Abstract
Aerobic life is based on a molecular machinery that utilizes oxygen as a terminal electron sink. The membrane-bound cytochrome c oxidase (CcO) catalyzes the reduction of oxygen to water in mitochondria and many bacteria. The energy released in this reaction is conserved by pumping protons across the mitochondrial or bacterial membrane, creating an electrochemical proton gradient that drives production of ATP. A crucial question is how the protons pumped by CcO are prevented from flowing backwards during the process. Here, we show by molecular dynamics simulations that the conserved glutamic acid 242 near the active site of CcO undergoes a protonation state-dependent conformational change, which provides a valve in the pumping mechanism. The valve ensures that at any point in time, the proton pathway across the membrane is effectively discontinuous, thereby preventing thermodynamically favorable proton back-leakage while maintaining an overall high efficiency of proton translocation. Suppression of proton leakage is particularly important in mitochondria under physiological conditions, where production of ATP takes place in the presence of a high electrochemical proton gradient.
Keywords: cell respiration, gating mechanism, proton leak, proton translocation
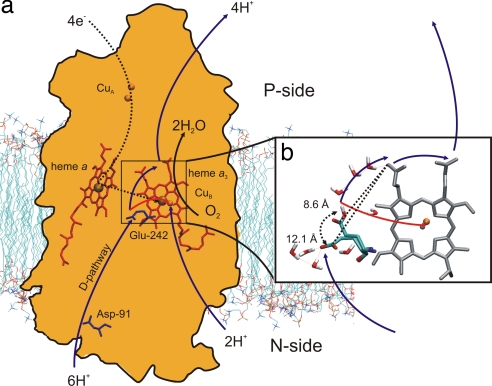
Cytochrome c oxidase drives the aerobic electron transport chain. It receives electrons from cytochrome c and transfers them through a sequence of metal centers to molecular oxygen. For each electron transferred, two protons are taken up from the negatively charged N-side of the membrane. One proton is pumped across the membrane to the positively charged P-side, and the other “chemical” proton is transferred to the oxygen intermediate to form the equivalent of water (Fig. 1a). Proton (and electron) translocation generates an electrochemical gradient that is used to drive the synthesis of ATP (1–3).
Fig. 1.
Structure and function of CcO. (a) Contour of subunits I and II together with the four redox-active centers CuA, heme a, and heme a3/CuB. The dashed black line shows the path of the electrons. Protons taken up by the D-pathway are transferred to Glu-242, from which they are either pumped across the membrane (blue arrows) or transferred to the binuclear site (red arrow). (b) Active site of CcO. Glu-242 is drawn in down and up configurations, with water molecules both below and above. The corresponding mean distances to the D-propionate of heme a3 are shown.
The bimetallic CuA center is the first electron acceptor and passes electrons further to the low-spin heme a, from where they are transferred, one at a time, to the binuclear heme a3/CuB site of O2 binding (Fig. 1a). Protons are taken up from the N-side by two proton-conducting routes, the D- and K-pathways (named after Asp-91 and Lys-319,§ respectively). The former is used to conduct all of the pumped protons and two of the chemical protons; the latter is active only in the last steps of the catalytic cycle (4–6). The D-pathway leads to a conserved glutamic acid, Glu-242, located at approximately equal distances from the two heme groups (Fig. 1a). Recent research has suggested that reduction of heme a is followed by proton donation from Glu-242 to an unknown proton-loading site (PLS), in conjunction with transfer of the electron to the binuclear site. Subsequently, this proton is ejected to the P-side of the membrane driven by uptake of a second proton that forms water at the binuclear site (7, 8).
For proton translocation to occur, Glu-242 has been proposed to undergo a cyclic conformational isomerization (Fig. 1b) (9–12). In the available x-ray structures, this residue points “down” toward the D-pathway (Fig. 1) (13–16), from which it can accept protons, but there is no obvious conduit for the proton to move further toward the P-side. The nonpolar cavity “above” Glu-242 appears empty in most x-ray structures but has been suggested to contain water molecules on the basis of computational studies (10, 11, 17). The most recent structure of CcO from Rhodobacter sphaeroides (18) indeed shows one water molecule in this cavity. The proposed conformational switch to an up position (Fig. 1b) would allow Glu-242, via water molecules in the cavity, to donate protons both to the D-propionate group of heme a3, which is a likely transient acceptor of the proton to be pumped (13, 19–21), and to the binuclear center for consumption.
Proton transfer from Glu-242, either to the PLS or to the binuclear site, yields the anionic glutamate, which has been subject of some previous computational studies (9, 22, 23). Olsson et al. (24) suggested that the Glu-242 anion cannot form for energetic reasons, but recent time-resolved FTIR data in a mutant enzyme demonstrated deprotonation and later reprotonation in the reaction sequence (25). We therefore decided to analyze the dynamics of the anionic Glu-242, and to extend our initial analysis of the protonated form (12).
