Abstract
Water reservoirs formed by the leaf axils of bromeliads are a highly derived system for nutrient and water capture that also house a diverse fauna of invertebrate specialists. Here we investigate the origin and specificity of bromeliad-associated insects using Copelatinae diving beetles (Dytiscidae). This group is widely distributed in small water bodies throughout tropical forests, but a subset of species encountered in bromeliad tanks is strictly specialized to this habitat. An extensive molecular phylogenetic analysis of Neotropical Copelatinae places these bromeliadicolous species in at least three clades nested within other Copelatus. One lineage is morphologically distinct, and its origin was estimated to reach back to 12–23 million years ago, comparable to the age of the tank habitat itself. Species of this clade in the Atlantic rainforest of southern Brazil and mountain ranges of northern Venezuela and Trinidad show marked phylogeographical structure with up to 8% mtDNA divergence, possibly indicating allopatric speciation. The other two invasions of bromeliad water tanks are more recent, and haplotype distributions within species are best explained by recent expansion into newly formed habitat. Hence, bromeliad tanks create a second stratum of aquatic freshwater habitat independent of that on the ground but affected by parallel processes of species and population diversification at various temporal scales, possibly reflecting the paleoclimatic history of neotropical forests.
Keywords: aquatic arthropods, evolution
The Bromeliaceae represent an outstanding adaptive radiation of vascular plants (1, 2). They are the largest (nearly) exclusively Neotropical family of flowering plants, with >2,600 species and 56 genera in a great variety of habitats, from granitic outcrops, coastal dune fields, and tropical rainforests to high-altitude cloud forests. Despite this great diversity, the major adaptive diversification of bromeliads derives from a common ancestor dated to only some 20 million years ago (MYA), possibly related to the evolution of CAM photosynthesis, epiphytism, and impounding leaves (2–4). More than half of the species in 26 genera are epiphytic (1), and particularly species with rosulate water and debris impounding tanks (phytotelmata) strongly contribute to the characteristic appearance of the Neotropical forest canopy (5). Bromeliad phytotelmata can be impressive, holding up to 45 liters of water each (6) and up to 50,000 liters per hectare (7). They often represent the only abundant lentic habitat in Neotropical forests (8).
Not surprisingly, bromeliad water tanks harbor a diverse aquatic fauna of >400 species including insects and amphibians, many of which are strictly dependent on this habitat (9, 10). Whereas most species merely undergo their larval development in the tanks, including bromeliad-breeding frogs, odonates, mosquitoes, and marsh beetles (Scirtidae) (9, 11), various rotifers, crustaceans, and diving beetles (Dytiscidae) are associated with these specialized habitats throughout their entire life cycle. The latter include several species in the subfamily Copelatinae, a group of medium-sized (≈5 mm) beetles dominating small standing-water habitats throughout the tropics including some 140 species from the Neotropics. Six species currently placed in the genera Aglymbus and Copelatus have been described or are here newly reported from water tanks in at least seven bromeliad genera, Aechmea, Brocchinia, Guzmania, Hohenbergia, Nidularium, Tillandsia, and Vriesea.
Bromeliads are an important feature of the Neotropical biota that have impacted biodiversity through time (12). However, the relevance of this water source for the evolutionary biology and diversification of their inhabitants remains unclear. Bromeliad tanks may simply add island-like freshwater habitat patches, easily available to organisms capable of flight such as Copelatus, which depend on small, ephemeral water bodies. The availability of bromeliads in addition to the typical surface water habitats may affect the spatial distribution, overall abundance, and dispersal patterns of the beetles. Alternatively, bromeliadicolous communities may have persisted in isolation without using other aquatic habitats, as suggested by a surprising species diversity and allopatric species distributions in ostracod crustaceans endemic to Jamaican bromeliads (12). Equally, a molecular phylogenetic analysis of a radiation of Jamaican land crabs included a single bromeliad-inhabiting species that separated from other lineages up to 3 MYA (13), also supporting the antiquity of bromeliad associations. The long-term persistence of bromeliad-dependent lineages may be expected specifically in flightless invertebrates, including ostracods, which show specific phoretic associations with amphibians for dispersal (14), whereas more dispersive, flighted groups may show a mixed utilization of bromeliad tanks and aquatic habitats on the ground (9). Although the Copelatinae found in bromeliads are generally assumed to be specific to this habitat (15, 16), the stringency and evolutionary persistence of these specialized associations in the presence of other freshwater bodies remain unclear (9) but have important implications for lineage evolution. We therefore investigated the origin of bromeliad associations and the evolution of diversity and endemism in this habitat in the context of a continental-scale molecular phylogeny of Neotropical Copelatinae.
