Abstract
p53-dependent G1 and G2 cell cycle checkpoints are activated in response DNA damage that help to maintain genomic stability. p53 also helps to protect cells from damage that occurs during S phase, for example, when the cells are starved for DNA precursors or irradiated with a low dose of UV. p53 is activated in normal cells starved for pyrimidine nucleotides by treatment with N-(phosphonacetyl)-l-aspartate (PALA). The treated cells progress through a first S phase with kinetics similar to those of untreated cells. However, the DNA of the treated cells begins to become damaged rapidly, within 12 h, as revealed by a comet assay, which detects broken DNA, and by staining for phosphorylated histone H2AX, which accumulates at sites of DNA damage. Because the cells survive, the damage must be reversible, suggesting single-strand breaks or gaps as the most likely possibility. The transiently damaged DNA stimulates activation of ATR and CHK1, which in turn catalyze the phosphorylation and accumulation of p53. Although PALA-induced DNA damage occurs only in dividing cells, the p53 that is activated is only competent to transcribe genes such as p21 and macrophage inhibitory cytokine 1 (whose products regulate G2 and G1 or S phase checkpoints, respectively) after the cells have exited the S phase during which damage occurs. We propose that p53 is activated by stimulation of mismatch repair in response to the misincorporation of deoxynucleotides into newly synthesized DNA, long before the lack of pyrimidine nucleoside triphosphates causes the rate of DNA synthesis to slow appreciably.
Keywords: macrophage inhibitory cytokine 1, p21, N-(phosphonacetyl)-l-aspartate, S phase
The tumor suppressor p53 is essential for the normal responses to DNA damage that help to maintain genomic stability. Extensive DNA damage leads to the irreversible arrest or apoptosis of normal cells, preventing cells in which mutations are likely to have occurred from propagating. In most human tumors, the normal negative regulatory function of p53 is compromised, either by direct mutation, deletion or inactivation of p53 itself, or disruption of signaling pathways that activate p53 or respond to activated p53, the “p53 network” (1). In contrast, arrest of DNA or RNA synthesis or depletion of pyrimidine nucleotide pools leads to reversible arrest, appropriate to protecting cells, rather than disposing of them to protect the organism. Cell-protective p53-dependent cell cycle checkpoints are activated in response to arrest of DNA synthesis (2, 3) or arrest of mitosis (4, 5). Fibroblasts from normal individuals or Li-Fraumeni patients arrest stably when treated with N-(phosphonacetyl)-l-aspartate (PALA), an inhibitor of pyrimidine nucleotide synthesis (6), and remain viable for weeks at a time (7). However, this property is lost in post-crisis Li-Fraumeni fibroblasts when the remaining wild-type p53 allele is lost (8). In one such cell line, MDAH041, restoration of wild-type p53 restores the normal response to PALA (8). As shown by several laboratories, (7–9), the ability of human and mouse cells to give rise to PALA-resistant colonies depends on inactivation of the p53 response pathway. Thus, normal cells that can arrest stably and reversibly in PALA do not give resistant colonies, whereas most cells defective in p53 die rapidly in PALA but do give rise to rare PALA-resistant cells through amplification of the CAD gene (10).
Linke et al. (2) showed that normal human fibroblasts do not incur extensive DNA damage as indicated by the absence of chromosomal aberrations such as chromatid breaks, chromosome exchanges, and double minute chromosomes upon treatment with PALA. Rodent and human fibroblasts with wild-type, functional p53 arrest in the cell cycle, preventing them from synthesizing DNA under adverse conditions (8, 11). Previously, we showed that p53-dependent arrest within S phase is revealed in cell lines that either have relatively low levels of p53 or lack expression of p21, a major mediator of arrest in G1 or G2 (8, 12). In contrast, cells lacking p53 continue DNA synthesis in the presence of PALA, thereby incurring DNA damage that activates a signaling cascade involving caspase activation, cytochrome c release, and finally apoptosis (13). Furthermore, activated p53 stimulates the secretion of the cytokine macrophage inhibitory cytokine 1 (MIC-1), which mediates reversible and protective cell cycle arrest in S phase (13). However, it is not known how p53 is activated in cells starved for nucleotides. We now find that cells starved for pyrimidine nucleotides, while attempting to progress through S phase, incur limited, reversible DNA damage that provides a signal for the phosphorylation and activation of p53 through the ATR–CHK1 pathway.