Because the protons are actively pumped against the electrochemical gradient, it is not trivial mechanistically how they are prevented from leaking back. Control of such back-leaks have been discussed (26–32), but definite answers are lacking. Here, we present molecular dynamics simulations, which suggest that the key role of Glu-242 in proton transfer includes a unique function as a “valve” that minimizes such back-leakage of protons.
Results
The structure of subunits I and II of the bovine heart enzyme (Protein Data Bank entry 1V54) (14) was used with four water molecules added into the nonpolar cavity above Glu-242 (see Models and Methods) and initially employing the Glu-242 anion. After minimization and equilibration, an unexpected behavior was encountered. After ≈2.3 ns the glutamate, originally positioned in an up position toward the nonpolar cavity, flipped down toward the D-pathway (the x-ray position) but brought with it all four water molecules from the cavity (data not shown). Inspection of the 1V54 structure shows that there is, in fact, no protonic connection between the end of the D-pathway and Gludown. In contrast, the structure of the Rh. sphaeroides enzyme (18) has four water molecules in this domain, which provide a potential proton transfer path from the D-pathway to Glu-242. Thus, our initial approach remarkably “corrected” the deficiency of water molecules below Glu-242 in the 1V54 structure, bringing it at least transiently to the state observed for the Rh. sphaeroides enzyme. It is indeed known that molecular dynamics can accurately predict the position of water molecules inside of protein cavities that appear empty in x-ray structures (33).
This behavior of the Glu-242 anion appears to be significant for function. When the nonpolar cavity was replenished with four water molecules (Fig. 1b) and the water molecules that had moved below Glu-242 were retained, such water transfer was no longer observed, and the dynamics of the glutamate anion were changed dramatically (see below). We conclude that simulations of the Glu-242 dynamics should be performed with water molecules in the cavities both above and below this residue. A hydrated state is particularly relevant in studies of the active enzyme, in which water is continuously produced. All further simulations were performed under such conditions. To assess possible effects of the redox states of heme a and the binuclear site, we performed the simulations in two conditions: either with heme a reduced and the binuclear site in the so-called PM state (ferryl heme a3, cupric CuB, and Tyr-244 as the neutral radical) (34, 35) or with heme a oxidized with the binuclear site in the PR state (Tyr-244 as the tyrosinate; heme a3 and CuB being the same as in PM) (36).
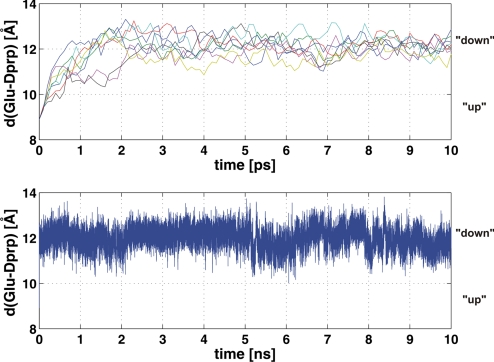
With water both in the active-site cavity and in the narrow space above the D channel, the anionic Glu-242, initially positioned up toward the nonpolar cavity, now flipped to the down conformation remarkably fast, with a t1/2 of ≈1 ps [0.54 ± 0.18 (SD) ps; eight independent simulations], breaking the hydrogen-bonded contact to the water molecules above it (Fig. 2a). These simulations were continued up to 10 ns, but no return to the up conformation was observed during this time (Fig. 2b). To test the hypothesis that the rapid down-flip is at least in part induced by electrostatics, we artificially switched off the charges of all heme propionate groups and found that the deprotonated Glu-242 remained in the up conformation for at least 500 ps. To explore whether protonation of the PLS above the hemes has an effect on the Glu-242 conformational equilibrium, we switched off the charges of only the D- or the A-propionate of heme a3. In both cases the down-flip was fast, suggesting that it is not prevented by a proton in the PLS. This result is important for pumping: If the Glu-242 anion remained in the up position for a prolonged time, back-transfer of the proton from the PLS would be possible, compromising the pump efficiency (see Discussion).
Fig. 2.
Molecular dynamics of the anionic Glu-242. (Upper) Distance between deprotonated Glu-242 (δ-carbon) and the Δ-propionate of heme a3 (O2D; see Fig. 1b) as a function of time for eight independent simulations. The side chain of Glu-242 flips from an up to a down configuration with a half-time of ≈1 ps (see text). (Lower) Same distance as a function of time up to 10 ns. Glu-242 remains in the down configuration during the whole simulation. These simulations were done for the PM state with heme a reduced (see text).