Results
Habitats.
We obtained confirmation of strict associations with bromeliads for Aglymbus bromeliarum, which was abundant in Guzmania water tanks up to 45 m high in the canopy of montane forest in Rancho Grande, Maracay, Venezuela. These forests were rather dry, and no other Copelatus, or indeed any other diving beetle, was found outside of the water tanks during an intensive search conducted by H. Escalona and M.B. in 2004. The species had previously been found in Tillandsia sp. in Trinidad (15) and in Tillandsia fendlerii at Altos de Pipe, Caracas (by A.V. in 2004). Detailed studies on Aglymbus bimaculatus in Salesópolis, São Paulo, recovered adults and larvae abundantly from tanks of Vriesea jonghei, Nidularium innocentii, and Aechmea pectinata, growing at a height of 0–13 m in the Atlantic Rainforest, but never outside bromeliads (16). A. bimaculatus (described from São Paulo, Brazil) and A. bromeliarum (from Trinidad) are morphologically readily separated from all other Copelatinae and thus easy to identify. Both species have never been found outside bromeliads and were absent among otherwise rich museum collections of Neotropical Copelatus sampled from surface water habitats. Two possibly undescribed species from Venezuela (Copelatus sp. 1) and Brazil (Copelatus sp. 2) are here reported from bromeliads for the first time. They were found in water tanks of Guzmania cylindrica and Guzmania lingulata as well as N. innocentii, respectively. We conducted intensive searches of puddle habitats in the adjacent areas but did not find these species outside bromeliads. Finally, two Copelatus species, not available for sequencing, were reported from Brocchinia cordylinoides in the Kaieteur National Park of Guyana (15) but not from any other habitat. This further suggests that bromeliad associations are a species-specific trait acquired by only a minority of copelatines, which, however, are widely distributed in the Neotropical region.
Molecular Phylogenetics.
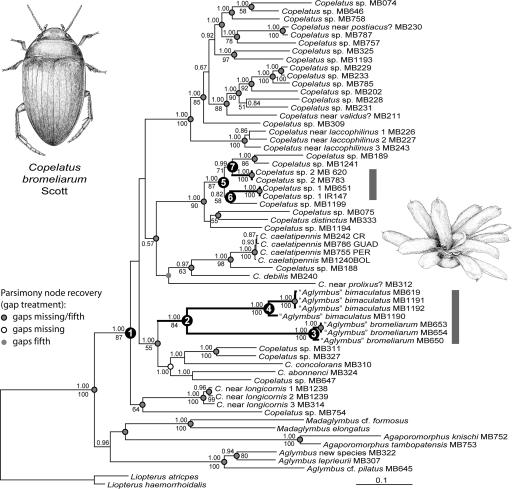
To provide the evolutionary context for the bromeliadicolous species, 57 specimens representing all major lineages of Neotropical Copelatinae were included in a combined analysis of one nuclear and three mtDNA loci. Among 2,283 characters in the aligned matrix 43.1–49.3% of positions were variable in the three mtDNA loci versus 33.8% in histone 3 (H3), resulting in 791 parsimony-informative characters, of which 82 were in H3. Sequence divergence was also consistently higher for the mtDNA markers (Table 1). Alignment of the length-variable rrnL marker was conducted with a variety of alignment algorithms, including a tree-generating alignment implemented in POY (17). Simultaneous analysis using parsimony and Bayesian methods of tree construction produced trees of very similar topology (Fig. 1). Different alignments of rrnL had minimal effect on the topology [supporting information (SI) Fig. S1]. Node support was high except for the basal branching within the Copelatus clade, although the monophyly of Copelatus (Fig. 1, node 1) was strongly supported [Bayesian posterior probability (PP) = 1.0; parsimony bootstrap (BS) = 87].
Table 1.
Marker performance and tree statistics
| Gene fragment | Variable characters | Informative characters | Tree length | No. of trees | Confidence interval | Retention index | P distance average* | P distance range* |
|---|---|---|---|---|---|---|---|---|
| cob | 176 | 159 | 1,281 | 16 | 0.219 | 0.549 | 0.161 | 0–0.232 |
| cox1 | 317 | 294 | 2,116 | 7 | 0.245 | 0.569 | 0.133 | 0–0.180 |
| nad1-rrnL | 354 | 248 | 1,182 | 55 | 0.412 | 0.685 | 0.074 | 0–0.107 |
| H3 | 108 | 90 | 371 | 594 | 0.442 | 0.730 | 0.063 | 0–0.106 |
| Total | 955 | 791 | 5,184 | 6 | 0.280 | 0.580 | 0.111 | 0–0.181 |
*Ingroup only.
