Abstract
Previous work identified the Rap1 GTPase-activating protein Sipa1 as a germ-line-encoded metastasis modifier. The bromodomain protein Brd4 physically interacts with and modulates the enzymatic activity of Sipa1. In vitro analysis of a highly metastatic mouse mammary tumor cell line ectopically expressing Brd4 demonstrates significant reduction of invasiveness without altering intrinsic growth rate. However, a dramatic reduction of tumor growth and pulmonary metastasis was observed after s.c. implantation into mice, implying that activation of Brd4 may somehow be manipulating response to tumor microenvironment in the in vivo setting. Further in vitro analysis shows that Brd4 modulates extracellular matrix gene expression, a class of genes frequently present in metastasis-predictive gene signatures. Microarray analysis of the mammary tumor cell lines identified a Brd4 activation signature that robustly predicted progression and/or survival in multiple human breast cancer datasets analyzed on different microarray platforms. Intriguingly, the Brd4 signature also almost perfectly matches a molecular classifier of low-grade tumors. Taken together, these data suggest that dysregulation of Brd4-associated pathways may play an important role in breast cancer progression and underlies multiple common prognostic signatures.
Keywords: gene expression signatures, metastasis, mouse models
The majority of deaths attributable to solid cancers result from the pathophysiological impact of metastasis. This is starkly illustrated when one considers that the median survival of patients with metastatic breast cancer is ≈2–4 years (1), compared with an ≈80% survival rate for women whose disease remains nonmetastatic. Advanced disseminated breast cancer thus remains an incurable condition regardless of new treatments (2). It is therefore important to develop a comprehensive understanding of the metastasis biology to identify patients at higher risk of tumor dissemination. This in turn may permit development of therapies and initiation of more aggressive treatment in women with poorer prognoses to reduce the incidence and extent of metastatic disease. Conversely, it may also prove possible to identify women at low risk of metastasis, thus sparing them needless adjuvant therapy.
Our laboratory has demonstrated that germ-line genetic variation influences tumor progression. Specifically, in a model system, the F1 progeny of the highly metastatic polyoma middle-T (PyMT) transgenic mouse and different inbred laboratory mouse strains display wide variations in metastatic efficiency after mammary tumor development (3). The most likely explanation for this observation is that germ-line variation modulates tumor progression. Subsequent identification of heritable loci modulating metastatic efficiency support this hypothesis (4, 5). Positional cloning subsequently identified Sipa1, a GTPase activating protein (GAP) that negatively regulates Rap-GTPases, as the first polymorphic metastasis efficiency gene in mice (6). Studies of human cancer have suggested that polymorphisms in human SIPA1 are associated with indicators of poor outcome in breast cancer (7), validating the utility of this mouse model to identify relevant human metastasis modifying genes.
Sipa1 interacts both in vitro and in vivo with the mammalian bromodomain protein BRD4 (8), which regulates cell growth by acting at different stages of the cell cycle (9, 10). This interaction modulates the enzymatic activity of SIPA1, with a consequent increase in the RAP1GAP activity (8). Therefore, we chose to investigate whether Brd4 is a modulator of metastasis given the apparent regulation of Sipa1 and investigate its involvement in a number of other prominent cellular systems, including transcriptional regulation (reviewed in ref. 11). Using a variety of experimental approaches, we demonstrate that activation of Brd4 in mice represses both tumor growth and metastasis and that Brd4 activation in human breast carcinomas induces a gene expression signature that predicts outcome in multiple breast cancer datasets. These data implicate Brd4 as a functional contributor to cancer metastatic potential.
Results
Ectopic Expression of Brd4 in a Highly Metastatic Mouse Mammary Tumor Cell Line Alters Cell Invasiveness and Cell Mobility but Not Cellular Proliferation Rates.
To investigate the role of Brd4 in a model system of mouse mammary tumorigenesis, cell lines ectopically expressing Brd4 were generated. The highly metastatic mouse mammary tumor cell line Mvt-1 (12) was stably transfected with either a mammalian expression vector encoding the full length Brd4 (13) or β-Galactosidase (β-Gal) cDNA (13), and individual clones were generated by serial dilution. Quantitative real-time PCR (qPCR) was used to confirm Brd4 ectopic expression.
