Abstract
Water–biomolecule interactions have been extensively studied in dilute solutions, crystals, and rehydrated powders, but none of these model systems may capture the behavior of water in the highly organized intracellular milieu. Because of the experimental difficulty of selectively probing the structure and dynamics of water in intact cells, radically different views about the properties of cell water have proliferated. To resolve this long-standing controversy, we have measured the 2H spin relaxation rate in living bacteria cultured in D2O. The relaxation data, acquired in a wide magnetic field range (0.2 mT–12 T) and analyzed in a model-independent way, reveal water dynamics on a wide range of time scales. Contradicting the view that a substantial fraction of cell water is strongly perturbed, we find that ≈85% of cell water in Escherichia coli and in the extreme halophile Haloarcula marismortui has bulk-like dynamics. The remaining ≈15% of cell water interacts directly with biomolecular surfaces and is motionally retarded by a factor 15 ± 3 on average, corresponding to a rotational correlation time of 27 ps. This dynamic perturbation is three times larger than for small monomeric proteins in solution, a difference we attribute to secluded surface hydration sites in supramolecular assemblies. The relaxation data also show that a small fraction (≈0.1%) of cell water exchanges from buried hydration sites on the microsecond time scale, consistent with the current understanding of protein hydration in solutions and crystals.
Keywords: biomolecular hydration, buried water molecules, Escherichia coli, Haloarcula marismortui, in vivo NMR
Water, the ubiquitous biosolvent, mediates or modulates the intermolecular forces that govern the self-assembly of biological cells, it controls the rates of substrate diffusion and conformational transitions, and it participates in molecular recognition and enzyme catalysis (1–4). It is therefore imperative to characterize and understand any differences between cell water and bulk water. Biopolymers and other solutes make up one-third of the mass of a typical cell, so this difference could be substantial. The few experimental techniques that can monitor the molecular properties of water in vivo have suffered from interpretational ambiguities, allowing widely discordant views about cell water structure and dynamics to coexist for a long time (5–7). NMR spectroscopy can provide information about cell water via the spin relaxation times of the dominant water–1H signal (8, 9). In fact, tissue-specific variations in water relaxation times provided the impetus for developing magnetic resonance imaging (10). Unfortunately, the interpretation of water–1H relaxation data from biological samples is confounded by cross-relaxation, intermolecular paramagnetic couplings, and proton-exchange modulation of the nuclear shielding (11, 12). Here, we circumvent these complications by measuring the relaxation rate of the longitudinal water–2H magnetization from cells cultured in D2O. We have chosen to study the bacterium Escherichia coli because of the wealth of information available about this organism and the extreme halophilic archaeon Haloarcula marismortui because of reports of unusual hydration behavior of halophilic proteins (13) and of anomalously slow water diffusion in H. marismortui cells (14).
Cell water is often said to be more “structured” or “ordered” than bulk water (6, 7), but this loosely formulated hypothesis is not easily tested. Water structure is a multifaceted property that has not been fully characterized experimentally even for bulk water (15, 16). Moreover, water is extensively hydrogen-bonded and therefore structurally robust. (The cohesive energy density of water, 23 kbar, is 1 order of magnitude higher than for most organic liquids.) Although the structure of hydration water usually differs only subtly from bulk water, the kinetic (and thermodynamic) effects of the solute may be substantial. Dynamical properties are therefore expected to be sensitive probes of intracellular water perturbations. Here, we monitor single-molecule water rotation via the water–2H spin relaxation rate R1 induced by rotational modulation of the anisotropic nuclear electric quadrupole coupling (11, 17).
In the heterogeneous intracellular environment, water molecules rotate at widely different rates depending on how they interact with biopolymers. To separate contributions to R1 from different water populations, it is essential to perform measurements over a wide range of Larmor frequencies. (The Larmor frequency ω0 is proportional to the strength of the applied magnetic field.) By using the fast field-cycling (FC) technique as well as conventional superconducting magnets, we have measured R1 over 5 orders of magnitude in frequency. Such magnetic relaxation dispersion (MRD) data have not been reported for as well as microorganism, and the few earlier water–2H relaxation studies of excised tissue have been limited to a single (MHz) frequency.
Results and Discussion
Subnanosecond Cell Water Dynamics.