Fig. 1.
Phylogeny of Neotropical Copelatus. The tree is the majority rule consensus of the stationary phase of the Bayesian analysis based on the implied alignment from POY. Values above branches are posterior probabilities (>0.5) as a measure of branch support; numbers below are parsimony bootstrap values above 50% where the same nodes were recovered in either analysis. Bromeliad-associated lineages are marked with thick branches. Nodes labeled 1–7 are discussed in the text.
The bromeliadicolous A. bimaculatus and A. bromeliarum were sisters and unambiguously nested within Copelatus (Fig. 1, node 2; PP = 1.0, BS = 84). These species had been placed in Aglymbus because they lack metacoxal lines on their ventral side, a suggested diagnostic [but homoplastic (18)] character of this genus. Reassessment of this trait has already resulted in the transfer of the Afrotropical and Malagasy Aglymbus to Copelatus and Madaglymbus, respectively (18). Hence, we transfer here both bromeliadicolous species to the genus Copelatus, with the following taxonomic changes: Copelatus bimaculatus (16), new combination, and Copelatus bromeliarum (Scott, 1912), new combination.
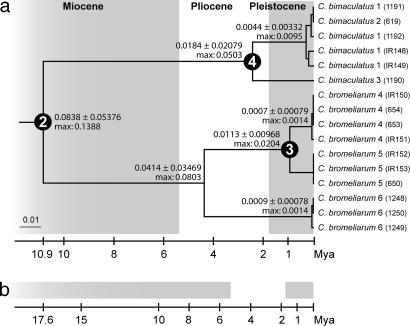
The sequence information was used to establish a temporal framework for the evolution of this group by using the standard insect mtDNA molecular clock of 2.3% divergence my−1 (19), supported by evidence from Fijian Copelatus (20). The crown age of the bromeliad-associated clade, and therefore the minimum age for the origin of this association, was dated to between 12.3 and 14.1 MYA depending on the model of branch length distribution (each with wide confidence intervals; Table 2). These values increase to 16.1–23.6 MYA if the date is inferred from the age of the common ancestor with the nonbromeliadicolous sister taxon (stem group node; Fig. 1, node 1). Different alignment procedures had only a very minor impact on these estimates, as is evident from measures of sequence divergence. The divergence of C. bimaculatus and C. bromeliarum (relevant to assessing the minimum age of the bromeliad clade) under the four alignment procedures produced distance estimates in a very narrow range (average uncorrected patristic distances P = 0.108–0.111; corrected under a GTR model = 0.118–0.122). A larger sample of this clade including 16 specimens (six specimens for C. bimaculatus and 10 for C. bromeliarum, from three localities each) was sequenced for cox1, and the resulting clock-constrained tree was calibrated by using the estimated age interval for node 2. Each sampling region formed a monophyletic clade, and genetic distance increased with geographical distance. Total cox1 sequence divergence (Fig. 2) between populations of C. bromeliarum from Maracay and Caracas (≈60 km apart) was 2%, and between these and samples from Trinidad (≈600 km) a maximum of 8%. Similarly, cox1 divergence between individuals of C. bimaculatus from Florianópolis and Nova Friburgo (≈800 km) was 5%. Specimens from Florianópolis and Santo Amaro (≈30 km) were in the same haplotype cluster, suggesting the lack of phylogeographical structure at this small geographical scale (Fig. 2, MB619 and MB1191).
Table 2.
Age of colonization of bromeliads and diversification of bromeliad-associated lineages
| Node | Constant |
Lognormal |
Exponential |
|||
|---|---|---|---|---|---|---|
| Mean | 95% C.I. | Mean | 95% C.I. | Mean | 95% C.I. | |
| 1 | 16.1 | 14.0–18.2 | 16.4 | 14.2–18.5 | 23.6 | 16.7–32.7 |
| 2 | 14.1 | 10.9–17.6 | 12.8 | 9.5–16.3 | 12.3 | 5.8–19.0 |
| 3 | 0.8 | 0.5–1.1 | 0.9 | 0.5–1.3 | 3.2 | 0.3–7.3 |
| 4 | 2.9 | 1.6–4.3 | 3.0 | 1.5–4.8 | 4.3 | 1.0–8.8 |
| 5 | 5.9 | 4.9–6.9 | 5.9 | 4.6–7.1 | 10.2 | 4.5–15.8 |
| 6 | 4.3 | 3.0–5.5 | 4.5 | 2.8–6.3 | 7.1 | 1.3–12.9 |
| 7 | 5.9 | 4.9–7.0 | 5.5 | 4.2–7.0 | 7.5 | 2.7–12.5 |
Three different age estimates [mean and 95% confidence interval (C.I.)] are given based on different clock assumptions.
