Abstract
Aminoacyl-tRNAs are generally formed by direct attachment of an amino acid to tRNAs by aminoacyl-tRNA synthetases, but Gln-tRNA is an exception to this rule. Gln-tRNAGln is formed by this direct pathway in the eukaryotic cytosol and in protists or fungi mitochondria but is formed by an indirect transamidation pathway in most of bacteria, archaea, and chloroplasts. We show here that the formation of Gln-tRNAGln is also achieved by the indirect pathway in plant mitochondria. The mitochondrial-encoded tRNAGln, which is the only tRNAGln present in mitochondria, is first charged with glutamate by a nondiscriminating GluRS, then is converted into Gln-tRNAGln by a tRNA-dependent amidotransferase (AdT). The three subunits GatA, GatB, and GatC are imported into mitochondria and assemble into a functional GatCAB AdT. Moreover, the mitochondrial pathway of Gln-tRNAGln formation is shared with chloroplasts as both the GluRS, and the three AdT subunits are dual-imported into mitochondria and chloroplasts.
Keywords: amidation, aminoacylation, GatCAB, glutamyl-tRNA synthetase, protein trafficking
The formation of aminoacylated tRNAs (aa-tRNAs) plays a central role in protein synthesis. Although most of the 20 aa-tRNAs that supply the translation apparatus are synthesized by direct attachment of each amino acid to its cognate transfer RNAs (tRNAs) by aminoacyl-tRNA synthetases (aaRSs), some can be synthesized by an indirect and nonconventional pathway. This is particularly the case for the formation of Asn-tRNAAsn and Gln-tRNAGln. In the cytosol of eukaryotes and in a small subset of bacteria, Gln-tRNAGln is formed directly by a glutaminyl-tRNA synthetase (GlnRS). In archaea and in the majority of bacteria, Gln-tRNAGln is synthesized by a two-step indirect pathway. The tRNAGln is first mischarged with glutamate (Glu) by a nondiscriminating glutamyl-tRNA synthetase (ND-GluRS). The glutamate attached to tRNAGln is then amidated into glutamine (Gln) by a tRNA-dependent amidotransferase (AdT) generating Gln-tRNAGln. Two types of AdT have so far been identified. The first one, found in bacteria and archaea, corresponds to the heterotrimeric GatCAB enzyme able to transamidate both Asp-tRNAAsn and Glu-tRNAGln. The second type of AdT, restricted to archaea, is the heterodimeric GatDE which can only transamidate Glu-tRNAGln (1).
Because mitochondria and chloroplasts have their own translation machinery, a complete set of aa-tRNAs is expected in these organelles. The pathway of Gln-tRNAGln formation has been explored in a few organelles. In the trypanosamatids Leishmania tarentolae and Trypanosoma brucei, both nuclear-encoded tRNAGln and GlnRS are imported from the cytosol into mitochondria (2, 3). The same situation is expected in Tetrahymena mitochondria (4). In the yeast Saccharomyces cerevisiae, nuclear-encoded tRNAsGln are imported into mitochondria and coexist with mitochondrial-encoded tRNAGln. The cytosolic GlnRS is also imported into these mitochondria (5). In these four cases, mitochondrial Gln-tRNAGln is therefore formed directly.
The present work explores the mitochondrial Gln-tRNAGln formation in plant. No mitochondrial GlnRS activity can be detected. By contrast, the dual-targeted mitochondrial-chloroplastic GluRS can attach Glu to both tRNAGlu, and tRNAGln thus appears to be nondiscriminating. Moreover, AdT activity is detected in a mitochondrial extract. The corresponding enzyme is encoded by three nuclear genes, gatA, gatB, and gatC, and each polypeptide is imported into mitochondria to form the active GatCAB enzyme. The transamidation route has been shown to be used in barley and spinach chloroplasts (6). Interestingly, we show here that the three Gat subunits are also imported into chloroplasts. Therefore, in plants, the indirect transamidation pathway serves to form Gln-tRNAGln in both mitochondria and chloroplasts, and the same enzymes GluRS and GatCAB are shared between the two organelles.
Results
Cytosolic tRNAGln Are Not Present in Mitochondria.