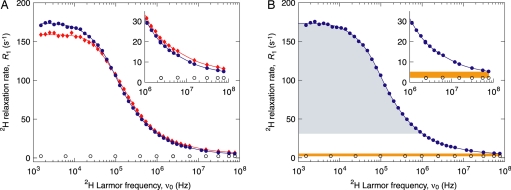
Fig. 1 shows the water–2H MRD profiles from living E. coli and H. marismortui cells. The MRD profile R1(ω0) is essentially a mapping in the frequency domain of the distribution of rotational correlation times τ of all water molecules in the sample (11, 18, 19). At a given frequency ω0, R1 samples water molecules with correlation times shorter than 1/ω0. The highest accessed frequency ω0* corresponds to a correlation time τ* ≡ 1/ω0* = 2 ns. We consider first water dynamics on time scales shorter than τ*, as reflected in R1(ω0*). The finding that R1(ω0*) exceeds the relaxation rate R10 in a bulk D2O reference sample (Fig. 1) implies that the average correlation time is longer in the cell sample than in bulk water. To eliminate the trivial dependence of R1 on the nuclear quadrupole coupling (11, 17), we focus on the relative excess relaxation rate ρ(ω0*) ≡ [R1(ω0*) − R10]/R10 (Table 1), which is a model-independent global measure of cell water dynamics on time scales <2 ns.
Fig. 1.
2H MRD profiles of cell water. (A) The water–2H relaxation rate R1 was measured on E. coli cells (blue circles) and H. marismortui cells (red diamonds) in stationary growth phase at 27°C and pD 8.0. Control measurements were performed in parallel on a bulk D2O reference sample (open circles). The curves are multi-Lorentzian numerical representations used for the model-free analysis. (B) The colored areas represent, for the E. coli sample, the contributions to R1 from surface hydration water with rotational correlation time <2 ns (yellow) and from internal water molecules with residence time >160 ns (blue). In both images, Inset shows the high-frequency region on an expanded scale.
Table 1.
Sample composition, 2H MRD parameters, and derived results
| Quantity | E. coli | H. marismortui |
|---|---|---|
| NW†, mol (g DCM)−1 | 0.174 | 0.0759 |
| mW/mP‡ | 6.08 | 4.27 |
| ρ(ω0*) | 1.50 ± 0.03 | 2.06 ± 0.03 |
| ξcell | 3.02 ± 0.04 | 3.77 ± 0.04 |
| ξhyd | 15.6 ± 3 | 15.0 ± 3 |
| I(ω0#), s−1 | 26.5 ± 0.3 | 27.0 ± 0.3 |
| nintSint2, μmol (g DCM)−1 | 56 ± 5 | 25 ± 2 |
| νint | 3.6 ± 1.0 | 2.5 ± 0.7 |
DCM, dry cell mass.
†Water content of sample.
‡Water-to-protein mass ratio in sample.
The observed 2H magnetization derives mainly from water deuterons, but under certain conditions, there may also be a pH-dependent contribution from labile biopolymer deuterons exchanging with D2O deuterons on the relaxation time scale (11). For solutions of small proteins, labile deuterons may contribute substantially to R1(ω0*) at high pH (20). But for large or immobilized proteins, where the relaxation time scale is shorter, the labile-deuteron contribution to R1(ω0*) is insignificant near neutral pH (21). To verify that this applies to the intracellular environment, we measured R1(ω0*) for both the water nuclides 2H and 17O in an E. coli sample prepared in the same way (but with 10% higher water content) as the one used to record the data in Fig. 1. After correcting the 2H rate to the 12% lower 17O resonance frequency (at constant magnetic field), we obtained ρ(ω0*) = 1.44 ± 0.03 for both nuclides. Because 17O monitors water molecules exclusively, this result indicates that the labile-deuteron contribution to the 2H rate is negligibly small. For our cell samples at pD 8.0, we can therefore attribute R1(ω0*) entirely to D2O molecules. (For the definition of pD, see Materials and Methods.)
In our densely packed cell samples, most of the water is intracellular. Based on the water-accessible intracellular (inside the outer membrane) volume in E. coli cells under our osmotic conditions (22), we estimate that the fraction intracellular water is fcell = 0.74. As shown in supporting information (SI) Text, this estimate holds approximately also for the H. marismortui sample. Water exchange across the inner (cytoplasmic) and outer membranes is fast on the 2H and 17O relaxation time scales (SI Text), so ρ is a population-weighted average. The relaxation rate of extracellular water should be virtually the same as in bulk water, because the macromolecular concentration is low and inorganic salt has little effect. Thus, ρ = fcellρcell.
The dynamic perturbation factor (DPF) ξ for a particular class of water molecules is defined as the ratio of the mean correlation time 〈τ〉 for all water molecules in that class to the correlation time τ0 in bulk water. To a good approximation (Fig. S1), we can obtain the intracellular DPF as ξcell = 1 + ρcell = 1 + ρ(ω0*)/fcell. The resulting ξcell values (Table 1) show that water rotation inside E. coli and H. marismortui cells is, on average, a factor 3–4 slower than in bulk water, with little difference between the two organisms. Because it is derived from R1(ω0*), ξcell pertains to all intracellular water molecules with rotational correlation times shorter than τ* = 2 ns. Based on MRD studies of protein solutions (23), we expect that >99% of cell water belongs to this class. The more slowly rotating water molecules, responsible for the large increase of R1 at lower frequencies (Fig. 1), reside in partly or fully buried hydration sites (see below).