Fig. 2.
Phylogenetic tree of the C. bromeliarum/C. bimaculatus clade. One of nine equally parsimonious tree based on the cox1 gene is shown (all other trees differed in zero- or near zero-length branches). Two different tree calibrations (a and b) are given corresponding to the extreme values of the 95% confidence interval of node 2 age estimated in Fig. 1 and Table 2 assuming a constant clock. Localities: 1, Santo Amaro; 2, Florianópolis; 3, Nova Friburgo; 4, Maracay; 5, Altos de Pipe; 6, Trinidad.
Two further species of Copelatus from bromeliad water tanks fell within a clade otherwise containing species from forest floor puddles (Fig. 1, node 5, PP = 1.0, BS = 87). Copelatus sp. 1 from Venezuela diverged from its puddle-inhabiting sister species 4.3–7.1 MYA (Fig. 1, node 6). The two collecting sites for the bromeliad species are ≈380 km apart (Caracas, MB651; Península de Paraguaná, Cerro Santa Ana, IR147), but we found no haplotype differences between these populations. Copelatus sp. 2 from southern Brazil (Florianópolis island, MB620; Santo Amaro da Imperatriz, MB783; 30 km apart, same haplotype) was estimated to have diverged from a puddle-inhabiting sister clade containing species from Bolivia and Argentina 4.9–7.5 MYA (Fig. 1, node 7).
Discussion
Ancient Origin of Bromeliad Associations.
Literature records as well as new field observations made at all sites where individuals were collected from bromeliad tanks confirmed the absence of bromeliad-associated species in adjacent small pools or puddles on the ground. Copelatines are frequently found in ephemeral, very small water bodies, indicating their good dispersal abilities, which is also evident from their abundance at light traps. These traits may have facilitated the colonization of the bromeliad habitat initially and ultimately promoted the evolution of a specialized lifestyle. Although their propensity for dispersal is still evident, our findings argue against the suggestion (9) that bromeliad tanks are used only occasionally in combination with other habitats.
The broad taxonomic coverage of Neotropical copelatines allowed the placement of bromeliad-associated lineages relative to species not found in this habitat. Thus, bromeliadicolous taxa previously assigned to the genus Aglymbus were recognized as morphologically derived Copelatus. Dating of this lineage along the subtending branch places this group to anywhere between 12.3 MYA (dated for the minimum, i.e., crown group, age) and 23.6 MYA (maximum age for the stem group), although the value for the more realistic lognormal branch length distribution applied during rate smoothing would reduce this maximum age to 16.4 MYA. These estimates are similar to that for the earliest bromeliad lineages containing water impounding species (4) and indicate that copelatines began to use the bromeliad water tanks soon after this new ecosystem became available.
In two other cases, the associations with bromeliad tanks were inferred to be more recent, indicating repeated origins in a mixed clade of surface water- and bromeliad-inhabiting taxa. These newly discovered species, plus two other Copelatus exclusively reported from bromeliads but not available here for sequencing, morphologically resemble typical Copelatus and might therefore lack specific adaptations attributable to the associations with bromeliads. However, although we know little about the biology of these species, their tight association with bromeliads at the sampling localities was unequivocal. Detailed morphological studies will have to assess any consistent allometric shifts in body shape parameters or body size, in comparison to their respective sister taxa, to establish evolutionary trends at the early stages of bromeliad associations. Further biogeographic analyses of these species and their equally poorly known sisters will identify the source of bromeliad-associated species either from a local or distant pool of ancestral species to test evidence for ecological speciation in sympatry.
Intraspecific Variation and Phylogeography.
Bromeliad water tanks are spatially clearly delimited and therefore suitable to investigate diversification and endemism in narrowly defined habitat patches. In our analyses, intraspecific variation reflected physical distances and known biogeographical barriers. Thus, the estimated divergence between the populations of C. bimaculatus from the northern and southern Mata Atlântica (2.2–4 MYA; Fig. 2) is in agreement with phylogeographic evidence from pitviper populations in this region, showing a divergence of 3.8 MYA in the wake of orogenic events that caused changes in precipitation regimes and fragmentation of the Atlantic Forest (21, 22). The genetic split between these lineages is supported by the fixation of subtle morphological differences. C. bimaculatus from Nova Friburgo is completely black, whereas the southern populations have a characteristic reddish patch on the base of the otherwise black elytra, suggesting the possibility of further speciation within the group.