Results
DNA Damage in Cells Starved for Pyrimidine Nucleotides.
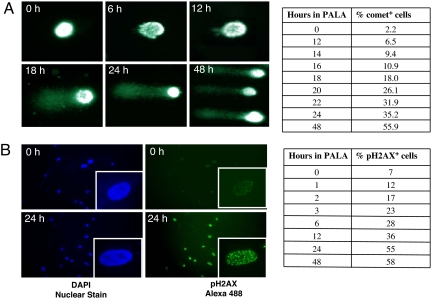
Unbalanced pools of nucleotides lead to misincorporation during DNA synthesis, activating the mismatch repair process, which includes the creation of a repair patch, a transient form of DNA damage (14). We used two sensitive assays to investigate whether treatment with PALA indeed leads to DNA damage capable of providing a signal that activates p53. Normal BJ fibroblasts treated with 250 μM PALA were analyzed by comet staining, an effective method for evaluating DNA damage. The denatured, cleaved DNA fragments migrate out of the cell under the influence of an electric field, resulting in a distinct tail, whereas undamaged DNA migrates more slowly and remains within the confines of the nucleus (15). This test detects both single- and double-stranded DNA breaks. A time-dependent increase in comet-positive cells was evident, beginning at ≈12 h after addition of PALA (Fig. 1A), and almost 56% of the cells were positive by 48 h. Because phosphorylated histone H2AX (γH2AX) accumulates at sites of broken DNA (16), we also examined the status of γH2AX after PALA treatment. BJ cells treated with 250 μM PALA for 1–48 h were stained for this protein and examined by confocal microscopy. As early as 2–3 h after PALA addition there was an increase in the number of γH2AX-positive cells and approximately half of these cells were positive by 24 h (Fig. 1B). Linke et al. (2) showed that pyrimidine nucleotide pools and the rate of RNA synthesis decline to ≈50% of their initial values 12 h after treatment of normal human fibroblasts with PALA. Together, these observations strongly support the conclusion that cells in which the pyrimidine nucleotide pools are just beginning to be depleted in response to PALA still do incur DNA damage that is detectable by two different sensitive assays.
Fig. 1.
DNA damage in cells starved for pyrimidine nucleotides. (A) (Left) DNA damage, measured by comet assay in BJ cells treated with 250 μM PALA. (Right) Represented are comet-positive cells. One hundred cells were counted for each time point. (B) pH2AX in BJ cells treated with 250 μM of PALA. (Left) A representative field of pH2AX-positive cells is shown. (Inset) At higher magnification showing distinct punctate staining. One hundred cells were counted for each time point. (Right) Represented are pH2AX-positive cells counted for each time point. (Magnifications: ×20; Insets, ×40.)
Activation and Phosphorylation of p53 in Response to PALA Uses ATR and CHK1.
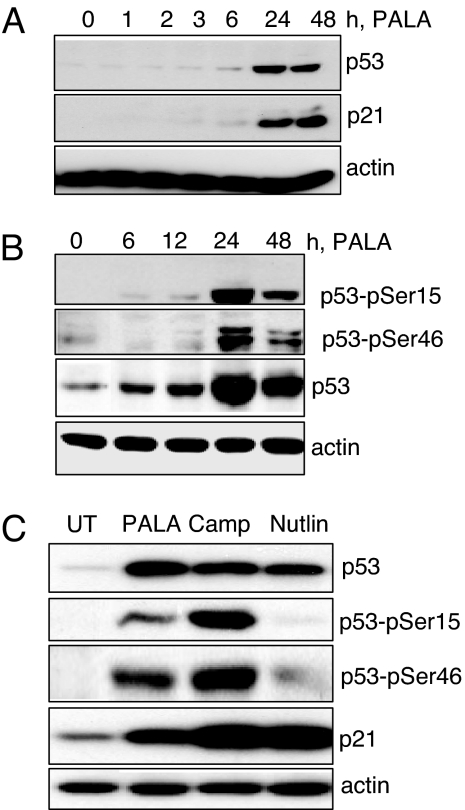
The observation that treatment with PALA does lead to detectable DNA damage made it important to investigate how such damage might lead to p53 induction. Total lysates from PALA-treated BJ cells were analyzed for the total amount of p53 and phosphorylation of serine residues of p53. There was an appreciable increase in the amount of p53 protein and in the amount of the p53-dependent gene product p21 (Fig. 2A). Time-dependent phosphorylation of p53 serine residues 15 and 46 was also observed upon treatment with PALA (Fig. 2B). As is well known, treatment of BJ cells with the DNA-damaging drug camptothecin resulted in p53 stabilization and phosphorylation of p53 (Fig. 2C). Treatment with nutlin, which induces p53 without phosphorylation by inhibiting its binding to the negative regulator mdm2 (17), served as a negative control. These observations demonstrate that a signal generated by starvation for pyrimidine nucleotides leads to the phosphorylation and accumulation of p53, confirming the observations of many laboratories.