The DPF ξcell is an integral measure of cell water dynamics, derived from the experimental data without any model assumptions. The ξcell values in Table 1 can therefore be used to rigorously rule out the possibility that a major fraction of the cell water is dynamically retarded by a factor ≫3–4. However, because ξcell depends on the relative amounts of water and solutes in the cell, it is not a suitable quantity for comparing cell water with water in protein solutions and other model systems. To do this, we note that studies of a wide range of aqueous model systems, from small solutes to proteins and membranes, show that the dynamic perturbation is essentially confined to water molecules in direct contact with the solute's surface (1, 2, 4, 23, 24). In other words, water outside the (first) hydration layer is practically indistinguishable from bulk water. We reasonably assume this is the case also within the cell. We can then use the fast-exchange relation ρ = ƒhydρhyd to derive the DPF ξhyd = 1 + ρhyd for the hydration water in the cell. Because it is independent of composition, the quantity ξhyd can be directly compared between cells and model systems. The fraction of hydration water in the cell sample can be calculated as fhyd = AS/(NW aW), where AS is the combined solvent-accessible surface area (SASA) of all solutes in the sample, NW is the number of water molecules in the sample, and aW = 10.75 Å2 is the mean SASA occupied by one water molecule in the hydration layer. This aW value was obtained by determining, by molecular dynamics simulations, the number of water molecules in the first hydration layer up to the first minimum in the water − (protein C, O, or N atom) radial distribution function (C. Mattea, J. Qvist, and B.H., unpublished work). [A larger value, aW = 15 Å2, has been used previously (23, 25), corresponding to a more conservative definition of the hydration layer that excludes most of the apolar hydration shells.]
Before estimating AS, we note that most solutes contribute negligibly to ρ. This is the case for phospholipids, saccharides, small molecules, and ions, which together account for only 5% (E. coli) or 8% (H. marismortui) of ρ (SI Text). If this small contribution is neglected, AS refers to the combined SASA of all proteins and nucleic acids. The macromolecular content and internal structure of the E. coli cell have been characterized in great detail (26). Proteins or nucleic acids occur in five principal locations: nucleoid, ribosomes, inner and outer membranes, cytoplasm and periplasm, and external organelles. For each of these locations, we use structural data to estimate the specific SASA ak. The total SASA is then obtained as As = Σk mkak, where mk is the known amount of protein or nucleic acid in location k for an E. coli cell in stationary phase. This calculation, described in detail in SI Text, yields AS = 10 (nucleoid) + 355 (ribosomes) + 125 (membranes) + 670 (other proteins) = 1,160 m2 (g DCM)−1 (DCM, dry cell mass). With NW from Table 1, we thus obtain fhyd = 0.103. In other words, 1 in 10 water molecules in the E. coli sample or 1 in 7 water molecules in the E. coli cell interacts directly with proteins or nucleic acids. Assigning 20% uncertainty to this geometric hydration fraction, we obtain ξhyd = 15.6 ± 3 for the hydration water of proteins and nucleic acids in E. coli. With τ0 = 1.74 ps for bulk H2O at 27°C, this DPF corresponds to a mean correlation time of 27 ps.
The hydration DPF ξhyd for H. marismortui cannot be obtained in the same way as for E. coli, because the information needed to estimate AS is unavailable. However, if the nucleic acid to protein mass ratio and the average specific SASAs of proteins and nucleic acids are the same in the two samples, then fhyd is inversely proportional to the known water/protein mass ratio (Table 1). Given this assumption, we obtain fhyd = 0.147 and ξhyd = 15.0 ± 3 for H. marismortui. There is thus no significant difference in hydration water dynamics (on time scales <2 ns) between H. marismortui and E. coli. The somewhat larger values of ρ and ξcell obtained for H. marismortui do not indicate slower hydration dynamics but merely reflect the higher protein/water ratio in this sample (Table 1).
The DPF is larger for cell hydration water than for the hydration layer of monomeric globular proteins in solution (23). Water–17O R1(ω0*) data with ω0*/(2π) = 49–81 MHz for 11 proteins (58–162 residues) yield ξhyd = 4.9 ± 0.6 (range 3.9–5.7). For a set of four larger proteins (rat intestinal fatty acid-binding protein, human carbonic anhydrase II, bovine β-trypsin, and BSA), which all have deep crevices with potentially large dynamic perturbations, ξhyd = 9 ± 2 is closer to the cell value. We therefore attribute the larger DPF in the cells to geometrically secluded, and thereby more dynamically perturbed, hydration sites at subunit interfaces in enzyme complexes, ribosomes, cytoskeleton, and other supramolecular assemblies.