An even greater divergence (up to 8%, corresponding to 2.9–4.3 MYA) and significant phylogeographic structure occurs in C. bromeliarum between three localities in Venezuela and Trinidad. Montane forests near Caracas and Maracay, in the western Cordillera de la Costa, are only ≈60 km apart, yet cox1 divergence amounts to 2%. Late Pleistocene forest fragmentation due to climatic change is well documented for the region (23–25), and possible marine transgressions along the northern coast dramatically changing the landscape were suggested for the late Pliocene (26). Most likely C. bromeliarum is present in the montane forests of the Península de Paria and the Turimikere Massif, which together with northern Trinidad form the isolated eastern Cordillera de la Costa, widely separated from the western Cordillera de la Costa and our localities, Caracas and Maracay, by the river plains west of Barcelona. The present range and genetic divergence of C. bromeliarum in these isolated ranges might be explained by rare successful dispersal events followed by allopatric differentiation, as suggested for other Copelatinae (27). The wide river plains of Barcelona never supported the type of montane forests occupied by bromeliadicolous Copelatus, but extensive fieldwork would be needed to establish their detailed phylogeographic structure.
The clear geographical structure and reciprocally monophyletic intraspecific subgroups (see ref. 20) would separate these entities under a diagnostic species concept (28). For example, the three populations of C. bromeliarum show between 55 (Venezuela vs. Trinidad) and a minimum of 14 (between close Venezuelan localities) unique differences in mtDNA (plus one consistent change in H3 between Venezuela and Trinidad). These strongly structured groups with great divergences also preclude the meaningful application of nested clade analysis (29). Morphological differences were subtle or absent between these groups, and a denser sampling would be necessary to confirm this possibility, but the finding of local differentiation supports the notion of great evolutionary persistence of these specialized lineages.
The deep nodal split between C. bromeliarum and C. bimaculatus as well as their highly disjointed geographical distribution some 4,500 km apart suggests the possibility of undiscovered species in the area between southeast Brazil and the north of Venezuela. The ancient formations of the Guyana Shield constitute the biogeographic origin of bromeliads and also harbor the center of diversity of the basal bromeliad genus Brocchinia, including some species with the tank habit (4). The Mata Atlântica coastal rainforest is also rich in tank bromeliads but is currently isolated from Amazonia by drier biomes of Caatinga and Cerrado. Connections between these areas in the past under changing climates (30) created different configurations of forest and dry formations in the Neotropics (31, 32). Present-day lowland Amazonia features a comparatively depauperate bromeliad flora. Although some large water impounding species exist, these tend to form more upright foliage, concealing the water (D. Benzing, personal communication). These considerations suggest that future searches for unknown bromeliadicolous copelatines should be focused in the northern Mata Atlântica, along the Andean slopes and especially in the Guyana highlands.
In contrast to the C. bimaculatus plus C. bromeliarum clade, the more recent origin of the bromeliad-associated Copelatus sp. 1 and sp. 2 is also reflected in their low intraspecific variation. Copelatus sp.1 was collected from two sites ≈380 km apart in Altos de Pipe, Caracas, and from the Cerro Santa Ana, Península de Paraguaná, but showed identical sequences. Montane forest of Cerro Santa Ana probably formed only during the Pleistocene, and present-day bromeliad habitats might be even younger. The Cerro Santa Ana mountain range is surrounded by an arid zone, and even the entire Cerro was very likely too dry to support moist forest during the last dry period only ≈12,000 years ago, as in the well documented formations on the opposite side of the Gulf of Venezuela (33). To colonize the Cerro, the beetles had to bridge an arid area to reach a suitable habitat in the Cordillera de la Costa and Sierra de San Luis, crossing a narrow, arid land connection to the Península, presumably by dispersal between water tanks. A striking aspect of the bromeliad associations is the high degree of endemism in several of these vagile beetles, which contrasts with other species with large areas and little genetic divergence. This appears to mirror the situation in puddle-associated species, some of which have huge ranges (including, e.g., Copelatus caelatipennis, for which we document here cox1 divergence of <0.2% between localities >3,000 km apart in Bolivia and Guadeloupe; Fig. 1) whereas others appear geographically limited.