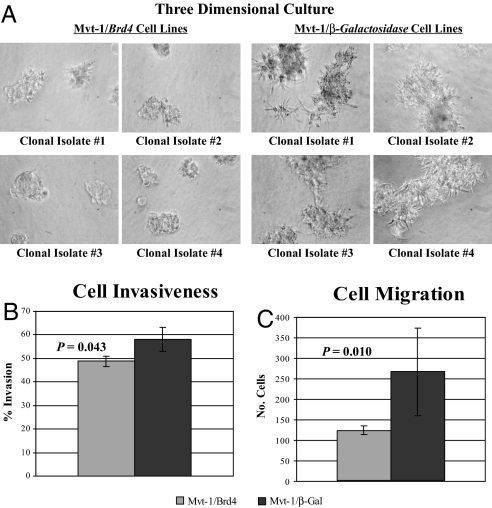
The invasiveness of Brd4- and β-Gal-expressing cells was determined by performing in vitro invasion assays. Three-dimensional cultures of the four Mvt-1 clonal isolates, using a basement membrane extract, showed that ectopic expression of Brd4 reduced the invasiveness of the Mvt-1 cells and the ability of these cells to form cell extensions compared with control cell lines (Fig. 1A). Furthermore, Matrigel invasion and Transwell migration assays demonstrated that both the invasiveness and the ability of Mvt-1 cells to migrate through a membrane were significantly reduced by ectopic expression of Brd4 (Fig. 1 B and C, respectively). However, the growth rate of those cell lines ectopically expressing Brd4 was not significantly different from that of the β-Gal-expressing control cell lines [supporting information (SI) Fig. S1]. These data imply that ectopic expression of Brd4 appears to reduce the invasion and migration properties of Mvt-1 cells without affecting intrinsic cellular properties, such as growth rate.
Fig. 1.
Ectopic expression of Brd4 positively regulates cell invasion and migration. (A) Four separate clonal isolates of Mvt-1/Brd4 and Mvt-1/β-Gal cells were grown on the top of a solidified basement membrane extract. Both the invasive properties and the ability to form cell extensions were significantly reduced in the Brd4 cells compared with the control cells. (B and C) Matrigel invasion and transwell migration assays of the four clonal isolates of Mvt-1/Brd4 and Mvt-1/β-Gal cells. Both the invasion and the migration properties of the Mvt-1 cells were reduced by ectopic expression of Brd4.
Tumor Growth and Metastatic Potential Are Reduced by Ectopic Expression of Brd4.
Spontaneous metastasis assays were performed to assess the effect of ectopic expression of Brd4 on tumor growth and metastasis in the highly metastatic Mvt-1 cell line. Mvt-1 clones ectopically expressing either Brd4 or β-Gal were s.c. implanted into virgin FVB/NJ female mice, and tumor weight and lung surface metastasis quantified after a four-week incubation period. Both tumor growth (Fig. 2A) and lung surface metastasis counts (Fig. 2B) were significantly reduced in the four Mvt-1 clonal isolates ectopically expressing Brd4. These data imply that activation of Brd4 is associated with a less malignant phenotype in the mouse, an observation that is consistent with the finding that the Mvt-1/Brd4 cells exhibit a less malignant phenotype in vitro (i.e., they are less invasive).
Fig. 2.
Ectopic expression of Brd4 in the highly metastatic mouse mammary tumor cell line Mvt-1 reduces tumor growth and pulmonary surface metastasis after s.c. implantation of cells into FVB/NJ mice. (A) A significant reduction in tumor growth was observed in the Mvt-1/Brd4 cells with the average tumor weight for the Mvt-1/Brd4 clones being 91 mg ± 42 mg compared with 595 mg ± 308 mg for the two Mvt-1/β-Gal clones (P < 0.001). (B) A similar reduction in pulmonary surface metastasis was observed with average lung surface metastasis count being 1.4 ± 2.5 for the Mvt-1/Brd4 clones compared with 11.1 ± 5.8 for the Mvt-1/β-Gal clones (P < 0.001).
Ectopic Expression of Brd4 Modulates ECM Gene Expression.
Studies have demonstrated that differential ECM gene expression is an important maker of metastatic capacity. Briefly, extracellular matrix (ECM) genes are common components of metastasis-predictive expression signatures in both human breast tumor tissue (14–16) and PyMT-induced mouse mammary tumors (17, 18). Expression quantitative trait loci (eQTL) mapping in AKXD recombinant inbred mice has shown that this relationship between tumor metastatic potential and differential ECM gene expression is at least partially controlled by genetic loci (18, 19). The most significant ECM eQTL in these mice is located on proximal chromosome 17 locus (peak region of linkage ≈29.5 Mb) and colocalizes with a described metastasis efficiency and tumor growth kinetics QTL (peak region of linkage ≈29.1 Mb) (5). Because both the eQTL and metastasis loci colocalize to the genomic region containing Brd4 (physical location ≈31.9 Mb) and examination of the mouse SNP databases reveals >50 SNPs between AKR/J and DBA2/J strains (consistent with the possibility that Brd4 is a candidate modifier gene), we chose to characterize the effect of differential Brd4 expression on metastasis-predictive ECM gene expression.