Fig. 2.
Induction and phosphorylation of p53 in cells starved for pyrimidine nucleotides. Western analysis of p53 in BJ cells treated with 250 μM PALA is shown. Treatment with camptothecin (a known DNA-damaging agent) and nutlin (activator of p53 with out DNA damage) served as controls.
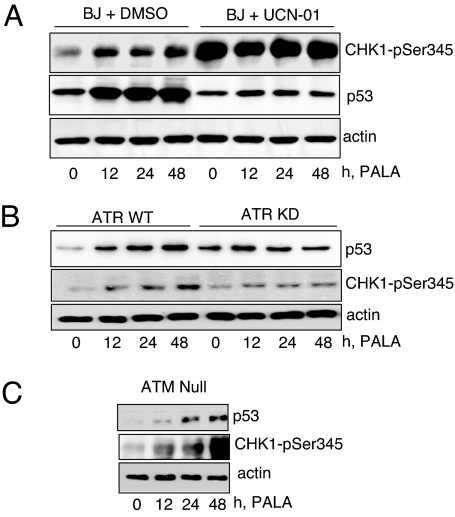
Because the signal from damaged DNA can lead to the phosphorylation of CHK1, to gain further insight into the mechanism of stabilization and activation of p53 by PALA we analyzed the phosphorylation of serine residues 345 and 317 of CHK1 by using specific antibodies. PALA treatment did lead to the phosphorylation of serine residues 345 (Fig. 3A) and 317 (data not shown). Therefore, modification and stabilization of p53 in response to transient DNA damage caused by PALA treatment is likely to involve the CHK1–ATR pathway. To test the involvement of ATR and ATM, we treated ATR WT, ATR kinase-dead (KD) cells (Fig. 3B), and ATM-null primary fibroblasts (Fig. 3C) with PALA for 12, 24, or 48 h. Exposure to PALA led to increased expression of total p53 (Fig. 3 B and C) and phosphorylation of p53 at serine 6 (data not shown) in ATR WT and ATM-null cells but not in ATR KD cells. Phosphorylation of CHK1 at serine 317 (data not shown) and serine 345 (Fig. 3 B and C) was also observed in ATR WT and ATM-null cells but not in ATR KD cells. To investigate further the involvement of CHK1, cells were pretreated with 300 nM UCN-01, a CHK1 inhibitor (18), for 45 min before treating them with 250 μM PALA (Fig. 3A). p53 induction and phosphorylation of CHK1 occurred only in control cells, and there was no significant increase in p53 levels in UCN-01-treated cells. The higher levels of CHK1-pSer345 seen in the presence of UCN-01 are caused by stabilization of this phosphorylation by this inhibitor (18). We conclude that p53 induction and phosphorylation in response to PALA are mediated primarily through ATR and CHK1 and that ATM has a minor role at most.
Fig. 3.
ATR- but not ATM-dependent increase in the phosphorylation of p53 and CHK-1. (A) Western analysis of phosphorylated CHK1 and p53. BJ cells were pretreated with UCN-01 or solvent (DMSO) for 45 min and then treated with 250 μM PALA for 12–48 h. (B) Total p53 and phosphorylated CHK1 in ATR WT (U2OS) and ATR KD cells treated with 250 μM PALA. (C) Total p53 and phosphorylated CHK1 in ATM-null primary fibroblasts treated with 250 μM PALA.
DNA Damage and p53 Activation only in Actively Dividing Cells.