Buried Water Molecules.
The dramatic increase in R1 in the kHz–MHz frequency range (Fig. 1) constitutes direct model-independent evidence for water dynamics on the 0.1 to 10 μs time scale. Similar low-frequency 2H relaxation dispersions have been observed in biopolymer gels (but not in solutions of freely tumbling proteins) and have been quantitatively linked to exchange of internal water molecules in rotationally immobilized biopolymers (21, 27, 28). The R1 dispersion below ≈1 MHz is a frequency mapping of the residence time distribution of these internal water molecules. Thus, whereas R1(ω0*) reflects rotational motions of surface hydration and bulk water, the much larger R1 at low frequencies is produced by water molecules buried in cavities inside rotationally immobilized proteins (and other macromolecules). At the high-frequency ω0*, spin relaxation is induced by local water rotation slowed down to varying degrees by interactions with macromolecular surfaces. At low frequencies, spin relaxation is instead induced by exchange-mediated orientational randomization (EMOR) of internal water molecules (18, 21, 27, 28).
In the range 1–100 MHz, the (weaker) frequency dependence of R1 is produced by orientational randomization of long-lived (residence time >1 ns) water molecules in freely tumbling protein molecules. This mechanism has been thoroughly studied by 2H and 17O MRD (11). In this frequency range, R1 primarily reflects protein tumbling rather than water dynamics. Moreover, we expect a substantial contribution to R1 from labile deuterons in this frequency range (11, 20). For these reasons, we focus here on the additional relaxation enhancement ΔR1(ω0) ≡ R1(ω0)–R1(ω0#) below a cutoff frequency ω0#/(2π) = 1 MHz, corresponding to a correlation time τ# = 1/ω0# = 160 ns. Because τ# corresponds to the tumbling time of a 260 kDa globular protein (29), this cutoff should eliminate nearly all contributions from freely tumbling macromolecules in the cell. The EMOR mechanism is effective for water exchange rates comparable to, or higher than, the water–2H nuclear quadrupole frequency (11) ωQ = 8.7 × 105 s−1. Therefore, ΔR1 monitors internal water molecules with residence times up to a few microseconds. Labile biopolymer deuterons, which all have residence times >1 ms at pD 8 (SI Text), do not contribute to ΔR1 (21).
To fully interpret ΔR1, we must specify the form of the residence time distribution. However, to obtain results that are as general as possible, we shall use a model-free analysis that rigorously separates the static (number of internal water molecules) and dynamic (their residence times) information contained in ΔR1 (19). The static information can be extracted from the integral I(ω0#) of ΔR1 from 0 to ω0#, which is proportional to the amount nint of internal water molecules and to their mean-square orientational order parameter Sint2 but is independent of the residence times (SI Text). Calculating I(ω0#) by numerical integration of a multi-Lorentzian representation of the R1 data in Fig. 1 (Table S1), we thus obtain the product nintSint2 (Table 1). In the following analysis, we use the estimate Sint2 = 0.6 ± 0.1, based on previous MRD studies of internal water molecules in protein solutions (11) and gels (28). The large difference in nintSint2 between the two cell samples is caused by a trivial “dilution” effect. According to our elemental analysis (Table S2), Na and K make up 20% of the DCM in the H. marismortui sample but only 3% in the E. coli sample.
Our model-free analysis shows that the large increase in R1 below 1 MHz is caused by a very small water fraction: nint/nW = (5.4 ± 1.0) × 10−4 for both samples (Table 1). If the analysis is correct, this fraction must be consistent with the expected amount of internal water molecules in the cell. Assuming that all internal water molecules reside in proteins, we can obtain the number, νint, of internal water molecules per 100 amino acid residues as 100 (nint/ximmob)(Mres/mP), where mP is the known protein mass in the sample, and Mres = 108 g mol−1 is the mean residue molar mass.
The fraction, ximmob, of rotationally immobilized protein was determined from the principal quadrupolar peak in the 1H MRD profiles from samples identical to those used for the 2H MRD experiments, except that H2O was used instead of D2O (Fig. 2). The quadrupolar peaks are caused by magnetization transfer from water protons to 14N in immobilized peptide NH groups at the 1H Larmor frequencies where the 14N quadrupolar energy splitting is matched (9, 30). The peak amplitude (the maximum R1Q) is proportional to the NH/water mole ratio and can therefore be used to determine ximmob when the total protein concentration is known (31). The proportionality constant was obtained from similar measurements on chemically cross-linked (ximmob ≈ 1) gels of bovine pancreatic trypsin inhibitor (28) or BSA (31), which differ by an order of magnitude in molar mass. The two sets of calibration data yield similar results, ximmob = 0.5 ± 0.1, with no significant difference between E. coli and H. marismortui. Thus, half of the cell protein mass is rotationally immobilized by incorporation into large supramolecular assemblies.