To achieve this, expression of the transcripts used to define the proximal chromosome 17 ECM eQTL (Col1a1, Col1a2, Col3a1, Fbn1, Mmp2, Nid1, and Serping1) (18, 19) and five related ECM genes (Col5a3, Col6a2, Fbln2, Mfap5, and Serpinf1) was quantified in the Mvt-1/Brd4 and Mvt-1/β-Gal cells using qPCR. Of the 12 ECM genes quantified in the cell lines ectopically expressing Brd4, five (Col1a1, Col5a3, Col6a2, Fbn1, and Serping1) displayed significantly altered expression (see Table 1). Two metastasis-predictive ECM genes (Col3a1 and Mmp2) were insufficiently expressed to be quantifiable, using qPCR. These results support the hypothesis that Brd4 is a causative factor in the transcriptional regulation of at least some ECM gene family members.
Table 1.
Ectopic expression of Brd4 in the highly metastatic mouse mammary tumor cell line Mvt-1 modulates expression of various metastasis-predictive ECM genes
| ECM Gene | Relative expression (±SD), transfected vs. control |
|---|---|
| Col1a1 | 7.94 ± 2.18* |
| Col1a2 | 0.41 ± 0.51 |
| Col3a1 | — |
| Col5a3 | 0.01 ± 0.01* |
| Col6a2 | 0.15 ± 0.20* |
| Fbn1 | 0.21 ± 0.09* |
| Fbln2 | 0.20 ± 0.23 |
| Mfap5 | 0.26 ± 0.29 |
| Mmp2 | — |
| Nid1 | 1.17 ± 0.40 |
| Serpinf1 | 0.40 ± 0.20 |
| Serping1 | 0.12 ± 0.18* |
Expression of each ECM gene was quantified in Mvt-1 cells by comparing ECM expression in four clonal isolates ectopically expressing Brd4 and three Mvt-1 isolates ectopically expressing β-galactosidase. The table shows the average expression ratio across the four Mvt-1/Brd4 clonal isolates. Adjusted Mann–Whitney U tests were performed to compare expression of ECM genes between Brd4-transfected and control cell lines. Bold-faced type indicates statistical significance at P ≤ 0.05. Dashes indicate that ECM gene expression was too low to quantify in the cell line.
Microarray Analysis of Mvt-1 Cell Lines Ectopically Expressing Brd4.
Affymetrix microarrays were used to compare gene expression in four Mvt-1/Brd4 clonal isolates and three Mvt-1/β-galactosidase clonal isolates. CEL files were analyzed by using the Affymetrix GeneChip probe level data RMA option of BRB ArrayTools software, Version 3.5.0. Genes with a <1.5-fold change from the gene's median value in 50% of samples or a log-ratio variation P > 0.01 were eliminated from analyses. To identify a Brd4 expression signature, we used the class comparison tool of BRB ArrayTools with a two-sample t test with random variance univariate test. P-values for significance were computed based on 10,000 random permutations, at a nominal significance level of each univariate test of 0.0001. A total of 2,577 probe sets passed these criteria.
Probe sets significantly up-regulated and down-regulated according to these criteria are listed in Dataset S1 and Dataset S2, respectively. Gene ontological analysis was performed by using BRB ArrayTools, and revealed that 149 classes of genes were modulated in response to ectopic expression of Brd4 at the nominal 0.005 level of the LS permutation test or KS permutation test (Dataset S3). Examination of this list reveals that consistent with its previously described functions, ectopic expression of Brd4 in Mvt-1 cells modulates expression of genes involved in processes, such as cellular proliferation, cell cycle, progression and chromatin structure. Furthermore, it is apparent that, at least in this cell line, Brd4 also regulates a number of processes that are critical to metastasis (e.g., cytoskeletal remodeling, cell adhesion, and extracellular matrix expression).
Mvt-1/Brd4 Signature Predicts Outcome in Multiple Breast Cancer Expression Datasets.