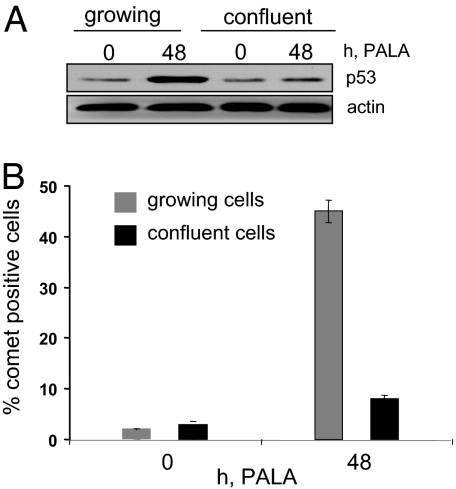
Because treatment with PALA inhibits de novo pyrimidine nucleotide synthesis, cells are most likely to be vulnerable during S phase, when they require large amounts of dCTP and dTTP for DNA synthesis. Consequently, it is logical that cells should be growing for PALA to induce DNA damage. To test this point, actively growing BJ cells were compared with confluent cells. After PALA treatment, only the growing cells showed an increased level of p53 (Fig. 4A). Actively growing cells did incur DNA damage (46% comet-positive cells; Fig. 4B). In contrast, there was very little DNA damage (4–6% comet-positive cells) and no induction of p53 in confluent cells. Actively growing cells were shown to incorporate much more BrdU than confluent BJ cells, as expected (data not shown). Moreover, BJ cells grown in complete media with 250 μM PALA plus 100 mM uridine did not induce p53 or p21 (data not shown), as expected because uridine bypasses the PALA-mediated block to pyrimidine nucleotide biosynthesis. These observations support the conclusion that PALA induces DNA damage only in actively dividing cells that are starved for pyrimidine nucleotides but not in arrested cells or cells that are not starved.
Fig. 4.
DNA damage and p53 activation in actively dividing cells. (A) Expression of p53 in actively growing or confluent BJ cells treated with 250 μM PALA. The Western procedure was used for analysis. (B) DNA damage, measured by comet assay in actively growing or confluent BJ cells treated with 250 μM PALA. One hundred cells were counted for each time point.
Induction and Activation of p53 in Cells Traversing Through S Phase.
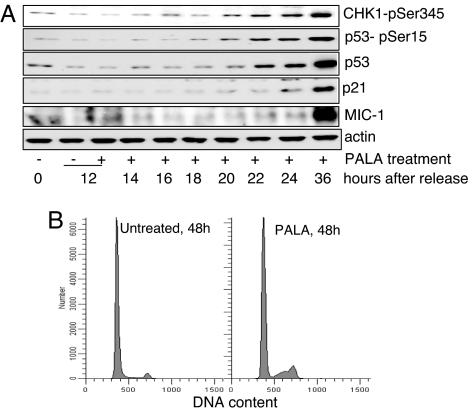
Based on the above findings, we propose that PALA causes DNA damage only when cells are in S phase. BJ cells were synchronized by serum starvation for 36 h and, at time 0, the block was released by adding fresh medium and uridine-free serum. The cells were treated with 250 μM PALA 12 h after release, harvested 0, 12, 14, 16, 18, 20, 22, 24, and 36 h after release and analyzed for the expression of p53 and two p53 target proteins. Cell cycle analysis showed that ≈30% of the cells were in S phase after 20 h, and ≈60% were in G1 after 36 h (data not shown). The amount of p53 increased 22 h after release and remained high up to 36 h (Fig. 5). Phosphorylation of p53 on serine 15 and CHK1 on serine 345 was also elevated, in parallel with total p53 (Fig. 5). Interestingly, protein (Fig. 5A) and RNA (data not shown) expression of the p53 target genes p21 and MIC-1 was increased only 36 h after synchronization and not after 22 h. We conclude that PALA induces DNA damage in S phase, when unbalanced pools of deoxynucleoside triphosphates are likely to stimulate mismatch repair. However, p53 target genes are not up-regulated in S phase, where p53 is transcriptionally inactive (19). Only after cells reach G2 can p53 induce the expression of p21 and MIC-1, which, in turn, are responsible for G1/G2 and S phase arrest, respectively, as discussed below.
Fig. 5.