Fig. 2.
1H MRD profiles and quadrupolar peaks. The water–1H relaxation rate R1 was measured on E. coli cells at 27°C (blue circles) and H. marismortui cells at 12°C (green triangles) or 27°C (red diamonds). The samples were prepared as in Fig. 1, but with H2O at pH 7.6. The shoulder at 10–30 MHz is due to paramagnetic ions. Inset shows the quadrupolar peaks on an expanded scale for E. coli (blue circles) and H. marismortui (red diamonds) at 27°C. The plotted quantity R1Q was obtained by subtracting the baseline MRD profile. The curves serve only to guide the eye.
The νint values deduced in this way (Table 1) are similar to the value νint = 3.4 obtained from analysis of 842 protein crystal structures (32). We do not expect our νint values to agree precisely with the structural νint value, because some internal water molecules have residence times outside the investigated MRD window, 0.16–10 μs (Fig. S2). But MRD studies of protein solutions (11, 24) and gels (28) indicate that most internal water molecules have residence times in this range. Furthermore, in the cell, water molecules are also trapped within rRNA and at the subunit interfaces of supramolecular assemblies. The close agreement between the MRD-derived νint value for E. coli and the crystallographic νint value suggests these differences are small or nearly compensating.
Concluding Remarks.
To assess the divergent estimates of cell water dynamics reported in the literature, it is important to understand precisely what is being measured. The diffusional dynamics of cell water can be probed via translational or rotational motions. The long-range (1 to 10 μm) translational apparent diffusion coefficient (ADC) of water (33) or macromolecules (34) (dynamically coupled to the solvent via frictional forces) is usually governed by obstruction (crowding) and confinement effects. The ADC is thus primarily a probe of tissue morphology or cell ultrastructure and provides little information about local water mobility. Even in the absence of obstruction effects, as in axons or muscle fibers, the water ADC is insensitive to dynamic perturbations of hydration water. The ADC represents a spatially averaged (translational) mobility, whereas the rotational correlation time measured by NMR is a spatially averaged inverse (rotational) mobility. In a heterogeneous system, therefore, the most strongly retarded hydration water molecules dominate τcell, whereas they hardly affect Dcell. For example, if fhyd = 0.01 and ξhyd = 100, then the translational cell DPF is ξcellT ≡ D0/Dcell = [1 − fhyd (1 − 1/ξhyd)]−1 = 1.01, whereas the rotational cell DPF is ξcellR ≡ τcell/τ0 = 1 + fhyd (ξhyd −1) = 1.99.
The rotational motion of fluorescent probe molecules (5, 35) and proteins (36), because it is insensitive to obstruction and confinement, has been used to infer local cytoplasmic viscosities. Such measurements may rule out large dynamic perturbations of a majority of cell water, but the deduced apparent viscosity ηapp is not simply related to water dynamics. If the probe interacts directly with cytoplasmic biopolymers, ηapp may become very large. Conversely, if the probe is excluded from the hydration layer, it will mainly sample bulk-like water. Therefore, rotational probe experiments do not necessarily reflect the state of hydration water.
Cell water dynamics are best characterized by techniques such as nuclear spin relaxation and quasielastic neutron scattering (QENS), which directly probe the rotational or short-range translational diffusive motions of individual water molecules. But a QENS study with a given neutron spectrometer can access only a narrow space-time window (37). Moreover, for a system as complex as a cell, the decomposition of the measured incoherent structure factor (ISF) into different motional modes and proton populations is highly model-dependent (37). Whereas the ISF primarily reflects the most abundant proton population with motions in the sampled space-time window, the spin relaxation rate R1(ω0) is dominated by the most slowly rotating water molecules up to a correlation time of order 1/ω0. The MRD technique is therefore uniquely suited for detecting even small populations of strongly perturbed water. With current instrumentation, QENS can detect neither the long-lived internal water molecules responsible for the low-frequency 2H relaxation dispersion nor the small population of secluded hydration sites that dominate the high-frequency (<2 ns) DPF ξ. On the basis of the present MRD results, we would expect QENS data from cell samples to report mainly on the dominant (≈90%) bulk-like intracellular water fraction, yielding ξ ≈ 1.