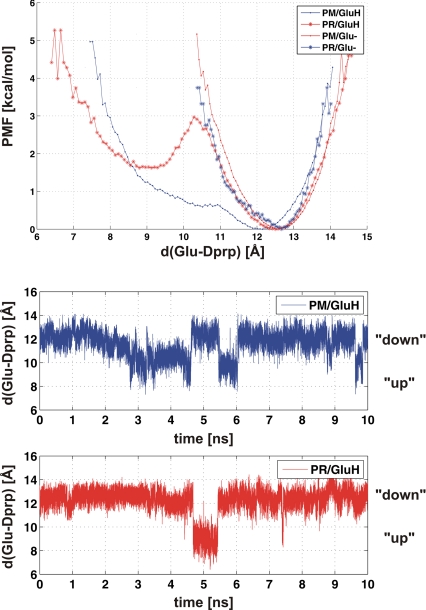
After reprotonation of Gludown−, an up-flip readily occurred, as described in ref. 12. Fig. 3 shows the free energy profiles for Glu-242 isomerization for both the protonated and the deprotonated case (Upper), as well as 10-ns trajectories of the protonated form in both redox states, both of which demonstrate transitions between the up and down conformations (Lower). In the current simulations there is approximately an equal preference for the up and down conformations in the PM state, but in PR the down state is preferred by almost 2 kcal/mol (Fig. 3). The standard errors were estimated as 0.5 and 0.85 kcal/mol, respectively. In contrast, the deprotonated Glu-242 strongly prefers the down conformation in both oxidation states; no case of switching back to the up configuration was sampled in simulations up to 10 ns (Fig. 2). Based on this, and on the time of down-flip (≈1 ps), we can give a lower boundary value of 104 (≈5.5 kcal/mol) for the down/up equilibrium, which will be used in the analysis below.
Fig. 3.
Thermodynamics of the Glu-242 isomers. (Top) Free energy profile, or potential of mean force, as a function of the distance between Glu-242 (OE2) and the Δ-propionate of heme a3 (O2D) for the protonated (GluH) and deprotonated (Glu−) forms of Glu-242. The results are shown for both the PM and PR redox states (see text). All profiles were calculated from 10-ns simulations, except for Glu− in the PR state (1 ns). (Middle and Bottom) Ten-nanosecond simulations of the protonated Glu-242 in the PM and PR states, respectively, depicting the Glu-242–Δ-propionate (heme a3) distance as a function of time.
Discussion
The conformational preference of the Glu-242 side chain depends on its charge state, the oxidation state of the enzyme, and the hydration states of the cavities above and below. While the up and down configurations of the protonated Glu-242 have approximately equal probabilities when heme a is reduced and the binuclear site is oxidized (PM state), moving the electron from heme a to the binuclear site to yield the PR state changed the distribution to an ≈20-fold preference of the down state. This finding may explain observations by infrared spectroscopy, where the characteristic absorption of the protonated Glu-242 near 1,740 cm−1 is shifted in a manner that depends on the redox state of heme a (37–41).
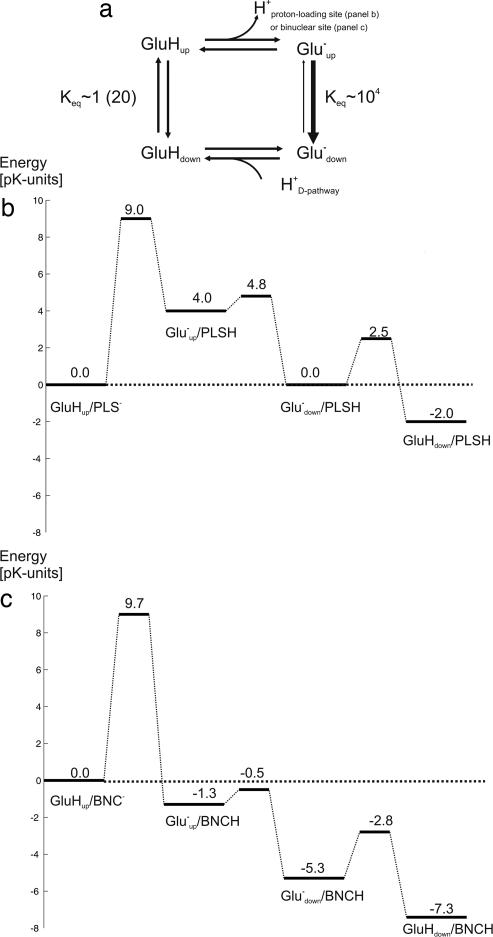
Fig. 4a shows a thermodynamic cycle of the conformational changes of Glu-242 in its protonated and deprotonated forms. By inserting boundary values of the equilibrium constants for rotational isomerization one obtains the ΔpKa between the up and down conformations, suggesting a pKa of Gluup of ≈4 and ≈2.7 pKa units higher than that of Gludown in the PM and PR states, respectively. The pKa of Glu-242 has been estimated from kinetic experiments to be ≈9.5 (42), but FTIR data shows no deprotonation at pH 10 (37, 43), and electrostatics calculations suggest a much higher pKa, near 14 (29, 44–46). We suggest that this difference is the result of an electrostatic interaction between Glu-242 and the PLS, which may be absent in the kinetic experiments because of a protonated (neutral) state of the PLS. For the purpose of discussion, we assume that the pKa is near 9 for Gludown because assessment of the energetics of internal proton transfer between Glu-242 and the PLS requires the pKa values of each in the absence of mutual interaction. It follows that pKa for Gluup would be ≈13 (PM) or ≈11.7 (PR). If we instead had assumed pK = 9 for Gluup, the pKa would be ≈5 in the down position prevalent in all crystal structures, which would be inconsistent with the observed hydrogen bonding (47). The pKa of the PLS has been estimated to be ≈9 when the electron is at heme a and ≈11.2 after transfer of the electron to the binuclear site (ref. 31; see also ref. 30). Ignoring possible differences between kinetic and thermodynamic pKa values and local electrostatic interactions, we combine this data to approximately illustrate the role of Glu-242 in preventing proton leakage (Fig. 4 b and c).