Induction and activation of p53 in cells traversing through S phase. (A) BJ Tert cells were serum-starved for 36 h and, after synchronization, fresh medium with dialyzed serum was added. Ten hours after release, the cells were treated with 250 μM PALA and harvested after 12–36 h. Lysates of treated cells were analyzed by Western analysis for phosphorylated CHK1, p53, p53-ser15, p21, and MIC-1. (B) Growing BJ cells (60–70% confluent) were stained with 20 μM BrdU for 30 min followed by 100 μM thymidine chase for an additional 30 min. Cells were then treated with 250 μM PALA and harvested for cell cycle analysis at different time points.
A different experiment was done to determine how soon after PALA treatment perturbation of the cell cycle became evident. Growing BJ cells were pulse-labeled with BrdU for 30 min, followed by a thymidine chase and administration (or not) of 250 μM PALA. Cells were trypsinized and fixed at various times, followed by staining with anti-phospho-S780-Rb (to mark mitotic cells), anti-BrdU, and DAPI, and analyzed kinetically (20, 21). For the first 24 h there was no significant difference between the PALA-treated and untreated cell populations: The S-phase transit times were 6.5 and 6.6 h (not statistically different) for the treated and untreated cells, respectively; nor were there differences in G1, G2, or M phase times (data not shown). Note that the activation of p53 in PALA-treated cells is very strong at 24 h (Fig. 2), clearly showing that p53 activation occurred well before the rate of DNA synthesis is affected by PALA. However, at 48 h there were many more cells in S phase in the PALA-treated samples than in the control samples, with a skewing toward late S phase (Fig. 5B), indicating an abnormal S phase in which cells are accumulating late. These results are consistent with a model in which, in normal cells with wild-type p53, the skewing of deoxynucleoside triphosphate pools in response to PALA creates reversible DNA damage, sufficient to activate p53 and thus to activate secondarily the expression of proteins that provide protective arrest at multiple points in the cell cycle. Taken together, these events occur before the deoxynucleoside triphosphate pools become depleted enough in response to PALA to cause a substantial decrease in the rate of DNA synthesis and related changes in S phase transit. As shown by earlier studies (13), in cells in which p53 is absent or in which the p53 pathway is compromised, failure to arrest DNA synthesis when pyrimidine nucleoside triphosphate pools are depleted leads to irreversible DNA damage that eventually causes apoptosis.
Discussion
Treatment of cells with PALA blocks de novo pyrimidine nucleotide biosynthesis and induces growth arrest of normal mammalian fibroblasts (8, 10). When nucleotide pools become unbalanced, a signal is activated that leads to the induction and activation of p53. The activated p53, in turn, leads to reversible, long-term cell cycle arrest. By analyzing metaphase spreads, Linke et al. (2) found no evidence for DNA breaks in PALA-treated normal human fibroblasts. However, several questions remain unanswered: How is p53 activated in response to starvation for pyrimidine nucleotides? Which p53-dependent signals trigger protective arrest in S phase? By what mechanisms are cell growth and DNA replication arrested for many days without irreversible damage to DNA in normal cells? Although the expression of MIC-1 has been reported to be both p53-dependent and -independent (22, 23), our recent work has identified MIC-1 as a p53 target that has a major role in protecting cells in S phase (13). The detailed mechanism of MIC-1-dependent protective arrest remains to be explored. The results of the current study deal with the issue of how p53 is activated in response to treatment with PALA.
We studied whether PALA treatment causes any DNA damage by using analyses different from those used by Linke et al. (2). Comet and γH2AX assays revealed that starvation for pyrimidine nucleotides does lead to DNA damage that is generated only during S phase. The damage then triggers the phosphorylation of p53 at serine residues, prompting us to examine proteins known to play a role in the activation of p53 in response to DNA damage. The serine/threonine kinases CHK1 and CHK2 share overlapping functions in controlling both the S and G2/M checkpoints. CHK1 is activated by ATR through the phosphorylation of serine residues 317 and 345. On the other hand, CHK2 is activated by the related kinase ATM. ATM is activated primarily after double-strand breaks in DNA, whereas ATR seems to be critical for cellular responses to the arrest of replication forks and single-strand breaks in DNA (24). Signals from broken DNA use the ATM/ATR–CHK1/2–cdc25 pathway to stabilize and phosphorylate p53. In the current study, by using a specific inhibitor of CHK1 and ATR KD cells, we found that the signal from the DNA damage induced in response to PALA uses the ATR/CHK1 pathway to modulate p53 activity. There is no obvious difference in the level of expression of p53 protein in ATM-null cells, ruling out a substantial role of ATM in the activation of p53 under the conditions studied. Consistent with the absence of double-strand breaks is the observation that starvation for pyrimidine nucleotides leads to limited DNA damage, which then serves as a signal for p53 induction and activation. It is significant that cells that express p53 survive this limited damage, again arguing for a lack of double-strand breaks. Cells lacking p53, when starved for pyrimidine nucleotides, attempt to synthesize DNA, as revealed by strong incorporation of BrdU, under highly unfavorable conditions when pyrimidine deoxynucleoside triphosphate pools are very low and thus incur irreversible, extensive DNA damage, triggering apoptosis (13).