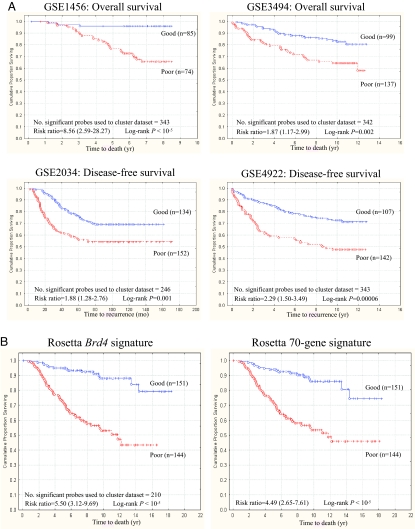
A high confidence human Brd4 gene expression signature was generated by mapping the most significantly differentially regulated genes (P < 10−7) from mouse array data to human Affymetrix and the Rosetta probe set annotations. The resulting gene signature for the five datasets (Dataset S4 and Table S1) consequently varied from 209 to 346 probe sets. Human Brd4 profiles were then used for unsupervised clustering of publicly available datasets into two groups representing high and low levels of Brd4 activation in patient samples. Kaplan–Meier survival analysis was then performed to investigate whether there was a survival difference between the two groups. Brd4 signature gene expression accurately predicted survival in each of the four Affymetrix-based breast cancer datasets, implying that the level of activation of Brd4 or Brd4-associated pathways within a tumor, presumably because of either somatic mutation or germ-line polymorphism, is an important determinant of the overall likelihood of relapse and/or survival (Fig. 3A). These findings were replicated in the Dutch Rosetta cohort, implying that the Brd4 signature possesses not only a cross-study but also a cross-platform predictive ability (Fig. 3B).
Fig. 3.
The Mvt-1/Brd4 microarray expression signature predicts survival and/or relapse in multiple breast cancer microarray datasets. (A) The Brd4 signature consistently and robustly predicts survival and/or relapse in four separate breast cancer microarray datasets performed on Affymetrix GeneChips. A significant difference in the overall likelihood of survival was observed in the GSE1456 dataset with 8-year survival being 95.9% vs. 65.5% for the good and poor prognosis Brd4 signatures, respectively (Upper Left). A similar effect was observed in the GSE3494 dataset with 12-year survival being 80.6% vs. 57.5% for the good and poor prognosis Brd4 signatures, respectively (Upper Right). The end point for the GSE2034 and GSE4922 differ in that disease-free survival was measured. A similar effect was seen in both cohorts with 10-year disease-free survival being 68.9% vs. 54.2% in the GSE2034 dataset (Lower Left) and 71.3% vs. 47.6% in the GSE4922 dataset (Lower Right) for the good and poor prognosis Brd4 signatures, respectively. (B) The Brd4 signature was also highly predictive of overall survival in the Dutch Rosetta dataset, with the overall survival being estimated to be 78.5% vs. 45.1% for the good and poor prognosis Brd4 signatures, respectively (Brd4 signature hazard ratio = 5.50, 95% C.I. = 3.12–9.69) (Left) compared with 72.6% vs. 47.0%, respectively (70 gene signature hazard ratio = 4.49, 95% C.I. = 2.65–7.61) for the same analysis, using the Rosetta 70 gene signature.
Characterization of Brd4 signature genes associated with survival in each of the breast cancer datasets revealed overlapping but not identical gene expression signatures (Dataset S5). The vast majority of Brd4 signature probes were predictive of survival in at least two of the four Affymetrix cohorts, and hazard ratios displayed the same directionality of effect for >99% of probes when a probe was predictive of survival in more than one cohort. The Dutch Rosetta cohort did have a number of unique predictive signature genes. Such variations likely reflect microarray platform differences, and population and tumor heterogeneity. Nevertheless, we argue that, because of the overlapping nature of the Brd4 signatures in the five cohorts and the finding that the Brd4 signature was the only consistent predictor of outcome on multivariate Cox proportional analysis in all of the cohorts (Table S2), the net effect of the Brd4 signature is both consistent and robust.
To determine whether the results might be explained by an artifact or underlying population structure permutation analysis was performed in the GSE1456 dataset. Ten thousand permutations were performed to determine whether similar results would be obtained by analysis of the dataset with similar sized gene lists of randomly selected genes. These 10,000 permutations demonstrated that it was highly unlikely that the survival difference observed because of BRD4 pathway activation was due to chance or population stratification (P <10−4). (Fig. S2).
The Mvt-1/Brd4 Signature Predicts Outcome in Lymph Node-Negative and Estrogen Receptor-Positive Breast Cancer.