Fig. 4.
Proton transfers involving Glu-242. (a) Thermodynamic cycle of Glu-242 shuttling between the up and down configurations. The down/up equilibrium constants given for GluH are ≈1 for the PM state and ≈20 for the PR state (see text). (b) Energy profile of the first proton transfer event from Gluup (pKa ∼ 13) to the proton-loading site (PLS) (pKa ∼ 9), followed by reprotonation from the N-side of the membrane (pH 7) via the D-pathway (pKa of Gludown ∼ 9); electron transfer from heme a to the binuclear site accompanies this sequence (data not shown). (c) Energy profile for the second proton transfer from Glu-242 (pKa ∼ 11.7) to the binuclear site (BNC) (pKa ∼ 13), and reprotonation via the D-pathway. The first proton is bound to the PLS (not indicated). Barrier heights are approximated from rates observed experimentally (8) or in the simulations, using transition-state theory. The activation barrier for protonation of Gludown− via the D-pathway was chosen arbitrarily for a rate of 50 ps.
Fig. 4b shows the energy profile of the primary proton transfer to the PLS, assuming the above pKa values and pH 7 in the aqueous N-phase. Proton transfer to the PLS is followed by flip-down of the glutamate side chain and protonation via the D-pathway. The sequence is also associated with isoenergetic electron transfer from heme a to the binuclear site (8, 31), which is omitted here for simplicity. These reactions will approach equilibrium in the steady state because the subsequent protonation of the binuclear site (Fig. 4c) is much slower (0.8 ms) (8, 31) than any reaction step in Fig. 4b. Under normal conditions, such reversibility is not detrimental for pump efficiency. However, protonation of the PLS by Glu-242 must be kinetically more facile than from the P-side of the membrane (see refs. 30 and 31).
After protonation of PLS, the chemical proton may also be delivered via Glu-242 (Fig. 1a) (4–6). Here, the observed dynamic behavior of Glu-242 becomes critical for preventing backflux of the proton in the PLS. The pKa of the binuclear site has been estimated to be ≈13 after receiving the electron from heme a but is much lower before this event (31). As shown in Fig. 4c, the GluHup → Gluup− transition, where the proton is transferred to the binuclear site (BNC), is followed by fast and exergonic down-flip of Glu−. This transition ensures that the proton in the PLS, which now has a pKa of ≈5 (31), will not be transferred back to Gluup−, which would compromise the pump. Both the high rate and the exergonicity of the down-flip are important. After reprotonation of Gludown−, the final step of the mechanism releases the proton in the PLS to the P-side of the membrane driven by uptake of the chemical proton (Fig. 1), usually on the milliseconds time scale (8, 48). This slow reaction creates a situation where prior steps (Fig. 4c) approach thermodynamic equilibrium in the steady state. As shown in Fig. 4c, the occupancy of the vulnerable Gluup− state is ≈10−6 relative to the lowest-energy state before release of the PLS proton. To effectively prevent leakage, the rate of proton transfer from PLS to Gluup− must now be at least two orders of magnitude slower than release of the PLS proton to the P-side. This requirement is fulfilled because of the energetics of the side chain isomerization, because even if the time constant for the leak were in microseconds, the leak rate would be 106 times slower (seconds), mainly because of the exergonicity of the down-flip. In addition, the picosecond down-flip of the Glu anion ensures that the leak will also not occur before the system reaches the steady state.