A careful examination of the progression of BrdU-incorporated cells through the cell cycle (Fig. 5), where cells accumulate in the second S phase after PALA treatment, is consistent with a model in which the skewing of deoxynucleoside triphosphate pools in response to PALA creates reversible DNA damage, sufficient to activate p53. This activated p53 then transactivates secondarily the expression of proteins that provide protective arrest at multiple points in the cell cycle. These events occur before the deoxynucleoside triphosphate pools become depleted enough to cause a substantial decrease in the rate of DNA synthesis. As shown by earlier studies (8, 13), in cells in which p53 is absent or in which the p53 pathway is compromised, failure to arrest DNA synthesis when pyrimidine nucleoside triphosphate pools are depleted leads to irreversible DNA damage that eventually causes apoptosis.
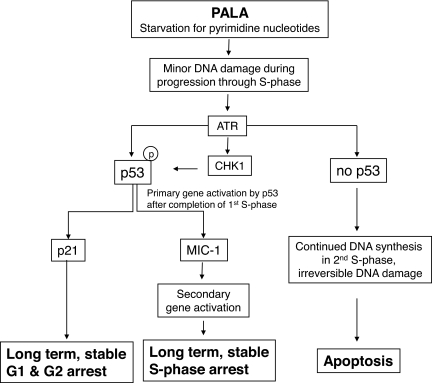
We propose a comprehensive model to explain the role p53 plays when pyrimidine nucleotide pools are depleted in response to PALA (Fig. 6). It is very likely that an increased frequency of misincorporation caused by unbalanced pools leads to an increase in mismatch repair, and that this reversible form of DNA damage occurs while cells progress through the first round of DNA synthesis. Our current observations indicate that the initial DNA damage occurs slowly and only in actively dividing cells. We propose that this form of DNA damage is sufficient to generate a signal for the phosphorylation and subsequent stabilization of p53 through the activities of ATR and CHK1, not ATM, which responds to more substantial DNA damage, such as double-strand breaks. As reported earlier (19), p53 remains transcriptionally inactive during S phase. From our analysis of the cell cycle and the kinetics of the activation of p53 and its two target proteins p21 and MIC-1, we conclude that p53 is activated well before DNA synthesis slows down because of depletion of pyrimidine deoxynucleoside triphosphates, and that only when the cells exit from the first S phase and enter the next cell cycle does the activated p53 becomes transcriptionally competent, eventually activating G2, G1, and S phase checkpoints by activating the expression of key regulatory proteins such as p21 and MIC-1.
Fig. 6.
Activation of p53 and its effects on cell cycle checkpoints in response to PALA.
PALA kills fibroblasts lacking p53 but not fibroblasts that express wild-type p53. Because virtually all cancer cells have some disruption of p53 function, the properties of PALA seem ideal for an antitumor drug, and early studies in cell culture and in animal models showed PALA to be an effective antitumor agent (25). However, because of dose-limiting gut toxicity, PALA was not useful in patients. Unfortunately normal human gut epithelial cells are much more sensitive to PALA than are normal human fibroblasts. The basis of this difference is not well understood. Our recent work and ongoing studies have clearly identified the secretory cytokine, MIC-1, as an important factor in effecting a p53-dependent protective mechanism and thus providing a new direction in re-evaluating the use of PALA in treating cancer (13).
Materials and Methods
Cell Lines and Treatments.
Normal human fibroblast BJ cells were obtained from the American Type Culture Collection. BJ-Tert cells were derived by retrovirus-mediated introduction of the Tert subunit of human telomerase. ATR WT and ATR KD cells and AT-null primary human fibroblasts were kind gifts of David Boothman (University of Texas Southwestern, Dallas) (26). Cell lines containing a neo marker were grown in the presence of 600 μg/ml G418 (Sigma). All cells were grown in 10% CO2 in DMEM supplemented with 10% FBS. The cells were treated with 250 μM PALA.