Data relating to lymph node status were available for four of the study cohorts (GSE2034, GSE3494, GSE4922, and Rosetta). Expression of the Brd4 signature predicted outcome in node-negative patients in each of the four cohorts (Fig. S3A), suggesting that this signature allows for accurate stratification of patients with node-negative breast cancer into good and poor prognosis subcohorts. In contrast, multivariate Cox proportional analysis demonstrated that nodal status was an independent predictor of outcome in only one of these four populations [GSE3494: RR = 2.74, 95% confidence interval (C.I.) = 1.56–4.82; P = 0.0004; Table S2].
Similarly, Brd4 signature gene expression predicted outcome in all datasets where sufficient numbers of patients with estrogen receptor (ER)-positive tumors were available (Fig. S3B). Statistical significance was not achieved in GSE4922, owing to the low number of ER-positive patients represented in this dataset (n = 38). As was the case with node-negative tumors, these analyses demonstrate that Brd4 signature gene expression in ER-positive primary breast tumors also allows patients to be stratified into good and poor prognosis subgroups. As was the case with nodal involvement, ER status was found to independently predict outcome in only one of the four datasets where tumor ER status data were available (Rosetta cohort: RR = 1.8, 95% C.I. = 1.08–1.76; P = 0.009; Table S2).
Discussion
Through implementation of an integrated approach involving genetics, functional genomics, and genome-wide expression analysis, we have demonstrated that Brd4 or Brd4-associated pathways may play an important role in tumor progression in both mice and humans. The physiological function of Brd4 is relatively well characterized, with it being shown to be a cell growth regulator (9, 20) that acts primarily to promote G2/M transition (9), with its role in G2/M transition appearing dependent upon the intracellular balance of BRD4 to its binding partner, the RAP1GAP SIPA1 (8). This is an especially noteworthy given that we have identified SIPA1 as a polymorphic germ line-encoded metastasis efficiency modifier (6, 7).
The primary reason for this study was to enhance our understanding of the role of Sipa1 in the modulation of metastatic efficiency. Because it is known that SIPA1 interacts with BRD4, we hypothesized that Brd4 itself is a modulator of metastatic progression either by virtue of this interaction or independently through its pleiotropic cellular functionality. In vitro analysis revealed that Brd4 activation reduces both the invasiveness and mobility of a highly metastatic cell line without affecting cellular proliferation rates. However, implantation of the same cell lines into mice profoundly reduced tumor growth and metastatic capacity, indicating that Brd4 has tumor and metastasis suppressor activity in the in vivo setting. At this point, it seems likely that Brd4 activation in the epithelial component of the primary tumor is somehow reprogramming responses to microenvironmental cues within the tumor, which in turn reduces overall tumor growth. However, the mechanism is complex, and the apparent disparity between the in vitro and in vivo growth characteristics of the Mvt-1/Brd4 cells will require substantial further analysis to elucidate its origins.
Earlier studies in this laboratory have also independently implicated Brd4 as a candidate metastasis modulator. Brd4 colocalizes to the peak region of linkage of a metastasis susceptibility QTL located on proximal mouse chromosome 17 (5). Additionally, Brd4 has been identified as an ECM eQTL candidate gene (18, 19), with eQTL mapping experiments demonstrating that Brd4 resides within the peak region of linkage of an ECM eQTL. Furthermore, these experiments also demonstrate that the expression of Brd4 correlates with the expression of ECM genes that constitute prominent components of metastasis-predictive gene expression signatures (14–16). Analysis of Mvt-1/Brd4 gene expression by qPCR proved that Brd4 modulates the expression of many ECM genes that are dysregulated in tumors more prone to dissemination. Microarray analysis of the same cell lines confirmed the role of Brd4 in ECM expression regulation and showed that activation of this gene impacts a wide variety of other cellular processes.
These microarray data were also used to address one of the central goals of our research: the translation of experimental data from mouse models of human breast cancer into potentially clinically relevant observations. Subsequently, by identifying human microarray probe sets equivalent to those impacted by ectopic expression of Brd4 in the Mvt-1 mouse mammary tumor cell line, we were able to identify a Brd4 gene expression signature that predicts outcome across five breast cancer datasets and two microarray formats. Therefore, this finding suggests that the Brd4 signature is both consistent and robust, which is a conclusion drawn from the following: (i) virtually all signature genes are predictive in at least two of the five cohorts with hazard ratios displaying the same directionality for >99% of probe sets, and (ii) the Brd4 signature predicted outcome in all cohorts analyzed. The impact of this is augmented when one considers the well documented interlaboratory and interplatform issues in reproducibly of microarray data (21). One could argue that the gene expression patterns simply reflect the well documented effect that Brd4 has on cellular proliferation. However, because the Brd4 activation signature was derived from the Mvt-1/Brd4 cell lines that we have shown to have an identical growth rate to that of the control cell line, we argue that this is not the case and that the expression signature presented here is truly a reflection of Brd4 activation.