Certain D-pathway mutants are unable to pump protons but have normal or enhanced turnover (49, 50). This phenotype has been associated with an increased pKa of Glu-242 (42). An alternative explanation would be lowering of the rate of proton conductance in the D-pathway (31, 32). A raised kinetic barrier for the Gludown− to GluHdown transition would increase the occupancy of the Gluup− state and may therefore enhance proton backflux. It is also worth noting that in the presence of an electrochemical proton gradient, the electrical membrane potential component will tend to oppose the down-flip of Gluup−. However, the down-flip occurs across a distance of ≈3 Å perpendicular to the membrane (Fig. 1b). Assuming a membrane thickness of ≈30 Å, and a smooth electric potential profile across it, the gradient only translates to a 2-fold preference of Gluup− relative to Gludown−, which is by far outweighed by the equilibrium constant of ≥104 in the opposite direction (Fig. 4a).
Our results suggest that Glu-242 shuttles between up and down conformations with rates compatible with CcO function (cf. ref. 9). Proton transfer to the proton-loading or binuclear sites occurs under conditions where proton accessibility toward the N-side of the membrane is absent. Conversely, during proton transfer to Glu-242 via the D-pathway, the connection toward the P-side is broken. There is at no point in time continuous proton conductivity across the entire membrane. Such a property is of fundamental mechanistic importance for any pump. We find that the molecular basis for this function is the disruption of the connection toward the P-side by flipping the side chain of the anionic Glu-242 to the down position in a fast and thermodynamically favorable reaction. This property is at least partially governed by electrostatic repulsion between Gluup− and the heme propionates. Such a valve is of particular importance at the point in the reaction sequence when proton transfer to the binuclear site (Fig. 4c) lowers the pKa of the PLS (30, 31), and assures that the proton is ejected to the P-side.
Interestingly, there are members of the heme-copper oxidase superfamily, and mutant enzymes, where a tyrosine occupies the position of Glu-242 in the structure, apparently with retained efficiency of proton translocation (51–54). The phenol ring of tyrosine is bulkier than the glutamic acid side chain, and these heme-copper oxidases might therefore lack the control valve suggested here. These enzymes might not sustain high-efficiency proton translocation in the presence of a sizable proton electrochemical gradient. However, this possibility is so far unexplored because proton-pumping assays are typically performed in the absence of a counterforce.
Conclusion
Our simulations have shown that water molecules in the active-site cavity (12), and in the narrow space connecting Gludown to the D-pathway, profoundly affect both the equilibrium and dynamics of the Glu-242 valve. As CcO turns over, water is continuously produced at the active site so that the hydration state of the active enzyme is likely to be out of equilibrium. We find that the shuttling of Glu-242 between the two conformers is particularly facile in the fully hydrated state, allowing maintenance of fast turnover. In contrast, the relatively dry state inferred from the lack of water in the crystal structures would be more consistent with the “slow” enzyme, as isolated (55). Water thus plays a central role in CcO function, not only as a product of the reaction and a mediator in proton transfer (56), but also by affecting the conformational preferences of ionizable groups, in particular Glu-242.
By definition, active transport takes place against a thermodynamic gradient. Mitchell's original concept of CcO functioning as a proton-motive half-loop (57) meant that the charges of electrons derived from the P-side and protons from the N-side of the membrane are annihilated at the active site in the reduction of O2 to water, thus creating a proton-motive force across the membrane. In such a case there is no immediate risk of leakage. But the function as a true proton pump (1) where protons are translocated across the entire membrane dielectric creates the problem of how leakage is avoided because there has to be proton access all through the membrane (32). Moreover, the proton pump of CcO is known to be reversible at high proton-motive force (58), which adds further constraints. Mitochondria produce ATP at a 170-mV proton-motive force (59). Measurements of the ratio of ATP produced per O2 consumed, together with the H+/ATP stoichiometry of the ATP synthase, show that CcO functions at maximum efficiency in such conditions (60), which puts very high demands on the proton pump mechanism. We suggest that the valve function of Glu-242 described here is essential to the fulfillment of these demands.
Models and Methods
The coordinates of CcO subunit I and II from Bos taurus were taken from Protein Data Bank entry 1V54 (14). Structures for MD simulations with both protonated and deprotonated Glu-242 were built in the MD package CHARMM (61), using the CHARMM27 force field (62) and our own density functional theory (DFT)-based parametrization set for the redox active metal centers (63). The simulation setup was similar to that of ref. 12. Initially, four modeled water molecules were added above Glu-242 (11). As described, in most simulations additional water molecules were added between Glu-242 and the D channel, consistent with the Rh. sphaeroides structure (18). In all reported simulations, the system was in the PM state with CuA oxidized, with heme a reduced, with ferryl heme a3, with CuB in its cupric state, and with Tyr-244 as a neutral tyrosyl radical. Alternatively, heme a was oxidized and the binuclear site in the PR state (see above). The energy difference between the two conformations of protonated Glu-242 was evaluated by counting the number of “up” (defined as d < 10 Å between Glu-242 and D-propionate of heme a3) and “down” (defined as d > 10 Å) conformations sampled during 10-ns simulations of the PR and PM states. The simulations were performed by using the program NAMD (64) with a 1-fs integration step, the CHARMM 27 force field, and Langevin dynamics to control the temperature at 310 K. Typical simulation times were 1–10 ns after an initial minimization procedure. The trajectories were monitored by using the program VMD (65).
