Abstract
Protein misfolding is intimately associated with devastating human neurodegenerative diseases, including Alzheimer's, Huntington's, and Parkinson's. Although disparate in their pathophysiology, many of these disorders share a common theme, manifested in the accumulation of insoluble protein aggregates in the brain. Recently, the major disease protein found in the pathological inclusions of two of these diseases, amyotrophic lateral sclerosis (ALS) and frontal temporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U), was identified as the 43-kDa TAR–DNA-binding protein (TDP-43), providing a molecular link between them. TDP-43 is a ubiquitously expressed nuclear protein that undergoes a pathological conversion to an aggregated cytoplasmic localization in affected regions of the nervous system. Whether TDP-43 itself can convey toxicity and whether its abnormal aggregation is a cause or consequence of pathogenesis remain unknown. We report a yeast model to define mechanisms governing TDP-43 subcellular localization and aggregation. Remarkably, this simple model recapitulates several salient features of human TDP-43 proteinopathies, including conversion from nuclear localization to cytoplasmic aggregation. We establish a connection between this aggregation and toxicity. The pathological features of TDP-43 are distinct from those of yeast models of other protein-misfolding diseases, such as polyglutamine. This suggests that the yeast model reveals specific aspects of the underlying biology of the disease protein rather than general cellular stresses associated with accumulating misfolded proteins. This work provides a mechanistic framework for investigating the toxicity of TDP-43 aggregation relevant to human disease and establishes a manipulable, high-throughput model for discovering potential therapeutic strategies.
Keywords: amyotrophic lateral sclerosis, frontal temporal dementia, TAR–DNA-binding protein, Saccharomyces cerevisiae
Protein folding is critically important for life, from microbes to man (1). Not unexpectedly, problems in protein folding are the root cause of many of the most devastating diseases, challenging public health worldwide. These protein-misfolding disorders include disastrous neurodegenerative diseases such as Alzheimer's, Parkinson's, amyotrophic lateral sclerosis (ALS), and the frontal temporal dementias (2). Identifying and characterizing the specific disease proteins associated with these disorders may provide insight into disease pathogenesis and aid the development of biomarkers and therapeutic strategies. Indeed, some of the landmark discoveries in neurodegenerative disease research over the last 20 years have been the identification of the misfolded disease proteins in Alzheimer's disease [β-amyloid and tau (3, 4)], Parkinson's disease [α-synuclein (5)], prion diseases [PrP (6)], and polyglutamine diseases [expanded polyglutamine (7)]. Underscoring the significance of identifying the misfolded proteins in pathological inclusions, mutations in several of the genes that encode these proteins cause neurodegenerative disease. In some cases, mutations were discovered before the proteinaceous component of the disease inclusion was identified (8); in others the identification of the aggregated protein led to the discovery of mutations (9). Thus, there is strong evidence that proteins identified in a pathological inclusion will be causally linked to disease pathogenesis.
Frontal temporal dementia (FTD) is the second leading cause of dementia in people under the age of 65, exceeded only by Alzheimer's disease(10) and is characterized by severe changes in personality and abnormal social behavior (11). Notably, motor defects have also been associated with FTD (12). FTDs comprise a heterogeneous class consisting of several subtypes, based on histopathological characteristics (13, 14). Some forms lack ubiquitinated inclusions, and others contain tau pathology. But the most common subtype is frontal temporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U). Patients with this subtype contain tau- and α-synuclein-negative cytoplasmic inclusions in affected neurons (15). Interestingly, although ALS is traditionally considered a motor neuron disease, some patients also exhibit symptoms of FTD, and in these cases, the cytoplasmic ubiquitinated inclusions resemble those of FTLD-U (12). Thus, the clinicopathological features of ALS and FTLD-U raise the possibility of common underlying pathogenic mechanisms.
Recently a molecular connection between FTLD-U and ALS was revealed (16). Biochemical and immunohistochemical approaches lead to development of monoclonal antibodies against insoluble high-molecular-mass material from FTLD-U patient brains. These antibodies identified the 43-kDa TAR–DNA-binding protein (TDP-43) as the immunogen (17). TDP-43-specific monoclonal antibodies marked FTLD-U inclusions and detected a TDP-43 biochemical disease “signature” specific to FTLD-U and ALS, but not Alzheimer's disease or Parkinson's disease tissue. This signature included proteolytically cleaved, hyperphosphorylated and ubiquitinated C-terminal fragments of TDP-43. Interestingly, although TDP-43 is normally a nuclear protein, pathological inclusions contained predominantly cytoplasmic TDP-43 accumulations, suggesting that altered subcellular localization might be integral to disease pathogenesis. However, whether TDP-43 and the aggregates it forms are causally linked to cytotoxicity in FTLD-U and ALS or simply accumulate as a response to other pathogenic events, is unknown.
TDP-43 is a highly conserved, ubiquitously expressed protein, initially identified by virtue of its ability to bind the HIV-1 TAR DNA element and act as a transcriptional repressor (18). In addition to a glycine-rich C-terminal region, TDP-43 contains two RNA recognition motifs (RRM1 and RRM2) and is able to bind UG-repeats in RNA (19, 20). Some reports suggest that TDP-43 might play a role in regulating splicing (21), whereas others propose that it can act as a kind of bridge for nuclear bodies via an interaction with the survival motor neuron (SMN) protein (22). It is not known whether loss of one of these functions, or perhaps a novel pathological gain-of-function, contributes to disease. To date, no evidence directly links TDP-43 cytoplasmic accumulations to cellular dysfunction. In fact, it is controversial whether TDP-43 has a role in disease pathogenesis at all (23).