Our initial approach to identify a microarray signature predictive of survival, although unconventional, is by no means the only example of how in vitro data can be successfully used to develop an expression signature that holds meaningful value in human breast cancer. For example, Bild et al. (22) recently described an approach where microarray gene expression patterns were quantified in human mammary epithelial cell cultures after adenovirus-induced activation of various oncogenic pathways. By combining signature-based predictions across several pathways, they were able to distinguish different cancers and tumor subtypes and to define prognosis based on these signatures. It would not be unreasonable to assume, however, that the composition of gene signatures derived from cell culture experiments might vary greatly in composition compared with signatures derived from the more complex primary tumor samples. Rather intriguingly, this is not the case with the Brd4 signature. For example, Dai et al. (23) characterized a 50-gene cell proliferation expression signature, which correlates with extremely poor outcome in a subpopulation of breast cancer patients. Twenty-five of the up-regulated genes were identified as being involved in G2/M transition, the same cell cycle phase regulated by Brd4 (9). When compared with the Brd4 signature, we find that all but one of the genes of the Dai et al. signature (23) can be identified as components of the Brd4 signature. We also see overlap with other prognosis signatures, with 6 of the 70 gene signature described by van't Veer et al. (15) and 4 of the 21 RT-PCR Oncotype DX assay (Genomic Healthcare) being components of the Brd4 signature. However, the most compelling example of signature convergence comes from a recent study in which a 19-gene expression signature was defined by correlating tumor gene expression, histological grade, and survival (24). Eighteen of the 19 signature genes are components of the Brd4 signature, with 15 having P < 10−7 for their fold-change in Mvt-1/Brd4 cells. Furthermore, the directionality of expression changes of these 18 Brd4 signature probes exactly matched the directionality of expression change observed in low-grade tumors (24), suggesting a conversion from a malignant to a more benign phenotype. This observation is completely consistent with the reduced tumor growth of these cell lines in our mouse model. Overall, our interpretation of all of these data is that Brd4-pathway dysregulation, either as a consequence of somatic mutation, germ-line polymorphism, or epigenetic silencing, may both drive the expression of many of the genes present in breast cancer gene expression signatures and be a central event in tumor progression.
The potential clinical impact of tumor gene expression profiling lies in the possibility of improving the classification of different breast cancer subtypes. This in turn could enable clinicians to tailor the treatment of individual patients. Indeed, the utility of tumor expression profiling is currently being investigated in clinical trials [e.g., TAILORx and MINDACT (25)], the results of which will be eagerly anticipated. One of the main questions these trials seek to answer is whether tumor molecular profiling can be used prospectively to identify those node-negative/ER-positive patients who eventually relapse, to initiate adjuvant therapy that would not be administered under current treatment protocols. One of the criticisms leveled at microarray tumor gene expression analyses has been that it is a “black-box” methodology that provides results without a coherent biologic explanation and that dysregulated expression of any particular gene as a marker of poor prognosis does not indicate a clear-cut condition (26). The current study allows us to further characterize the origins of poor prognosis signatures in breast cancer by demonstrating that dysregulated expression of a single gene, Brd4, can drive the expression of many genes that are frequently observed as components of metastasis-predictive gene expression signatures. In turn, this suggests that assessing BRD4 status in breast cancer might add additional sensitivity and accuracy to current clinical tools. At present, however, it is unclear whether BRD4 is a proximal factor or is an intermediary molecule of some other inherited factor that drives the progression of breast cancer. More than 90 putative polymorphisms, including potential missense mutations, have been identified for the human ortholog in dbSNP that might impact transcriptional efficiency and/or protein function. Efforts to evaluate a possible causative role of these SNPs in breast cancer progression are currently underway.
Materials and Methods
Cell Culture.
The Mvt-1 cell line was obtained as a gift from Lalage Wakefield (National Cancer Institute, Bethesda, MD). Cells were cultured in DMEM (Cellgro) containing 10% FBS (Cellgro) and 1% penicillin-streptomycin (Cellgro).
Development of Mvt-1 Clonal Isolates Ectopically Expressing Brd4.