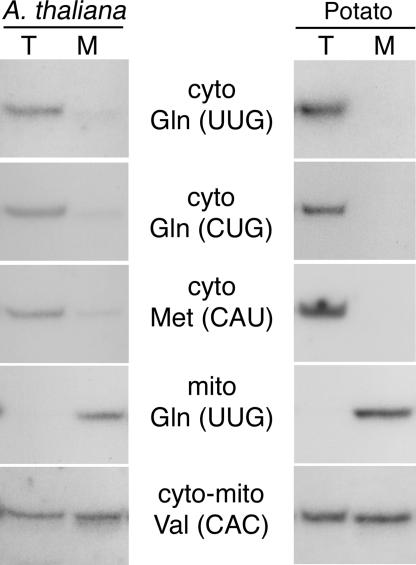
The mitochondrial tRNAGln gene is expressed in Arabidopsis thaliana and other plants (7) (Fig. 1). We verified that, unlike in yeast mitochondria, the two cytosolic tRNAGln are not imported into A. thaliana and potato mitochondria (Fig. 1) and into bean and tobacco mitochondria (data not shown). Therefore, the only tRNAGln used in plant mitochondria is mitochondrial-encoded.
Fig. 1.
Cytosolic tRNAGln are not imported into plant mitochondria. Hybridization of tRNA-specific probes to Northern blot of total (T) and mitochondrial (M) tRNAs from A. thaliana and potato. The mitochondrial tRNAGln gene is expressed [mito Gln(UUG)]. The probes corresponding to the cytosolic tRNAsGln [cyto Gln(UUG) and cyto Gln(CUG)] give similar signals to the probe corresponding to the cytosolic nonimported tRNAMet [cyto Met(CAU)]. As a comparison, the hybridization with a probe corresponding to the cytosolic mitochondrial-imported tRNAVal is shown [cyto-mito Val(CAC)].
Cytosolic GlnRS Is Not Imported into Mitochondria.
A unique GlnRS gene has been found in the A. thaliana nuclear genome. Although its sequence does not display any obvious organellar targeting sequence, we searched for its activity in A. thaliana mitochondrial extracts. A GlnRS activity can be detected only in the total but not the mitochondrial extract (Fig. 2A). Similar results were obtained with a potato mitochondrial extract (data not shown) and with barley and spinach chloroplastic extracts (6).
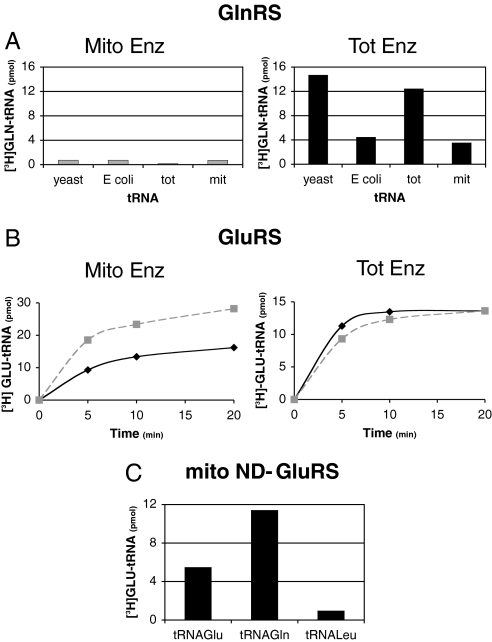
Fig. 2.
ND-GluRS and GlnRS activities in A. thaliana mitochondrial and total extracts. Aminoacylations were performed with A. thaliana mitochondrial (Mito Enz) or total (Tot Enz) enzymatic extracts. (A) Glutaminylation of tRNAs from yeast, E. coli, whole plant cells (tot) or plant mitochondria (mit). (B) Glutamylation of plant total tRNAs (black diamond) and plant mitochondrial tRNAs (gray square, dotted line). (C) Glutamylation of plant mitochondrial tRNAGlu and tRNAGln with A. thaliana mitochondrial enzymatic extract. The assays were performed in the presence of partially purified plant mitochondrial tRNAGlu, tRNAGln, or tRNALeu.
Mitochondrial GluRS Is Nondiscriminating.