The limited amount of in vivo QENS data available (14, 38) has been taken to support the view (6, 7) that most cell water is very different from bulk water. QENS data were recently reported for E. coli and H. marismortui cells under similar conditions as used here (14). Using an instrument that samples motions in the 10 ps and 1–3 Å window and interpreting the ISF with a combined jump diffusion and spherical surface diffusion model, these authors obtained ξcellT = 1.2 and, surprisingly, ξcellR = 0.23 for H. marismortui at 12°C. These DPFs were attributed to extracellular water and a minor fraction of cell water, with the implication that the major cell water fraction is too strongly retarded (≫10 ps) to be detected with this instrument. Longer time scales (≈1 ns) were accessed with another instrument, where the ISF was interpreted with jump diffusion (short range) or confined diffusion (at longer range) models yielding ξcellT = 260 and 39, respectively. A motional retardation by 2 orders of magnitude, attributed to 76% of the cell water (14), is grossly inconsistent with our MRD data (ξcellR = 3.8). If the QENS interpretation were correct, we would have measured R1 ≈ 420 s−1 at the highest frequency, instead of 6.7 s−1 (Fig. 1). Furthermore, the reported (14) qualitative difference in local water mobility between 27°C and 12°C is not apparent in the 1H MRD profiles at these temperatures (Fig. 2). Finally, whereas the QENS study inferred extremely slow cell water in H. marismortui but not in E. coli (14), there is little (ξcell) or no (ξhyd) difference between the MRD-derived DPFs for these organisms. We note that, in the present work but not in the QENS study (14), the two cell samples were subjected to identical experimental (NMR) protocols.
The ambiguities and fallacies in the interpretation of previously reported 1H NMR relaxation data from biological cells are avoided here by using the 2H nuclide to simplify the relaxation mechanism, by using an array of NMR instruments to cover a 2H frequency range of unprecedented width, by analyzing the low-frequency relaxation dispersion with rigorous spin relaxation theory, and by invoking as few model assumptions as possible. Each of these methodological advances is essential for the comprehensive dynamical characterization of cell water presented here. Our findings are fully consistent with the current understanding of protein hydration in solutions and crystals but contradict the view that a substantial fraction of cell water differs greatly from bulk water. The 2H MRD approach used here opens up new possibilities for studying water dynamics in vivo and for elucidating the origins of endogenous contrast in magnetic resonance images of soft tissue.
Materials and Methods
Preparation of Cell Samples.
E. coli strain K-12 RV308 [American Type Culture Collection (ATCC) 31608] and H. marismortui (ATCC 43049) were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Cultures were grown aerobically at 37°C in LB medium for E. coli and in DSMZ's Halobacteria Medium 372 for H. marismortui. Growth media were prepared either with distilled H2O or with 99.9% D2O (Spectra Stable Isotopes). In the text, pH of D2O-based samples is reported as pD = pH* + 0.41, where pH* is the value measured with a pH electrode calibrated in H2O buffers. The hydronium ion activity is thus the same in the D2O-based cell samples with pD 8.0 as in the H2O-based cell samples with pH 7.6. After 6 (E. coli) or 24 h (H. marismortui) of incubation, the cell suspensions were centrifuged and the cell pellets washed twice with a D2O–saline buffer. The cell mass was then centrifuged in a 10-mm NMR tube. For further details, see SI Text. The samples used for 1H MRD were prepared in the same way but using distilled H2O instead of D2O. Water content determination, elemental analysis, and amino acid analysis were performed on the cell mass after drying for 12 h at 130°C. The elemental composition of the samples is given Table S2.
To minimize cell death in the samples, the MRD measurements were completed within 6 h of centrifuging the cell suspension into the NMR tube. A control experiment on an E. coli sample, prepared as described above, was performed to assess cell viability. A portion of the cell mass was removed before and after the 6-h measurement period. After serial dilutions, the cell suspension was plated and left to grow overnight. Colony counts indicated that 70 ± 7% of the cells were viable at the end of the MRD measurements (SI Text). Consistent with this result, repeated 2H R1 measurements during the intervening 6-h period showed no significant (at 76.8 MHz) or only ± 5% (at 1.5 kHz) variation.
MRD Experiments.
The longitudinal relaxation rate R1 of the water–2H magnetization was measured from 1.5 kHz to 76.8 MHz by using six different NMR instruments: a Stelar Spinmaster 1 Tesla fast FC spectrometer (1.5 kHz to 6.4 MHz); a Tecmag Discovery spectrometer equipped with an iron-core magnet (Drusch EAR-35N), variable-field lock, and flux stabilizer (11.7 MHz); and five spectrometers equipped with conventional cryomagnets: Bruker Avance DMX 100 (15.4 MHz) and 200 (30.7 MHz), Varian Unity Plus 400 operated at 55.5 MHz, and Varian Unity Inova 500 (76.8 MHz). The relaxation rate R1 of the water–17O magnetization (at natural abundance) was measured at 67.8 MHz on the E. coli sample used for the cell viability control. The longitudinal relaxation rate of the water–1H magnetization was measured from 10 kHz to 500 MHz on the FC spectrometer (10 kHz–40 MHz) and on conventional spectrometers with cryomagnets (200, 360, and 500 MHz). For FC measurements, the prepolarized (PP) and nonpolarized (NP) sequences were used with polarization (for PP) and detection at 6.14 and 4.80 MHz 2H frequency, respectively. The recovery and polarization times were set to 4 T1. In the non-FC experiments, standard inversion recovery pulse sequences were used. Single-exponential recovery/decay curves were obtained throughout (Fig. S3), from which the relaxation rate was determined by a three-parameter fit. The estimated experimental error in R1 is <1%. The sample temperature was maintained at 27.0 ± 0.1°C by a thermostated air flow and was checked before and after each relaxation experiment with a thermocouple referenced to an ice-water bath. The 2H relaxation rate of a pure D2O reference sample (99.9% 2H) was also measured at 27°C, yielding 2.15 ± 0.02 s−1 (E. coli) or 2.18 ± 0.05 s−1 (H. marismortui).