Acknowledgments.
This work was supported by grants from the Sigrid Jusélius Foundation, Biocentrum Helsinki, and the Academy of Finland. CSC—Scientific Computing Ltd., is acknowledged for computational resources. V.R.I.K. is supported by the Graduate School of Biotechnology and Molecular Biology and the Finnish Cultural Foundation. G.H. is supported by the Intramural Research program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
The numbering of amino acids is based on subunit I of CcO from bovine heart mitochondria.
References
- 1.Wikström MKF. Proton pump coupled to cytochrome c oxidase in mitochondria. Nature. 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- 2.Babcock GT, Wikström M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 3.Wikström M. Cytochrome c oxidase: 25 years of the elusive proton pump. Biochim Biophys Acta. 2004;1655:241–247. doi: 10.1016/j.bbabio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Gennis RB. Coupled proton and electron transfer reactions in cytochrome oxidase. Front Biosci. 2004;9:581–591. doi: 10.2741/1237. [DOI] [PubMed] [Google Scholar]
- 5.Brzezinski P. Redox-driven membrane-bound proton pumps. Trends Biochem Sci. 2004;29:380–387. doi: 10.1016/j.tibs.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Konstantinov AA, Siletsky S, Mitchell D, Kaulen A, Gennis RB. The roles of the two proton input channels in cytochrome c oxidase from Rhodobacter sphaeroides probed by the effects of site-directed mutations on time-resolved electrogenic intraprotein proton transfer. Proc Natl Acad Sci USA. 1997;94:9085–9090. doi: 10.1073/pnas.94.17.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belevich I, Verkhovsky MI, Wikström M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature. 2006;440:829–832. doi: 10.1038/nature04619. [DOI] [PubMed] [Google Scholar]
- 8.Belevich I, Bloch D, Belevich N, Wikström M, Verkhovsky MI. Exploring the proton pump mechanism of cytochrome c oxidase in real time. Proc Natl Acad Sci USA. 2007;104:2685–2690. doi: 10.1073/pnas.0608794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pomès R, Hummer G, Wikström M. Structure and dynamics of a proton shuttle in cytochrome c oxidase. Biochim Biophys Acta. 1998;1365:255–260. [Google Scholar]
- 10.Hofacker I, Schulten K. Oxygen and proton pathways in cytochrome c oxidase. Proteins. 1998;86:100–107. [PubMed] [Google Scholar]
- 11.Riistama S, et al. Bound water in the proton translocation mechanism of the haem-copper oxidases. FEBS Lett. 1997;414:275–280. doi: 10.1016/s0014-5793(97)01003-x. [DOI] [PubMed] [Google Scholar]
- 12.Tuukkanen A, Kaila VRI, Laakkonen L, Hummer G, Wikström M. Dynamics of the glutamic acid 242 side chain in cytochrome c oxidase. Biochim Biophys Acta. 2007;1767:1102–1106. doi: 10.1016/j.bbabio.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 14.Tsukihara T, et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci USA. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svensson-Ek M, et al. The x-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 16.Abramson J, et al. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat Struct Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 17.Zheng X, Medvedev DM, Swanson J, Stuchebrukhov AA. Computer simulation of water in cytochrome c oxidase. Biochim Biophys Acta. 2003;1557:99–107. doi: 10.1016/s0005-2728(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 18.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puustinen A, Wikström M. Proton exit from the heme-copper oxidase of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:35–37. doi: 10.1073/pnas.96.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brändén G, et al. The protonation state of a heme propionate controls electron transfer in cytochrome c oxidase. Biochemistry. 2005;44:10466–10474. doi: 10.1021/bi0502745. [DOI] [PubMed] [Google Scholar]