The A. thaliana nuclear genome encodes two GluRSs. The protein encoded by one GluRS gene (At5g64050) has been shown to be imported into both plant mitochondria and chloroplasts, and the second GluRS (At5g26710) is expected to be cytosolic (8). Aminoacylation assays show that GluRS activities can be detected in the total (mostly representing the cytosolic GluRS activity) and mitochondrial extracts in A. thaliana (Fig. 2B) and potato (data not shown). Further, we performed aminoacylation tests with an A. thaliana mitochondrial enzymatic extract and partially purified mitochondrial tRNAGlu and tRNAGln. Both tRNAs were glutamylated (Fig. 2C), showing the nondiscriminating character of the organellar GluRS [see also supporting information (SI) Fig. S1]. Altogether, the absence of GlnRS activity and the presence of a nondiscriminating GluRS suggest that, like in chloroplasts (6), the indirect pathway for the formation of Gln-tRNAGln is used in plant mitochondria. The ND-GluRS allows formation of Glu-tRNAGln, which should then be modified into Gln-tRNAGln by an AdT.
Mitochondrial GatCAB Amidotransferase.
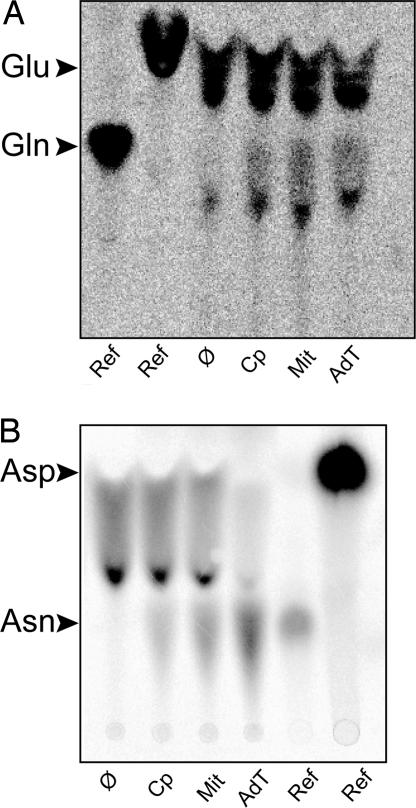
Plant mitochondrial Glu-tRNAs were incubated with Gln as amide donor and either an A. thaliana chloroplastic or mitochondrial enzymatic extract. As expected, the tRNA-dependent conversion of Glu into Gln was obtained with the chloroplastic extract (Fig. 3A) (6). A similar Glu-tRNAGln to Gln-tRNAGln conversion was observed with the mitochondrial extract, showing that a tRNA-dependent AdT also exists in mitochondria. Moreover, the conversion of a bacterial Asp-tRNAAsn into Asn-tRNAAsn was also obtained with both the chloroplastic and the mitochondrial extracts (Fig. 3B).
Fig. 3.
Plant organellar AdT activity. Transamidation assays were performed with A. thaliana chloroplastic (Cp) or mitochondrial (Mit) extracts or with the purified A. thaliana AdT overexpressed in E. coli (AdT). The tRNA-bound [14C]aa were fractionated by TLC. [14C]Glu and [14C]Gln and [14C]Asp and [3H]Asn were used as reference markers (Ref). (A) Transamidation of [14C]Glu-tRNAGln into [14C]Gln-tRNAGln. Plant mitochondrial tRNA were first glutamylated with [14C]Glu by partially purified plant mitochondrial ND-GluRS. Therefore, both [14C]Glu-tRNAGlu and [14C]Glu-tRNAGln were obtained (lane Ø) and further used in transamidation assays with different extracts (Cp, Mit, AdT). (B) Transamidation of [14C]Asp-tRNAAsn into [14C]Asn-tRNAAsn. Purified T. thermophilus tRNAAsn overexpressed in E. coli was first aspartylated by using T. thermophilus ND-AspRS (lane Ø), then used in transamidation tests with different extracts.
We found three genes encoding potential GatA (At3g25660), GatB (At1g48520), and GatC (At4g32915) subunits in the A. thaliana nuclear genome. Phylogenetic analysis of Gat sequences shows that plant GatA and GatB are related to cyanobacteria, by contrast, GatC sequences are poorly conserved (SI Text). Compared with their closest bacterial counterparts, the three A. thaliana proteins display N-terminal extensions that are predicted to be mitochondrial and/or chloroplastic targeting signals (Table 1). The three Gat-coding sequences depleted from their 5′ extension were used to construct an artificial operon for overexpressing the A. thaliana AdT in Escherichia coli. Purified AdT was capable to convert both Glu-tRNAGln into Gln-tRNAGln and Asp-tRNAAsn into Asn-tRNAAsn (Fig. 3) and is indeed of dual specificity. The kcat value (3 s−1) of the purified AdT for Thermus thermophilus Asp-tRNAAsn was determined within the same range of bacterial AdTs (9, 10).