Conclusions
Phytotelmata provide a natural model for the study of interactions in communities and food webs, as they represent simplified microcosms in island-like habitats (9). Phylogenetic analysis is required to bring these composite communities into an evolutionary, comparative framework. Bromeliad phytotelmata represent an ancient habitat, which, as shown here, maintained its specialized fauna over evolutionarily extended periods, in one case reaching back to the earliest origin of the tank habitat itself. This ecosystem is known to include poorly dispersive species such as ostracods, earthworms, and crabs (9). It is striking that these specializations are found also in the generally highly dispersive, winged species of Copelatinae, which might be expected to use other water bodies as well. However, the retention of dispersal abilities seems necessary to avoid local extinctions in the ephemeral bromeliad phytotelmata and therefore may be instrumental in the evolutionary persistence of these associations. Dispersal is an underexplored key factor relevant to virtually all aspects of the composition and evolution of the “tank islands” communities. In copelatines, the high dispersal ability sustains the formation of a network of aquatic habitats parallel to that on the ground, which also produced phylogeographic patterns affected by expansion and contraction of forest habitat in a way similar to the open-water lineages. Comparisons of bromeliad-associated lineages with their open-water sister groups, applied to taxa with various levels of dispersal abilities (e.g., ref. 12), will produce a predictive framework of their population structure, phylogeography, speciation/extinction dynamics, and evolutionary lineage persistence.
Materials and Methods
Sampling and Genetic Data.
We included all known genera of Neotropical Copelatinae (Copelatus, Aglymbus, and Agaporomorphus) (Table S1). Preliminary phylogenetic studies of Copelatinae from across their pantropical range revealed that Afrotropical and Malagasy species considered members of Aglymbus are monophyletic with Copelatus or Madaglymbus, respectively (18, 34), leaving the remaining Aglymbus as a monophyletic group confined to the Neotropics. Molecular clock estimates suggest that these groups separated from a common ancestor ≈60–40 MYA (27, 34), and their current distribution probably involved exchange between continents (27). Trees were rooted by using the Palearctic copelatines Liopterus haemorrhoidalis and Liopterus atriceps and the Malagasy Madaglymbus cf. formosulus and Madaglymbus elongatus (34). We sequenced four gene fragments previously used in the group (27), coding for mitochondrial cytochrome b (cob = 357 bp); cytochrome c oxidase subunit 1 (cox1 = 735 bp); NADH subunit 1 and large subunit ribosomal RNA (nad1-rrnL = 750–763 bp); and partial nuclear histone 3A gene (h3 = 320 bp), using PCR primers and well established procedures (35). Sequences have been deposited in the GenBank database under accession numbers AM947384–AM947434 (cob), AM945966–AM946014 (rrnL), AM945593–AM945649 (cox1), and AM945690–AM945739 (h3) (Table S1).
Phylogenetic Analyses.
The rrnL marker showed length variation of up to 13 bp between any two sequences. Alignment was performed by employing a range of recent alignment procedures implementing tree alignment (“direct optimization”) using POY version 3.0.11 (17) and “progressive” alignment procedures including Muscle (36), Prank (37), and Mafft (38). We also performed tree searches on a matrix (based on the Muscle alignment) from which all “gapped” characters had been removed. The POY alignments were generated from simultaneous optimization of nucleotide changes and indels conducted in combined analysis with the three other markers (see ref. 39). An “implied alignment” derived from ancestral character states at nodes can be used for subsequent model-based analyses on the branch length because it presents an explicit optimization of homologies (i.e., optimized for minimal conflict with the other data) that cannot be obtained with other alignment procedures (40). Bayesian analysis on the implied alignment was performed with MrBayes 3.1.2. Tree searches involved four runs and eight chains of 5 × 106 generations under a GTR+I+Γ model preferred by Modeltest (41), with and without setting a prior for uniform branch lengths (i.e., constant clock), discarding the initial 1 × 106 trees before reaching stationarity in likelihood values (burn-in). Parsimony heuristic searches were conducted in PAUP* version 4.0b10 (42) using 1,000 replicates of random sequence addition and alternative gap treatment as missing data or a fifth character state. Parsimony bootstrap support was obtained after 1,000 pseudoreplicates with five random sequence addition replicates each.
Tree Calibration and Node Dating.
We tested for rate constancy with a likelihood ratio test comparing the likelihood scores from the unconstrained and clock-constrained Bayesian analyses. The lack of fossils for Neotropical Copelatinae precluded a direct calibration of tree topologies. Instead, node dating was based on the arthropod standard mtDNA clock of 0.0115 substitutions per site per lineage per MY (= 2.3% pairwise sequence divergence per MY) (19). This rate was applied to the combined three mtDNA markers, and branch lengths as well as associated node ages for relevant nodes were coestimated in a Bayesian framework by using Beast 1.3 and analyzed by using Tracer 1.3 (http://evolve.zoo.ox.ac.uk/). Seven relevant nodes were dated (nodes 1–7 in Fig. 1). Three molecular clock models were tested, including a uniform clock model and two relaxed clock models (lognormal and exponential branch length distribution models) (Table 2). Bayesian searches implemented a GTR+I+Γ model and were based on a Markov chain Monte Carlo search with 1 × 107 generations.
Supplementary Material
Acknowledgments.