- 21.Wikström M, et al. Gating of proton and water transfer in the respiratory enzyme cytochrome c oxidase. Proc Natl Acad Sci USA. 2005;102:10478–10481. doi: 10.1073/pnas.0502873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cukier RI. Quantum molecular dynamics simulation of proton transfer in cytochrome c oxidase. Biochim Biophys Acta. 2004;1656:189–202. doi: 10.1016/j.bbabio.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Olkhova E, Hutter MC, Lil MA, Helms V, Michel H. Dynamic water networks in cytochrome c oxidase from Paracoccus denitrificans investigated by molecular dynamics simulations. Biophys J. 2004;86:1873–1889. doi: 10.1016/S0006-3495(04)74254-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsson MHM, Warshel A. Monte Carlo simulations of proton pumps: On the working principles of the biological valve that controls proton pumping in cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:6500–6505. doi: 10.1073/pnas.0510860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorbikova EA, Belevich NP, Wikström M, Verkhovsky MI. Time-resolved ATR-FTIR spectroscopy of the oxygen reaction in the D124N mutant of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2007;46:13141–13148. doi: 10.1021/bi701614w. [DOI] [PubMed] [Google Scholar]
- 26.Brzezinski P, Larsson G. Redox-driven proton pumping by heme-copper oxidases. Biochim Biophys Acta. 2003;1605:1–13. doi: 10.1016/s0005-2728(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 27.Brändén G, Pawate AS, Gennis RB, Brzezinski P. Controlled uncoupling and recoupling of proton pumping in cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:317–322. doi: 10.1073/pnas.0507734103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popovic DM, Stuchebrukhov AA. Proton pumping mechanism and catalytic cycle of cytochrome c oxidase: Coulomb pump model with kinetic gating. FEBS Lett. 2004;566:126–130. doi: 10.1016/j.febslet.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Popovic DM, Quenneville J, Stuchebrukhov AA. DFT/electrostatic calculations of pK(a) values in cytochrome c oxidase. J Phys Chem B. 2005;109:3616–3626. doi: 10.1021/jp046535m. [DOI] [PubMed] [Google Scholar]
- 30.Siegbahn PEM, Blomberg MRA. Energy diagrams and mechanism for proton pumping in cytochrome c oxidase. Biochim Biophys Acta. 2007;1767:1143–1156. doi: 10.1016/j.bbabio.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Wikström M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme-copper oxidases. Biochim Biophys Acta. 2007;1767:1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Kim YC, Wikström M, Hummer G. Kinetic models of redox-coupled proton pumping. Proc Natl Acad Sci USA. 2007;104:2169–2174. doi: 10.1073/pnas.0611114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.García AE, Hummer G. Water penetration and escape in proteins. Proteins Struct Funct Genet. 2000;38:261–272. [PubMed] [Google Scholar]
- 34.Babcock GT. How oxygen is activated and reduced in respiration. Proc Natl Acad Sci USA. 1999;96:12971–12973. doi: 10.1073/pnas.96.23.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabian M, Wong WW, Gennis RB, Palmer G. Mass spectrometric determination of dioxygen bond splitting in the “peroxy” intermediate of cytochrome c oxidase. Proc Natl Acad Sci USA. 1999;96:13114–13117. doi: 10.1073/pnas.96.23.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan JE, Verkhovsky MI, Palmer G, Wikström M. Role of the PR intermediate in the reaction of cytochrome c oxidase with O2. Biochemistry. 2001;40:6882–6892. doi: 10.1021/bi010246w. [DOI] [PubMed] [Google Scholar]
- 37.Lübben M, Prutsch A, Mamat B, Gerwert K. Electron transfer induces side-chain conformational changes of glutamate-286 from cytochrome bo3. Biochemistry. 1999;38:2048–2056. doi: 10.1021/bi981859k. [DOI] [PubMed] [Google Scholar]
- 38.Nyquist RM, Heitbrink D, Bolwien C, Gennis RB, Heberle J. Direct observation of protonation reactions during the catalytic cycle of cytochrome c oxidase. Proc Natl Acad Sci USA. 2003;100:8715–8720. doi: 10.1073/pnas.1530408100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okuno D, Iwase T, Shinzawa-Itoh K, Yoshikawa S, Kitagawa T. FTIR detection of protonation/deprotonation of key carboxyl side chains caused by redox change of the Cu(A)-heme a moiety and ligand dissociation from the heme a3-Cu(B) center of bovine heart cytochrome c oxidase. J Am Chem Soc. 2003;125:7209–7218. doi: 10.1021/ja021302z. [DOI] [PubMed] [Google Scholar]
- 40.Iwaki M, Rich PR. An IR study of protonation changes associated with heme-heme electron transfer in bovine cytochrome c oxidase. J Am Chem Soc. 2007;129:2923–2929. doi: 10.1021/ja067779i. [DOI] [PubMed] [Google Scholar]
- 41.McMahon BH, et al. FTIR studies of internal proton transfer reactions linked to inter-heme electron transfer in bovine cytochrome c oxidase. Biochim Biophys Acta. 2004;1655:321–331. doi: 10.1016/j.bbabio.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Namslauer A, Pawate AS, Gennis RB, Brzezinski P. Redox-coupled proton translocation in biological systems: Proton shuttling in cytochrome c oxidase. Proc Natl Acad Sci USA. 2003;100:15543–15547. doi: 10.1073/pnas.2432106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorbikova EA, Belevich NP, Wikström M, Verkhovsky MI. Protolytic reactions on reduction of cytochrome c oxidase studied by ATR-FTIR spectroscopy. Biochemistry. 2007;46:4177–4183. doi: 10.1021/bi602523a. [DOI] [PubMed] [Google Scholar]