Table 1.
Characteristics of A. thaliana Gat proteins
| Total length, aa | N-term extension, aa | Targeting predictions* |
|||
|---|---|---|---|---|---|
| MitoProt2 | Predotar | TargetP | |||
| GatA | 537 | 45 | mito | mito | chloro |
| GatB | 550 | 55 | mito | chloro | mito |
| GatC | 155 | 60 | mito | mito | chloro |
The SubCellular Proteomic Database (SUBA, www.suba.bcs.uwa.edu.au).
GatCAB Is Targeted to Mitochondria and Chloroplasts.
GatA, GatB, and GatC N-terminal extensions are predicted to be mitochondrial and/or chloroplastic targeting signals (Table 1). A few N-terminal sequences have been shown to allow targeting to both mitochondria and chloroplasts. These sequences are called dual or ambiguous targeting sequences, and prediction programs are usually inefficient to determine the precise localization of proteins with such an extension (11).
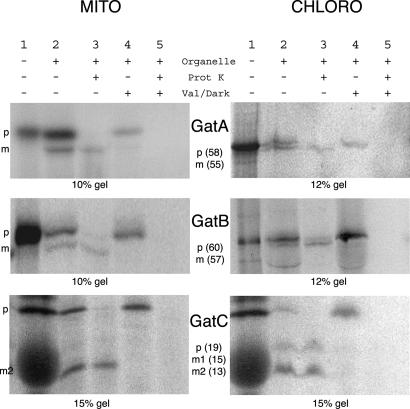
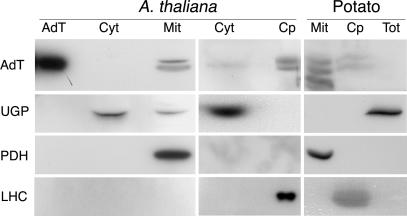
In vitro import assays showed that GatA, GatB, and GatC can be imported into both mitochondria and chloroplasts (Fig. 4), and the mitochondrial and chloroplastic localization of GatA and GatB was confirmed by Western blot (Fig. 5). Thus, the A. thaliana subunits responsible for AdT activity are dual-localized in both mitochondria and chloroplasts.
Fig. 4.
GatA, GatB, and GatC are in vitro imported into isolated mitochondria (MITO) and chloroplasts (CHLORO). Radioactive full-length GatA–C preproteins (p) were obtained by in vitro transcription/translation (lanes 1), then incubated with isolated mitochondria or chloroplasts (lanes 2 and 3). Smaller proteinase K-resistant peptides appear, showing that each preprotein is imported into both organelles and processed into the mature form (m) by cleavage of the targeting sequence. When mitochondria or chloroplasts are preincubated with valinomycine (Val) or in the dark (Dark), respectively (lanes 4 and 5), protein import is inhibited, and the formation of mature proteins is prevented. Moreover, in lanes 5, all of the radioactive signals are digested by proteinase K (Prot K), showing that the signals observed in lanes 3 represent genuine imported proteins protected by mitochondrial or chloroplastic membranes. The molecular mass of preproteins (p) and mature proteins (m) was evaluated on gel and is indicated (in kDa). Two mature GatC proteins, m1 and m2, are obtained in chloroplasts and one in mitochondria (m2). Two matured products have already been observed for other chloroplastic proteins (11).
Fig. 5.
GatA and GatB are immunodetected in plant mitochondrial and chloroplastic protein extracts. Immunodetections were performed with antibodies directed against A. thaliana AdT (AdT; this work), cytosolic UDP-glucose pyrophosphorylase (UGP; Agrisera AB), mitochondrial pyruvate dehydrogenase (PDH; GT Monoclonal Antibodies), and chloroplastic light-harvesting complex II (LHC; C. de Vitry, Institut de Biologie Physico-Chimique, Paris). Although raised against the whole purified AdT, the AdT antibodies recognize only GatA and GatB subunits but not GatC. Two bands were detected with the AdT antibodies in A. thaliana extracts, at 55 and 57 kDa and correspond to GatA and GatB subunits. Another band of lower molecular mass corresponding to an unknown protein is also observed in the potato mitochondrial extract. Mit, mitochondrial; Cp, chloroplastic; Cyt, cytosolic; Tot, total proteins extracts; AdT, overexpressed and purified A. thaliana AdT.