PALA and CHK1 Inhibitor.
PALA (NSC224131), an inhibitor of the aspartate transcarbamylase activity of the multifunctional enzyme CAD, was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, National Cancer Institute (Bethesda, MD). Cells were treated with PALA as described by Agarwal et al. (8). For each cell type, the ID50 was determined, and selection of 2 × 105 cells per 10-cm plate was done in the presence of 10% (vol/vol) dialyzed FCS, using PALA at 3 × ID50. The selective medium was changed every 4–5 days. The CHK1 inhibitor UCN-01 was a kind gift of Robert Shultz (Department of Developmental Therapeutics, National Cancer Institute). BJ cells were pretreated with 300 nM UCN-01 for 45 min.
Comet Assay.
BJ cells were treated with 250 μM of PALA in dialyzed serum for 24 or 48 h, and the assay was performed according to the manufacturer's protocol (Trevigen). Briefly, cells were gently scraped into the medium, counted, and washed in ice-cold 1× PBS (free of Ca2+ and Mg2+). After washing, the cells (1 × 105/ml) were suspended in cold PBS and combined with molten LMAgarose (Trevigen) at 37°C at a ratio of 1:10 (vol/vol). Seventy-five microliters of the mixture was spread onto a slide and allowed to solidify at 4°C in the dark for a minimum of 10 min. Cells were lysed by immersing the slides in prechilled lysis solution for 30–60 min, after which the cells were immersed in a freshly prepared alkaline solution (pH >13) for 20–60 min. The slides were then washed with 1× Tris-borate-EDTA buffer and run on a horizontal electrophoresis apparatus (1 V/cm) for 10 min. The slides were washed in 70% ethanol, air-dried, and kept in the dark until analysis. Cells were visualized by adding 50 μl of diluted SYBR green 1 (excitation and emission, 494 nm and 521 nm, respectively) and observed by epifluorescence microscopy.
γH2AX Staining.
BJ cells (1,500 cells per chamber) were plated overnight in DMEM with dialyzed serum and then treated with 250 μM of PALA for 0, 1, 2, 3, 6, 12, 24 or 48 h. The following controls were used: primary antibody alone, isotype control, secondary antibody alone, and 1 μM etoposide for 3 h as a positive control. After treatment the cells were fixed in 4% paraformaldehyde in TBS buffer and stained overnight in primary anti-γH2AX (1:500 dilution; Cell Signaling) After overnight incubation, the cells were washed a few times with TBS/BSA and incubated for 1 h with anti-rabbit secondary antibody conjugated with Alexa 488 (Invitrogen). The cells were washed and fixed in Prolong Gold Antifade with DAPI (Invitrogen) and stored at 4°C for 24 h (as recommended) before examining the slides. Pictures were taken by using a Nikon Eclipse E 800 microscope with attached camera and using Spot Software (Diagnostic Instruments). Images were taken at ×20 (×20/0.75 DIC M Plan APO lens), except that the images in the Fig. 1 Insets were taken with a ×40 lens (×40/1.0 DIC H Plan APO oil immersion lens). Objectives were set at wavelengths of 488 nm for Alexa 488 and 350 nm for DAPI.
Western Analyses.
Total cellular protein was isolated by lysing cells in 20 mM Tris·HCl (pH 7.5), 2% (wt/vol) SDS, 2 mM benzamidine, and 0.2 mM phenylmethanesulfonyl fluoride. Protein concentrations were determined by the Bradford method (Bio-Rad). Proteins (25 μg per lane) were separated by SDS/10% PAGE) and electro-blotted onto polyvinylidene difluoride membranes (Stratagene). The membranes were probed with antibodies against p53, p21 (Santa Cruz Biotechnology), phosphoserine p53, phosphoserine CHK1 (Cell Signaling Technology), or actin (Labvision), followed by incubation with anti-mouse or anti-rabbit secondary antibody-horseradish peroxidase conjugate (Amersham Life Sciences). Chemiluminescence was developed by using an ECL kit (Amersham Life Sciences).
Cell Cycle Analyses.