- 44.Kannt A, Lancaster CR, Michel H. The coupling of electron transfer and proton translocation: Electrostatic calculations on Paracoccus denitrificans cytochrome c oxidase. Biophys J. 1998;74:708–721. doi: 10.1016/S0006-3495(98)73996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Y, Michonova-Alexova E, Gunner MR. Calculated proton uptake on anaerobic reduction of cytochrome c oxidase: Is the reaction electroneutral? Biochemistry. 2006;45:7959–7975. doi: 10.1021/bi052183d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olkhova E, Helms V, Michel H. Titration behavior of residues at the entrance of the D-pathway of cytochrome c oxidase from Paracoccus denitrificans investigated by continuum electrostatic calculations. Biophys J. 2005;89:2324–2331. doi: 10.1529/biophysj.105.062091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 Å resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody Fv fragment. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salomonsson L, Faxén K, Ádelroth P, Brzezinski P. The timing of proton migration in membrane-reconstituted cytochrome c oxidase. Proc Natl Acad Sci USA. 2005;102:17624–17629. doi: 10.1073/pnas.0505431102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfitzner U, et al. Tracing the D-pathway in reconstituted site-directed mutants of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2000;39:6756–6762. doi: 10.1021/bi992235x. [DOI] [PubMed] [Google Scholar]
- 50.Pawate AS, et al. A mutation in subunit I of cytochrome oxidase from Rhodobacter sphaeroides results in an increase in steady-state activity but completely eliminates proton pumping. Biochemistry. 2002;41:13417–13423. doi: 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]
- 51.Soulimane T, et al. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereira MM, Verkhovskaya ML, Teixeira M, Verkhovsky MI. The caa(3) terminal oxidase of Rhodothermus marinus lacking the key glutamate of the D-channel is a proton pump. Biochemistry. 2000;39:6336–6340. doi: 10.1021/bi992848+. [DOI] [PubMed] [Google Scholar]
- 53.Backgren C, Hummer G, Wikström M, Puustinen A. Proton translocation by cytochrome c oxidase can take place without the conserved glutamic acid in subunit I. Biochemistry. 2000;39:7863–7867. doi: 10.1021/bi000806b. [DOI] [PubMed] [Google Scholar]
- 54.Gomes C, et al. Heme-copper oxidases with modified D- and K-pathways are yet efficient proton pumps. FEBS Lett. 2001;497:159–164. doi: 10.1016/s0014-5793(01)02431-0. [DOI] [PubMed] [Google Scholar]
- 55.Brunori M, Antonini G, Malatesta F, Sarti P, Wilson MT. Cytochrome c oxidase. Subunit structure and proton pumping. Eur J Biochem. 1987;169:1–8. doi: 10.1111/j.1432-1033.1987.tb13572.x. [DOI] [PubMed] [Google Scholar]
- 56.Wikström M, Verkhovsky MI, Hummer G. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim Biophys Acta. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. [DOI] [PubMed] [Google Scholar]
- 57.Mitchell P. Chemiosmotic Coupling and Energy Transduction. Bodmin, UK: Glynn Research; 1968. [Google Scholar]
- 58.Wikström M. Energy-dependent reversal of the cytochrome oxidase reaction. Proc Natl Acad Sci USA. 1981;78:4051–4054. doi: 10.1073/pnas.78.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicholls D. The influence of respiration and ATP hydrolysis on the proton-electrochemical gradient across the inner membrane of rat-liver mitochondria as determined by ion distribution. Eur J Biochem. 1974;50:305–315. doi: 10.1111/j.1432-1033.1974.tb03899.x. [DOI] [PubMed] [Google Scholar]
- 60.Wikström M, Verkhovsky MI. Proton translocation by cytochrome c oxidase in different phases of the catalytic cycle. Biochim Biophys Acta. 2002;1555:128–138. doi: 10.1016/s0005-2728(02)00267-0. [DOI] [PubMed] [Google Scholar]
- 61.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 62.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 63.Johansson MP, Kaila VRI, Laakkonen L. Charge parameterization of the metal centers in cytochrome c oxidase. J Comput Chem. 2007;29:753–767. doi: 10.1002/jcc.20835. [DOI] [PubMed] [Google Scholar]
- 64.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]