Discussion
In this article, we show that the formation of Gln-tRNAGln is achieved by an indirect pathway in plant mitochondria. The mitochondrial-encoded tRNAGln, which is the only mitochondrial tRNAGln, is first charged with Glu by the mitochondrial ND-GluRS prior to its conversion into Gln-tRNAGln by a mitochondrial AdT. We have identified three genes whose products GatA, GatB, and GatC are imported into mitochondria and assemble into a functional AdT. GatCAB, like all bacterial AdTs, appears to be a dual-specific AdT that displays both Glu-AdT and Asp-AdT activities. However, the formation of Asn-tRNAAsn is probably achieved by the direct pathway in plant organelles, because a chloroplastic-mitochondrial AsnRS (At4g17300) has been identified in A. thaliana (12). Last, the mitochondrial indirect pathway of Gln-tRNAGln formation is shared with chloroplasts, because both GluRS and the three subunits of AdT are dual-imported into mitochondria and chloroplasts.
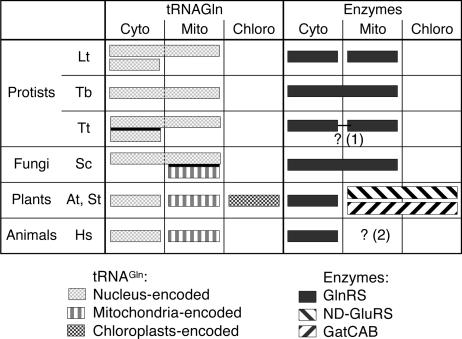
These results contrast with those obtained in protists and fungi mitochondria, where the direct pathway is used for Gln-tRNAGln formation (Fig. 6). Mitochondria and chloroplasts are expected to arise via endosymbiosis of α-proteobacteria and cyanobacteria, respectively. In these two types of bacteria, GatCAB have been identified but not GlnRS. In the protists and fungi mitochondria, at least some of the mitochondrial tRNAGln are shared with the cytosol, and the direct pathway of Gln-tRNAGln formation, also used in the cytosol, has supplanted the ancestral bacterial originating indirect route (Fig. 6).
Fig. 6.
Cytosolic and organellar tRNAGln and Gln-tRNAGln forming enzymes. Cross-compartment sharing of tRNA and enzymes is observed in L. tarentolae (Lt) (2), T. brucei (Tb) (3), T. thermophila (Tt) (4), and S. cerevisiae (Sc) (5). At, A. thaliana; St, S. tuberosum; Hs, H. sapiens. In Lt, the mitochondrial GlnRS is distinct from the cytosolic one; by contrast, the same GlnRS is used in the cytosol and mitochondria of Tb and Sc. In Tt (1), GlnRS activities have similar specificities in the two compartments, and it is not excluded that the same enzyme is shared between the cytosol and the mitochondria. No mitochondrial GlnRS gene has been identified in Hs (2) (22).
In plants, mitochondrial and chloroplastic tRNAGln are encoded by mitochondrial and chloroplastic genomes, respectively, and their sequences are close to bacterial tRNA (13). In parallel, the bacterial ancestral indirect pathway of Gln-tRNAGln synthesis was preserved in plant organelles by using a ND-GluRS and GatCAB. During evolution, most mitochondrial and chloroplastic protein genes have been lost or transferred to the nucleus. The present-time organellar GluRS, GatA, and GatB subunits are clearly related to their cyanobacterial counterparts (8) (Figs. S2 and S3), suggesting that the ancestral chloroplastic pathway was kept during evolution as the mitochondrial one was lost.
The plant GatC phylogenetic origin is difficult to rule out. This is not specific to the plant AdT subunit but is a general feature of GatC proteins (14). There is little or no sequence conservation in this short polypeptide along the tree of life. Therefore, the low degree of sequence conservation among GatC protein does not allow phylogenetic inference. This is probably a consequence of the structural role played by this subunit that is a linker in the heterotrimer, which, through its N- and C-terminal ends, interacts with parts of GatA and GatB that display only partial sequence conservations (15).