We used a construct encoding full-length Brd4 described in ref. 13. The control cell line was generated by using the vector pCMV-SPORT-β-Galactosidase (Invitrogen). Supercoiled plasmids were transfected into Mvt-1 cells, using Superfect transfection reagent (Qiagen) per the manufacturer's instructions. The Brd4-pFLAG-CMV2 and pCMV-SPORT-β-Galactosidase vectors were cotransfected with the vector pSuper.Retro.Puro (Oligoengine), containing no insert, as a selectable marker for transfectants. Cells in each culture vessel were transfected with a total of 20 μg of vector DNA, using Superfect at a 6:1 lipid-to-DNA ratio. Twenty-four hours after transfection, the cells were selected in normal growth medium containing 10 μg/ml puromycin (Sigma–Aldrich) and transferred to 96-well plates, and individual clones were selected by limiting dilution. Colonies were screened by quantitative PCR as described below to identify clones ectopically expressing Brd4.
Cell Invasion Assay.
The invasiveness of the Mvt-1 cells ectopically expressing either Brd4 or β-Gal was assayed by two different methods. The first assay was performed by culturing the cells in three dimensions, using Cultrex basement membrane extract with reduced growth factors (Trevigen). Chamber slides (Nalge Nunc; catalog no. 154534) were coated with 70 μl of cultrex per 0.7-cm2 well and incubated in 5% CO2 at 37°C for 30 min to solidify. Cells were then seeded at the density of 5,000 per well and incubated in 5% CO2 at 37°C for 8 days. Fresh culture media containing 10% FCS, and 4% basement membrane extract was added after 4 days. Pictures were taken after 8 days of incubation. In the second assay, invasion chambers coated with a thin layer of Matrigel basement membrane matrix were used (BD Biosciences; catalog no. 534480) and control inserts (BD Biosciences; catalog no. 354578). Cells were seeded at the density of 75,000 cells per well and incubated in 5% CO2 at 37°C for 20 h. Cells were then fixed with 100% methanol, stained with crystal violet, and mounted by using mineral oil. Cells were then counted, and both the invasion (percentage of invasion) and migration were determined according to the manufacturer‘s protocol.
Cell Growth Assay.
Mvt-1/Brd4 and Mvt-1/β-Gal cells were seeded on 24-mm plates at a density of 105 cells per plate and incubated in 5% CO2 at 37°C for the indicated periods of time. After incubation, the cells were harvested, and the number of cells was determined by using a cellometer.
Spontaneous Metastasis Assays.
Transfected cells proven to be stably expressing Brd4 were s.c. implanted into virgin FVB/NJ mice as described in ref. 6. Tumor growth and metastasis were compared with mice injected with 105 Mvt-1 cells stably cotransfected with pCMV-Sport-β-Gal and pSuper.Retro. Puro. These experiments were performed in compliance with the National Cancer Institute's Animal Care and Use Committee guidelines.
Microarray and Survival Analysis.
Microarray expression analysis of the Mvt-1/Brd4 and Mvt-1/β-Gal cells was performed by using methodology described in ref. 18. Hybridization cocktails were applied to the Affymetrix GeneChip Mouse Genome 430 2.0 arrays, processed on the Affymetrix Fluidics Station 400, and analyzed on an Agilent GeneArray Scanner with Affymetrix Microarray Suite software, Version 5.0.0.032. Normalization was performed with the BRB-Array Tools software (27, 28). To generate a high confidence human transcriptional signature of Brd4 expression, 638 probe sets whose differential expression demonstrated P < 10−7 were selected. A gene list representing the probes was developed and used to map to the probe sets of the human U133 Affymetrix GeneChip, using the Batch Search function of NetAffx (www.affymetrix.com/analysis/netaffx). A human signature of 971 probe sets representing 379 genes was identified (Dataset S5). The Brd4 signature for the Dutch Rosetta cohort was generated by matching the gene symbols from the mouse dataset to the published Hu25K chip annotation files.
Analysis of tumor gene expression from breast cancer datasets was performed by using BRB ArrayTools. Affymetrix datasets were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo). The Dutch dataset was downloaded from the Rosetta Company website (www.rii.com/publications/2002/vantveer.html). Expression data were loaded into BRB ArrayTools, using the Affymetrix GeneChip Probe Level Data option or the Data Import Wizard. Data were filtered to exclude any probe set that was not a component of the Brd4 signature, and to eliminate any probe set whose expression variation across the dataset was P ≥ 0.01.