The high salt content, 4.2 mol (Na+ + K+) (kg D2O)−1, of the H. marismortui sample could in principle interfere with the NMR experiments. Thus, the NMR sensitivity deteriorates, because thermal ionic motion induces noise in the receiver coil (39), and the sample is heated by electrolyte friction induced by the electric component of the oscillating radiofrequency field (40). However, the sensitivity loss is insignificant at the low frequencies used here, as also indicated by the negligible difference in 90° pulse length between the H. marismortui and E. coli samples. Substantial sample heating has been reported only in NMR experiments with high duty cycles (as in spin decoupling) at relatively high frequencies. At the low frequencies (< 80 MHz) and very low duty cycles (<10−4) used here, sample heating can be safely neglected even for samples of high conductivity.
Supplementary Material
Acknowledgments.
We thank Hanna Nilsson for help with bacterial cultures and Hans Lilja for NMR spectrometer maintenance. This work was supported by the Swedish Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709585105/DCSupplemental.
References
- 1.Makarov V, Pettitt BM, Feig M. Solvation and hydration of proteins and nucleic acids: A theoretical view of simulation and experiment. Acc Chem Res. 2002;35:376–384. doi: 10.1021/ar0100273. [DOI] [PubMed] [Google Scholar]
- 2.Raschke TM. Water structure and interactions with protein surfaces. Curr Opin Struct Biol. 2006;16:152–159. doi: 10.1016/j.sbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Levy Y, Onuchic JN. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 4.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 5.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: Volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 6.Shepherd VA. The cytomatrix as a cooperative system of macromolecular and water networks. Curr Top Develop Biol. 2006;75:171–223. doi: 10.1016/S0070-2153(06)75006-2. [DOI] [PubMed] [Google Scholar]
- 7.Chaplin M. Do we underestimate the importance of water in cell biology? Nat Rev Mol Cell Biol. 2006;7:861–866. doi: 10.1038/nrm2021. [DOI] [PubMed] [Google Scholar]
- 8.Hazlewood CF, Nichols BL, Chamberlain NF. Evidence for the existence of a minimum of two phases of ordered water in skeletal muscle. Nature. 1969;222:747–750. doi: 10.1038/222747a0. [DOI] [PubMed] [Google Scholar]
- 9.Koenig SH, Brown RD. In: NMR Spectroscopy of Cells and Organisms. Gupta RK, editor. Boca Raton, FL: CRC Press; 1987. pp. 75–114. [Google Scholar]
- 10.Damadian R. Tumor detection by nuclear magnetic resonance. Science. 1971;171:1151–1153. doi: 10.1126/science.171.3976.1151. [DOI] [PubMed] [Google Scholar]
- 11.Halle B, Denisov VP, Venu K. In: Biological Magnetic Resonance. Krishna NR, Berliner LJ, editors. New York: Kluwer Academic/Plenum; 1999. pp. 419–484. [Google Scholar]
- 12.Vaca Chávez F, Halle B. Molecular basis of water proton relaxation in gels and tissue. Magn Reson Med. 2006;56:73–81. doi: 10.1002/mrm.20912. [DOI] [PubMed] [Google Scholar]
- 13.Britton KL, et al. Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei. Proc Natl Acad Sci USA. 2006;103:4846–4851. doi: 10.1073/pnas.0508854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tehei M, et al. Neutron scattering reveals extremely slow cell water in a Dead Sea organism. Proc Natl Acad Sci USA. 2007;104:766–771. doi: 10.1073/pnas.0601639104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg D, Kauzmann W. The Structure and Properties of Water. Oxford, UK: Oxford Univ Press; 1969. [Google Scholar]
- 16.Errington JR, Debenedetti PG. Relationship between structural order and the anomalies of liquid water. Nature. 2001;409:318–321. doi: 10.1038/35053024. [DOI] [PubMed] [Google Scholar]
- 17.Abragam A. The Principles of Nuclear Magnetism. Oxford, UK: Clarendon; 1961. [Google Scholar]
- 18.Halle B. Spin dynamics of exchanging quadrupolar nuclei in locally anisotropic systems. Progr NMR Spectrosc. 1996;28:137–159. [Google Scholar]