Orthologs of the Gat subunits have been annotated in a few other eukaryotic genomes. In yeast, GatA and GatB orthologs (YMR293C and PET112, respectively) are essential for mitochondrial functions (16, 17), and mouse and human GatB are annotated as mitochondrial. The function of these Gats is still an open question. So far, no mitochondrial AdT has been identified in nonplant eukaryotes (5) (Fig. 6).
In plants, because their products have to be imported into organelles, GluRS, GatA, GatB, and GatC nuclear genes have acquired targeting signals during evolution. These dual-targeting sequences allow import of the proteins into both chloroplasts and mitochondria. In most cases, targeting sequences are specific to one organelle or the other, and dual targeting is rare. Approximately 40 examples of dual-targeted proteins of the thousands of organellar-imported proteins have been identified in A. thaliana, but half of these proteins correspond to aaRSs (11). Eighteen of 24 identified A. thaliana organellar aaRSs are shared between mitochondria and chloroplasts (8, 18). The dual localization of GatCAB gives an example of cross-compartment sharing of enzymes involved in aa-tRNA synthesis. A higher level of complexity, however, is reached, because GatCAB is an example of a multimeric enzyme with three subunits dual-imported into mitochondria and chloroplasts.
Materials and Methods
Cloning of A. thaliana Gat Sequences.
RNA was extracted from A. thaliana leaves by using the TRI-reagent (Molecular Research Center) according to the manufacturer's instructions. The GatC coding sequence was amplified by RT-PCR by using specific primers overlapping the initiation and stop codons and cloned into pCRII vector. The full-length cDNAs corresponding to GatA and GatB were obtained from the RIKEN BioResource Center (19).
Cloning and Purification of A. thaliana AdT Overexpressed in E. coli.
The gatC, gatA, and gatB genes from A. thaliana were amplified by PCR from RIKEN and RT-PCR clones to (i) remove the mitochondrial and chloroplastic targeting sequences (corresponding to the sequence encoding, respectively, the 58, 44, and 53 first aa), and (ii) introduce restriction sites that were subsequently used to ligate the three genes, organized in an artificial gatCAB operon, in the pCYB1 expression vector (New England Biolabs), as described in ref. 20. Overexpression was performed in E. coli BL21 Rosetta 2 strain. The AdT was purified from the S100 extract by chromatographies on DEAE–cellulose, Phosphocellulose (Whatman), and Hydroxyapatite (Bio-Rad). Pure enzyme (70 mg) was obtained from 40 g of cells.
Isolation of Mitochondria and Chloroplasts.
Mitochondria were extracted from potato tubers or A. thaliana cell culture growing in the dark. Chloroplasts were extracted from pea, potato, or A. thaliana leaves (see SI Text for detailed methods).
Transfer RNA Extraction.
Total and mitochondrial tRNAs were prepared as described in ref. 21. Because of sequence identity, the same probes were used for hybridizing with A. thaliana and potato tRNAs in Northern blots (13). For aminoacylation and amidation assays, tRNAs were extracted from potato tubers because of higher yield. Potato mitochondrial partially purified tRNAGln and tRNAGlu were obtained after 2D polyacrylamide gel electrophoresis (21). The purity obtained for these tRNA was ≈60%, which corresponds to a 30× enrichment in comparison with the unfractionated mitochondrial tRNAs. The spots corresponding to tRNAGln and tRNAGlu were not adjacent on the 2D gel, and cross-contamination could not be detected by Northern blot.
Aminoacylation Assays.
Enzymatic extracts were prepared from A. thaliana leaves, mitochondria or chloroplasts, or potato mitochondria (21). Aminoacylation reactions were performed with 15 μg of total enzymatic extract or 7.5 μg of organellar enzymatic extract, in the presence of 60 μM [3H]-Gln or [3H]-Glu (820 mCi/mmol) and 40 μM E. coli or yeast tRNAs or 8 μM plant total or mitochondrial tRNAs (21).
tRNA-Dependent Amidation Assays.
The preparation of [14C]Asp-tRNAs and [14C]Glu-tRNAs is described in SI Text and Tables S1–S3. The amidation reaction mixture containing 100 pmol of [14C]Asp-tRNA or 10 pmol of [14C]Glu-tRNA and 1 μM A. thaliana pure AdT or 15 μg of A. thaliana mitochondrial or chloroplastic crude extract was incubated during 15 min at 37°C. The reaction was stopped, and the tRNA-bound [14C]aa were analyzed by TLC (SI Text and Tables S1–S3).