We thank David Benzing, T. Givnish, Ernesto Medina, Hermes Escalona, J. H. Frank, J. C. Navarro, and J. Reichholf for information relevant to this work and helpful comments on the manuscript. We thank Daegan Inward, Rolf Beutel, and Kelly Miller for specimens. We are grateful to Ascanio Rincón (Caracas), who provided invaluable information on the paleoecology of Venezuela. Saul Flores, Jafet Nassar, Ernesto Medina, and Robert Wingfield provided help with identifying the Venezuelan bromeliad species. Processing of samples from Venezuela was possible thanks to the Agreement of Access to Genetic Resources contracted between the Instituto Venezolano de Investigaciones Científicas and the Ministry of the Environment and Natural Resources. We thank Ruth Kühbander for the drawings. Funding was provided by German Science Foundation Grant DFG BA 2152/3-2, SYNTHESYS Grant GB-TAF 2811, the Natural History Museum London travel fund, British Ecological Society Grant SEPG 2257(a), Spanish Science Ministry Grant CGL2004-00028, and the Alexander von Humboldt Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AM947384–AM947434 (cob), AM945966–AM946014 (rrnL), AM945593–AM945649 (cox1), and AM945690–AM945739 (h3)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0710368105/DCSupplemental.
References
- 1.Benzing DH, editor. Bromeliaceae: Profile of an Adaptive Radiation. Cambridge, UK: Cambridge Univ Press; 2000. [Google Scholar]
- 2.Crayn DM, Winter K, Smith JAC. Multiple origins of crassulacean acid metobolism and the epiphytic habit in the Neotropical family Bromeliaceae. Proc Natl Acad Sci USA. 2004;101:3703–3708. doi: 10.1073/pnas.0400366101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Givnish TJ, et al. Ancient vicariance or recent long-distance dispersal? Inferences about phylogeny and South American-African disjunctions in Rapateaceae and Bromeliaceae based on ndhF sequence data. Int J Plant Sci. 2004;165:S35–S54. [Google Scholar]
- 4.Givnish TJ, Millam KC, Berry PE, Sytsma KJ. In: Monocots: Comparative Biology and Evolution—Poales. Columbus JT, Friar EA, Porter JM, Prince LM, Simpson MG, editors. Claremont, CA: Rancho Santa Ana Botanic Garden; 2007. pp. 3–26. [Google Scholar]
- 5.Ozanne CMP, et al. Biodiversity meets the atmosphere: A global view of forest canopies. Science. 2003;301:183–186. doi: 10.1126/science.1084507. [DOI] [PubMed] [Google Scholar]
- 6.Zahl PA. Hidden worlds in the heart of a plant. Natl Geogr Mag. 1975;147:389–397. [Google Scholar]
- 7.Williams DD. The Biology of Temporary Waters. Oxford: Oxford Univ Press; 2006. [Google Scholar]
- 8.Thienemann A. Die Tierwelt der tropischen Pflanzengewässer. Arch Hydrobiol Suppl. 1934;13:1–91. [Google Scholar]
- 9.Kitching RL. Food Webs and Container Habitats: The Natural History and Ecology of Phytotelmata. New York: Cambridge Univ Press; 2000. [Google Scholar]
- 10.Greeney HF. The insects of plant-held waters: A review and bibliography. J Trop Ecol. 2001;17:241–260. [Google Scholar]
- 11.Lehtinen RM, Richards CM, Nussbaum RA. Origin of a complex reproductive trait: Phytotelm breeding in mantellid frogs. Misc Publ Univ Michigan Mus Zool. 2004;193:45–54. [Google Scholar]
- 12.Little TJ, Hebert PDN. Endemism and ecological islands: The ostracodes from Jamaican bromeliads. Freshwater Biol. 1996;36:327–338. [Google Scholar]
- 13.Schubart CD, Diesel R, Hedges SB. Rapid evolution to terrestrial life in Jamaican crabs. Nature. 1998;393:363–365. [Google Scholar]
- 14.Lopez LCS, Filizola B, Deiss I, Rios RI. Phoretic behaviour of bromeliad annelids (Dero) and ostracods (Elpidium) using frogs and lizards as dispersal vectors. Hydrobiologia. 2005;549:15–22. [Google Scholar]
- 15.Balfour-Browne J. On two new species of bromeliadicolous Copelatus (Col., Dytiscidae) Entomol Monthly Mag. 1938;74:100–102. [Google Scholar]
- 16.Resende CM, Vanin SA. Aglymbus bimaculatus, sp. n., a new bromeliadicolous beetle from the Atlantic forest (Coleoptera: Dytiscidae) Aquat Insects. 1991;13:123–128. [Google Scholar]
- 17.Wheeler WC, Gladstein DS, Laet JD. POY. 2002 Version 3.0, ftp.amnh.org/pub/molecular/poy.