Growing BJ cells were plated (2 × 106 cells per plate) and grown overnight (60–70% confluent). Cells were then incubated with 20 μM BrdU for 30 min followed by two washes with PBS. Ten milliliters of media with 100 μM thymidine was added, and cells were incubated for 30 min. At the end, the plates were again washed twice with 1× PBS. Then DMEM with 10% dialyzed serum was added for PALA-treated cells and normal DMEM with 10% FBS for untreated cells. Cells were then treated with 250 μM PALA and harvested for cell cycle analysis at different time points.
Acknowledgments.
We thank Dr. William Burhans for his advice on several experiments, Dr. Robert Shultz for UCN-01, and Dr. David Boothman for the ATR wild-type and kinase dead cell lines and ATM null primary fibroblasts. This work was supported by National Institutes of Health Grants CA973413 and CA043703 (to J.W.J.), GM 49345 (to G.R.S.), and R01 CA98916-02 (to M.L.A.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Linke SP, Clarkin KC, Di LA, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 3.Taylor WR, Agarwal ML, Agarwal A, Stacey DW, Stark GR. p53 inhibits entry into mitosis when DNA synthesis is blocked. Oncogene. 1999;18:283–295. doi: 10.1038/sj.onc.1202516. [DOI] [PubMed] [Google Scholar]
- 4.Cross SM, et al. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 5.Di LA, et al. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 6.Swyryd EA, Seaver SS, Stark GR. N-(phosphonacetyl)-l-aspartate, a potent transition state analog inhibitor of aspartate transcarbamylase, blocks proliferation of mammalian cells in culture. J Biol Chem. 1974;249:6945–6950. [PubMed] [Google Scholar]
- 7.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal ML, et al. A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides. Proc Natl Acad Sci USA. 1998;95:14775–14780. doi: 10.1073/pnas.95.25.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livingstone LR, et al. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 10.Chernova OB, Chernov MV, Agarwal ML, Taylor WR, Stark GR. The role of p53 in regulating genomic stability when DNA and RNA synthesis are inhibited. Trends Biochem Sci. 1995;20:431–434. doi: 10.1016/s0968-0004(00)89094-5. [DOI] [PubMed] [Google Scholar]
- 11.Khan SH, Wahl GM. p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res. 1998;58:396–401. [PubMed] [Google Scholar]
- 12.Stark GR, Taylor WR. Analyzing the G2/M checkpoint. Methods Mol Biol. 2004;280:51–82. doi: 10.1385/1-59259-788-2:051. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal MK, et al. Macrophage inhibitory cytokine 1 mediates a p53-dependent protective arrest in S phase in response to starvation for DNA precursors. Proc Natl Acad Sci USA. 2006;103:16278–16283. doi: 10.1073/pnas.0607210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc Natl Acad Sci USA. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemay M, Wood KA. Detection of DNA damage and identification of UV-induced photoproducts using the CometAssay kit. BioTechniques. 1999;27:846–851. doi: 10.2144/99274pf01. [DOI] [PubMed] [Google Scholar]
- 16.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 17.Thompson T, et al. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 18.Syljuåsen RG, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc Natl Acad Sci USA. 2001;98:1036–1041. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobberger JW, et al. A new biomarker for mitotic cells. Cytometry A. 2008;73:5–15. doi: 10.1002/cyto.a.20501. [DOI] [PubMed] [Google Scholar]
- 21.Begg AC, McNally NJ, Shrieve DC, Karcher H. A method to measure the duration of DNA synthesis and the potential doubling time from a single sample. Cytometry. 1985;6:620–626. doi: 10.1002/cyto.990060618. [DOI] [PubMed] [Google Scholar]
- 22.Tan M, Wang Y, Guan K, Sun Y. PTGF-β, a type transforming growth factor (TGF-β) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-β signaling pathway. Proc Natl Acad Sci USA. 2000;97:109–114. doi: 10.1073/pnas.97.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albertoni M, et al. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21:4212–4219. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- 24.Stiff T, et al. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin DS, Spriggs D, Koutcher JA. A concomitant ATP-depleting strategy markedly enhances anticancer agent activity. Apoptosis. 2001;6:125–131. doi: 10.1023/a:1009692631748. [DOI] [PubMed] [Google Scholar]
- 26.Nghiem P, Park PK, Kim YS, Desai BN, Schreiber SL. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J Biol Chem. 2002;277:4428–4434. doi: 10.1074/jbc.M106113200. [DOI] [PubMed] [Google Scholar]