Western Blot Analysis.
Western blot analysis was performed as described in ref. 8 (SI Text and Tables S1–S3). The purified overexpressed A. thaliana AdT was injected into rabbits to raise antibodies that were purified by affinity chromatography (CNBr-activated Sepharose 4B, Amersham Pharmacia Biotech).
In Vitro Import of Proteins.
In vitro import of proteins into isolated mitochondria or chloroplasts was performed as in ref. 8 (SI Text and Tables S1–S3).
Supplementary Material
Acknowledgments.
We thank Valérie Cognat for the phylogenetic analysis. C.P. has a fellowship from the French Ministère Délégué à l'Enseignement Supérieur et à la Recherche. M.B. is a fellow of the Association pour la Recherche sur le Cancer (ARC). This work was supported by the Université Louis Pasteur, Centre National de la Recherche Scientifique, ARC, and Action Concertée Incitative Biologie Cellulaire Moléculaire et Structurale.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712299105/DCSupplemental.
References
- 1.Ibba M, Becker HD, Stathopoulos C, Tumbula DL, Söll D. The adaptor hypothesis revisited. Trends Biochem Sci. 2000;25:311–316. doi: 10.1016/s0968-0004(00)01600-5. [DOI] [PubMed] [Google Scholar]
- 2.Nabholz C, Hauser R, Schneider A. Leishmania tarentolae contains distinct cytosolic and mitochondrial glutaminyl-tRNA synthetase activities. Proc Natl Acad Sci USA. 1997;94:7903–7908. doi: 10.1073/pnas.94.15.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinehart J, Horn EK, Wei D, Söll D, Schneider A. Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei. J Biol Chem. 2004;279:1161–1166. doi: 10.1074/jbc.M310100200. [DOI] [PubMed] [Google Scholar]
- 4.Rusconi C, Cech T. The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes Dev. 1996;10:2870–2880. doi: 10.1101/gad.10.22.2870. [DOI] [PubMed] [Google Scholar]
- 5.Rinehart J, Krett B, Rubio MA, Alfonzo JD, Söll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schon A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 7.Duchêne AM, Maréchal-Drouard L. The chloroplast-derived trnW and trnM-e genes are not expressed in Arabidopsis mitochondria. Biochem Biophys Res Commun. 2001;285:1213–1216. doi: 10.1006/bbrc.2001.5303. [DOI] [PubMed] [Google Scholar]
- 8.Duchêne AM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailly M, et al. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J Biol Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 11.Pujol C, Maréchal-Drouard L, Duchêne AM. How can organellar protein N-terminal sequences be dual targeting signals? In silico analysis and mutagenesis approach. J Mol Biol. 2007;369:356–367. doi: 10.1016/j.jmb.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Peeters NM, et al. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J Mol Evol. 2000;50:413–423. doi: 10.1007/s002390010044. [DOI] [PubMed] [Google Scholar]
- 13.Brubacher-Kauffmann S, Maréchal-Drouard L, Cosset A, Dietrich A, Duchêne AM. Differential import of nuclear-encoded tRNAGly isoacceptors into Solanum tuberosum mitochondria. Nucleic Acids Res. 1999;27:2037–2042. doi: 10.1093/nar/27.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curnow AW, et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 16.Mulero JJ, Rosenthal JK, Fox TD. PET112, a Saccharomyces cerevisiae nuclear gene required to maintain rho+ mitochondrial DNA. Curr Genet. 1994;25:299–304. doi: 10.1007/BF00351481. [DOI] [PubMed] [Google Scholar]
- 17.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 18.Small I, et al. The strange evolutionary history of plant mitochondrial tRNAs and their aminoacyl-tRNA synthetases. J Hered. 1999;90:333–337. [Google Scholar]
- 19.Seki M, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 20.Bailly M, et al. tRNA-dependent asparagine formation in prokaryotes. Characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNAAsn. Methods. 2008;44:146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Maréchal-Drouard L, Small I, Weil JH, Dietrich A. Transfer RNA import into plant mitochondria. Methods Enzymol. 1995;260:310–327. doi: 10.1016/0076-6879(95)60148-1. [DOI] [PubMed] [Google Scholar]
- 22.Bonnefond L, et al. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry. 2005;44:4805–4816. doi: 10.1021/bi047527z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.