- 19.Halle B, Jóhannesson H, Venu K. Model-free analysis of stretched relaxation dispersions. J Magn Reson. 1998;135:1–13. doi: 10.1006/jmre.1998.1534. [DOI] [PubMed] [Google Scholar]
- 20.Denisov VP, Halle B. Hydrogen exchange and protein hydration: The deuteron spin relaxation dispersions of bovine pancreatic trypsin inhibitor and ubiquitin. J Mol Biol. 1995;245:698–709. doi: 10.1006/jmbi.1994.0056. [DOI] [PubMed] [Google Scholar]
- 21.Vaca Chávez F, Hellstrand E, Halle B. Hydrogen exchange and hydration dynamics in gelatin gels. J Phys Chem B. 2006;110:21551–21559. doi: 10.1021/jp057567s. [DOI] [PubMed] [Google Scholar]
- 22.Cayley S, Lewis BA, Guttman HJ, Record MT. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolality. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 23.Halle B. Protein hydration dynamics in solution: a critical survey. Philos Trans R Soc London Ser B. 2004;359:1207–1224. doi: 10.1098/rstb.2004.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halle B. In: Hydration Processes in Biology. Bellisent-Funel M-C, editor. Dordrecht, The Netherlands: IOS; 1998. pp. 233–249. [Google Scholar]
- 25.Schröder C, Rudas T, Boresch S, Steinhauser O. Simulation studies of the protein-water interface. I. Properties at the molecular resolution. J Chem Phys. 2006;124:234907. doi: 10.1063/1.2198802. [DOI] [PubMed] [Google Scholar]
- 26.Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: Am Soc Microbiol; 1996. [Google Scholar]
- 27.Vaca Chávez F, Persson E, Halle B. Internal water molecules and magnetic relaxation in agarose gels. J Am Chem Soc. 2006;128:4902–4910. doi: 10.1021/ja058837n. [DOI] [PubMed] [Google Scholar]
- 28.Persson E, Halle B. Nanosecond to microsecond protein dynamics probed by magnetic relaxation dispersion of buried water molecules. J Am Chem Soc. 2008;130:1774–1787. doi: 10.1021/ja0775873. [DOI] [PubMed] [Google Scholar]
- 29.Halle B, Davidovic M. Biomolecular hydration: From water dynamics to hydrodynamics. Proc Natl Acad Sci USA. 2003;100:12135–12140. doi: 10.1073/pnas.2033320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter F, Kimmich R. Spin lattice relaxation of dipole nuclei (I = 1/2) coupled to quadrupole nuclei (S = 1) Mol Phys. 1982;45:33–49. [Google Scholar]
- 31.Jiao X, Bryant RG. Noninvasive measurement of protein concentration. Magn Reson Med. 1996;35:159–161. doi: 10.1002/mrm.1910350205. [DOI] [PubMed] [Google Scholar]
- 32.Park S, Saven JG. Statistical and molecular dynamics studies of buried waters in globular proteins. Proteins. 2005;60:450–463. doi: 10.1002/prot.20511. [DOI] [PubMed] [Google Scholar]
- 33.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- 34.Konopka MC, Shkel IA, Cayley S, Record MT, Weisshaar JC. Crowding and confinement effects on protein diffusion in vivo. J Bacteriol. 2006;188:6115–6123. doi: 10.1128/JB.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fushimi K, Verkman AS. Low viscosity in the aqueous domain of cell cytoplasm measured by picosecond polarization microfluorimetry. J Cell Biol. 1991;112:719–725. doi: 10.1083/jcb.112.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams S-P, Haggie PM, Brindle KM. 19F NMR measurements of the rotational mobility of proteins in vivo. Biophys J. 1997;72:490–498. doi: 10.1016/S0006-3495(97)78690-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bée M. Quasielastic Neutron Scattering. Bristol, UK: Adam Hilger; 1988. [Google Scholar]
- 38.Trantham EC, et al. Diffusive properties of water in Artemia cysts as determined from quasi-elastic neutron scattering spectra. Biophys J. 1984;45:927–938. doi: 10.1016/S0006-3495(84)84239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gadian D, Robinson F. Radiofrequency losses in NMR experiments on electrically conducting samples. J Magn Reson. 1979;34:449–455. [Google Scholar]
- 40.Led J, Petersen S. Heating effects in carbon-13 NMR spectroscopy on aqueous solutions caused by proton noise decoupling at high frequencies. J Magn Reson. 1978;32:1–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.