- 18.Shaverdo H, Monaghan MT, Lees DC, Ranaivosolo R, Balke M. A new genus of Malagasy endemic diving beetles and description of a highly unusual species based on morphology and DNA sequence data (Dytiscidae: Copelatinae) Syst Biodivers. 2008;6:43–51. [Google Scholar]
- 19.Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monaghan MT, Balke M, Pons J, Vogler AP. Beyond barcodes: Complex DNA taxonomy of a South Pacific Island radiation. Proc Biol Sci. 2006;273:887–893. doi: 10.1098/rspb.2005.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grazziotin FG, Monzel M, Echeverrigaray S, Bonatto SL. Phylogeography of the Bothrops jararaca complex (Serpentes: Viperidae): Past fragmentation and island colonization in the Brazilian Atlantic Forest. Mol Ecol. 2006;15:3969–3982. doi: 10.1111/j.1365-294X.2006.03057.x. [DOI] [PubMed] [Google Scholar]
- 22.Behling H, Negrelle RRB. Late Quaternary tropical rain forest and climate dynamics from the Atlantic lowland in southern Brazil. Quaternary Res. 2001;56:383–389. [Google Scholar]
- 23.Lin HL, Peterson LC, Overpeck JT, Trumbore SE, Murray DW. Late Quaternary climate change from δ18O records of multiple species of planktonic foraminifera: High-resolution records from the anoxic Cariaco Basin, Venezuela. Paleoceanography. 1997;12:415–427. [Google Scholar]
- 24.Curtis JH, Brenner M, Hodell DA. Climate change in the Lake Valencia Basin, Venezuela, ∼12,600 yr BP to present. Holocene. 1999;9:609–619. [Google Scholar]
- 25.González LA, Gómez R. High resolution speleothem paleoclimatology of Northern Venezuela: A progress report. Bol Soc Venez Espeleol. 2002;36:27–29. [Google Scholar]
- 26.Frailey CD. In: Dort W Jr, editor. Neogene Paleogeography of the Amazon Basin; TER-QUA Symposium Series; Lincoln, NE: Institute for Tertiary-Quaternary Studies; 2002. pp. 71–97. [Google Scholar]
- 27.Balke M, Pons J, Ribera I, Sagata K, Vogler AP. Infrequent and unidirectional colonization of hyperdiverse Papuadytes diving beetles in New Caledonia and New Guinea. Mol Phylogenet Evol. 2007;42:505–516. doi: 10.1016/j.ympev.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 28.Sites JW, Marshall JC. Delimiting species: A Renaissance issue in systematic biology. Trends Ecol Evol. 2003;18:462–470. [Google Scholar]
- 29.Templeton AR. Statistical phylogeography: Methods of evaluating and minimizing inference errors. Mol Ecol. 2004;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- 30.Santos AMM, Cavalcanti DR, da Silva JMC, Tabarelli M. Biogeographical relationships among tropical forests in north-eastern Brazil. J Biogeogr. 2007;34:437–446. [Google Scholar]
- 31.Haffner J. Speciation in Amazonian forest birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- 32.Simpson BB. The South American Herpetofauna: Its Origin, Evolution, and Dispersal. In: Duellman WE, editor. Lawrence: Univ of Kansas; 1979. pp. 157–188. [Google Scholar]
- 33.Sugden AM. The ecological, geographic, and taxonomic relationships of the flora of an isolated Colombian cloud forest, with some implications for island biogeography. J Arnold Arboretum. 1982;63:31–61. [Google Scholar]
- 34.Balke M, Ribera I, Vogler AP. MtDNA phylogeny and biogeography of Copelatinae, a highly diverse group of tropical diving beetles (Dytiscidae) Mol Phylogenet Evol. 2004;32:866–880. doi: 10.1016/j.ympev.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Balke M, Ribera I, Beutel RG. The systematic position of Aspidytidae, the diversification of Dytiscoidea (Coleoptera, Adephaga) and the phylogenetic signal of third codon positions. J Zool Syst Evol Res. 2005;43:223–242. [Google Scholar]
- 36.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucl Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler WC. Optimization alignment: The end of multiple sequence alignment in phylogenetics? Cladistics. 1996;12:1–9. [Google Scholar]
- 40.Wheeler WC. Implied alignment: A synapomorphy-based multiple-sequence alignment method and its use in cladogram search. Cladistics. 2003;19:261–268. [PubMed] [Google Scholar]
- 41.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 42.Swofford DL. Sunderland, MA: Sinauer; 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4.0b. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.