed] [Google Scholar]
- Greene R. C., Su C. H., Holloway C. T. S-Adenosylmethionine synthetase deficient mutants of Escherichia coli K-12 with impaired control of methionine biosynthesis. Biochem Biophys Res Commun. 1970 Mar 27;38(6):1120–1126. doi: 10.1016/0006-291x(70)90355-4. [DOI] [PubMed] [Google Scholar]
- Hirshfield I. N., Rosenfeld H. J., Leifer Z., Maas W. K. Isolation and characterization of a mutant of Escherichia coli blocked in the synthesis of putrescine. J Bacteriol. 1970 Mar;101(3):725–730. doi: 10.1128/jb.101.3.725-730.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsie A. W., Puck T. T. Morphological transformation of Chinese hamster cells by dibutyryl adenosine cyclic 3':5'-monophosphate and testosterone. Proc Natl Acad Sci U S A. 1971 Feb;68(2):358–361. doi: 10.1073/pnas.68.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunninghake D., Grisolia S. A sensitive and convenient micromethod for estimation of urea, citrulline, and carbamyl derivatives. Anal Biochem. 1966 Aug;16(2):200–205. doi: 10.1016/0003-2697(66)90147-3. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lawrence D. A., Smith D. A., Rowbury R. J. Regulation of methionine synthesis in Salmonella typhimurium: mutants resistant to inhibition by analogues of methionine. Genetics. 1968 Apr;58(4):473–492. doi: 10.1093/genetics/58.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAAS W. K. Studies on repression of arginine biosynthesis in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1961;26:183–191. doi: 10.1101/sqb.1961.026.01.023. [DOI] [PubMed] [Google Scholar]
- Maas W. K. Mapping of genes involved in the synthesis of spermidine in Escherichia coli. Mol Gen Genet. 1972;119(1):1–9. doi: 10.1007/BF00270439. [DOI] [PubMed] [Google Scholar]
- Morris D. R., Jorstad C. M. Growth and macromolecular composition of a mutant of Escherichia coli during polyamine limitation. J Bacteriol. 1973 Jan;113(1):271–277. doi: 10.1128/jb.113.1.271-277.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R., Jorstad C. M. Isolation of conditionally putrescine-deficient mutants of Escherichia coli. J Bacteriol. 1970 Mar;101(3):731–737. doi: 10.1128/jb.101.3.731-737.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R., Koffron K. L. Putrescine biosynthesis in Escherichia coli. Regulation through pathway selection. J Biol Chem. 1969 Nov 25;244(22):6094–6099. [PubMed] [Google Scholar]
- Morris D. R., Koffron K. L. Urea production and putrescine biosynthesis by Escherichia coli. J Bacteriol. 1967 Nov;94(5):1516–1519. doi: 10.1128/jb.94.5.1516-1519.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R., Pardee A. B. Multiple pathways of putrescine biosynthesis in Escherichia coli. J Biol Chem. 1966 Jul 10;241(13):3129–3135. [PubMed] [Google Scholar]
- Srinivason P. R., Young D. V., Maas W. Stable ribonucleic acid synthesis in stringent (rel+) and relaxed (rel-) polyamine auxotrophs of Escherichia coli K-12. J Bacteriol. 1973 Nov;116(2):648–655. doi: 10.1128/jb.116.2.648-655.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor H., Tabor C. W. Biosynthesis and metabolism of 1,4-diaminobutane, spermidine, spermine, and related amines. Adv Enzymol Relat Areas Mol Biol. 1972;36:203–268. doi: 10.1002/9780470122815.ch7. [DOI] [PubMed] [Google Scholar]
- Tabor H., Tabor C. W. Formation of 1,4-diaminobutane and of spermidine by an ornithine auxotroph of Escherichia coli grown on limiting ornithine or arginine. J Biol Chem. 1969 May 10;244(9):2286–2292. [PubMed] [Google Scholar]
- Tabor H., Tabor C. W. Partial separation of two pools of arginine in Escherichia coli; preferential use of exogenous rather than endogenous arginine for the biosynthesis of 1,4-diaminobutane. J Biol Chem. 1969 Dec 10;244(23):6383–6387. [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]