Unsupervised clustering of each dataset was performed by using the samples only clustering option of BRB ArrayTools. Clustering was performed by using average linkage, the centered correlation metric and center the genes analytical option. Samples were assigned into two groups based on the first bifurcation of the cluster dendogram, and Kaplan–Meier survival analysis performed by using the Survival module of the software package Statistica. Significance of survival analyses was performed by using the Cox F test.
Permutation Analysis of Brd4 Expression Signature to Detect Underlying Structure.
Survival data in the GEO dataset GSE1456 were used for the purpose of permutation analysis. The survival curve and the P value were computed by dividing the sample set into two clusters based on the Brd4 expression signature. The values of the cluster variable were permuted to generate two random clusters and the survival curves, and the P value were computed for each permutation. The permutation process was repeated 10,000 times to evaluate the significance of the survival curves for each transfect gene.
Supplementary Material
Acknowledgments.
We thank Drs. Doug Lowy, Pat Steeg, and Lalage Wakefield for critical comments on this manuscript and Dr. Dalit Barkan for her assistance with the invasion assays. This work was supported in part by the Intramural Research Program of the National Institutes of Health National Cancer Institute Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710331105/DCSupplemental.
References
- 1.Chung CT, Carlson RW. Goals and objectives in the management of metastatic breast cancer. Oncologist. 2003;8:514–520. doi: 10.1634/theoncologist.8-6-514. [DOI] [PubMed] [Google Scholar]
- 2.Guarneri V, Conte PF. The curability of breast cancer and the treatment of advanced disease. Eur J Nucl Med Mol Imaging. 2004;31(Suppl 1):149–161. doi: 10.1007/s00259-004-1538-5. [DOI] [PubMed] [Google Scholar]
- 3.Lifsted T, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–644. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Hunter KW, et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res. 2001;61:8866–8872. [PubMed] [Google Scholar]
- 5.Lancaster M, Rouse J, Hunter KW. Modifiers of mammary tumor progression and metastasis on mouse chromosomes 7, 9, and 17. Mamm Genome. 2005;16:120–126. doi: 10.1007/s00335-004-2432-y. [DOI] [PubMed] [Google Scholar]
- 6.Park YG, et al. Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat Genet. 2005;37:1055–1062. doi: 10.1038/ng1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford NP, et al. Polymorphisms of SIPA1 are associated with metastasis and other indicators of poor prognosis in breast cancer. Breast Cancer Res. 2006;8:R16. doi: 10.1186/bcr1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farina A, et al. Bromodomain protein Brd4 binds to GTPase-activating SPA-1, modulating its activity and subcellular localization. Mol Cell Biol. 2004;24:9059–9069. doi: 10.1128/MCB.24.20.9059-9069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dey A, et al. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruyama T, et al. A mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 12.Pei XF, et al. Explant-cell culture of primary mammary tumors from MMTV-c-Myc transgenic mice. In Vitro Cell Dev Biol Anim. 2004;40:14–21. doi: 10.1290/1543-706X(2004)40<14:ECOPMT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 15.van't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 16.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, et al. Caffeine suppresses metastasis in a transgenic mouse model: A prototype molecule for prophylaxis of metastasis. Clin Exp Metastasis. 2004;21:719–735. doi: 10.1007/s10585-004-8251-4. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, et al. Metastasis Predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis. 2005;22:593–603. doi: 10.1007/s10585-005-6244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford NP, et al. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet. 2007;3:e214. doi: 10.1371/journal.pgen.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houzelstein D, et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall E. Getting the noise out of gene arrays. Science. 2004;306:630–631. doi: 10.1126/science.306.5696.630. [DOI] [PubMed] [Google Scholar]
- 22.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 23.Dai H, et al. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005;65:4059–4066. doi: 10.1158/0008-5472.CAN-04-3953. [DOI] [PubMed] [Google Scholar]
- 24.Ivshina AV, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 25.Bogaerts J, et al. Gene signature evaluation as a prognostic tool: Challenges in the design of the MINDACT trial. Nat Clin Pract Oncol. 2006;3:540–551. doi: 10.1038/ncponc0591. [DOI] [PubMed] [Google Scholar]
- 26.Modlich O, Prisack HB, Bojar H. Breast cancer expression profiling: The impact of microarray testing on clinical decision making. Expert Opin Pharmacother. 2006;7:2069–2078. doi: 10.1517/14656566.7.15.2069. [DOI] [PubMed] [Google Scholar]
- 27.Hedenfalk I, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 28.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.