The baker's yeast, Saccharomyces cerevisiae, is a powerful experimental system for studying complex biological processes. Importantly, most of the key cellular pathways of yeast are very similar to those of mammalian cells, and strains with gain- or loss-of-function mutations in these core pathways are readily available. The yeast genome is well characterized and amenable to genetic manipulation, with methods available to overexpress or knock out every gene. In recent years, yeast cells revealed insights on the basic cellular mechanisms involved in a variety of protein-misfolding diseases, including Parkinson's (24, 25), Huntington's (26–29), Alzheimer's (30), and Creutzfeldt–Jakob diseases (31). Thus, the tools are available for yeast cells to serve as a robust system for investigating even complex human diseases such as ALS and FTLD-U (32).
Here, we describe a yeast TDP-43 proteinopathy model to investigate the molecular determinants of TDP-43 aggregation and its cellular consequences. Remarkably, this model recapitulated several key pathological features seen in humans, including nuclear localization, followed by cytoplasmic sequestration. We establish that cellular toxicity is associated with cytoplasmic accumulation of TDP-43 and define specific sequence determinants governing protein localization, aggregation, and toxicity. This model provides the foundation for screens for therapeutic approaches to the human disease.
Results
TDP-43 Expression in Yeast Cells: Nuclear Localization and Cytoplasmic Aggregation.
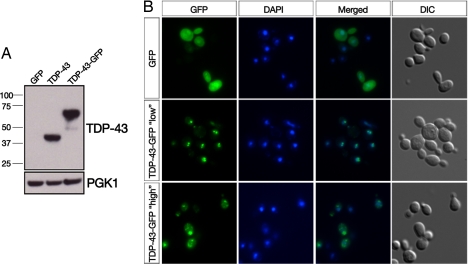
Cytoplasmic inclusions containing TDP-43 are the hallmark pathological feature of ALS and FTLD-U (17). Normally TDP-43 is a nuclear protein, but unknown factors initiate its sequestration to cytoplasmic accumulations. To determine whether yeast cells might serve as a model for investigating mechanisms of altered TDP-43 localization and its potential pathological consequences, we generated a yeast strain harboring an integrated single copy of human TDP-43 fused to GFP. We placed TDP-43-GFP expression under the control of a tightly regulated galactose-inducible promoter to prevent any potential deleterious effects to the cells during routine passage, and to allow an approximately synchronous induction of TDP-43 expression in all cells in the culture. Cells grown to log phase in raffinose medium (noninducing) were transferred to galactose medium to induce TDP-43 expression (Fig. 1A) and visualized by fluorescence microscopy. At early time points (3–4 h), GFP alone was diffusely distributed between the cytoplasm and nucleus (Fig. 1B Top), but the TDP-43-GFP fusion protein localized exclusively to the nucleus (Fig. 1B Middle), with many cells containing 2–3 intranuclear TDP-43-GFP foci. Intriguingly, intranuclear TDP-43 aggregation is a feature of TDP-43 pathology in select FTLD-U subtypes (33–35).
Fig. 1.
Yeast TDP-43 proteinopathy model. (A) TDP-43 or TDP-43-GFP expression in yeast cells was detected by immunoblotting with a mouse polyclonal TDP-43 antibody. Phosphoglycerate kinase 1 (PGK1) was used as a loading control. (B) Fluorescence microscopy was used to visualize the subcellular localization of C-terminally GFP-tagged TDP-43 fusion proteins. Cells were stained with DAPI to visualize nuclei. Whereas GFP alone was distributed between the cytoplasm and nucleus (Top), one integrated copy of TDP-43-GFP strongly localized to the nucleus (Middle) with the occasional formation of intranuclear foci. Expressing TDP-43-GFP from a 2-μm (2 μ) high-copy plasmid profoundly altered its localization because the majority of TDP-43 was now found in multiple cytoplasmic foci (Bottom).
We wondered whether increasing the level of TDP-43 expression might overwhelm the cell's protein quality-control mechanisms (as may well happen in affected neurons) and result in altered TDP-43 localization. Accordingly, we expressed TDP-43-GFP from a 2-μm (2 μ) high-copy plasmid. This profoundly altered protein localization, with the majority of TDP-43 now found in multiple cytoplasmic foci (Fig. 1B Bottom). A similar change in localization occurred with an N-terminal GFP fusion or fusion to an unrelated red fluorescent protein DsRed Express. We also expressed untagged TDP-43 and confirmed these findings by immunocytochemistry with a TDP-43 antibody. In all cases, we observed a similar subcellular localization and aggregation pattern [supporting information (SI) Fig. S1 and data not shown]. Thus, both nuclear localization and the formation of cytoplasmic TDP-43 “aggregates” are modeled in the genetically tractable yeast system. We refer here and below to these TDP-43-GFP foci as aggregates, however, we note that this classification is based primarily on their morphological properties as visualized by fluorescence microscopy. In the strictest sense, protein aggregates represent protein conformational states that are defined biochemically (36).
TDP-43 Accumulation Is Toxic to Yeast Cells.
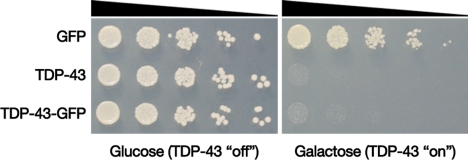
Accruing evidence links TDP-43 aggregation to disease pathology, but whether this is cause or consequence is unclear. The pathogenic conversion of TDP-43 from a normal to a disease-associated conformation likely requires years or even decades in human cells, but the yeast model allows us to monitor the process in only a few hours, providing a unique opportunity to investigate the effects of TDP-43 aggregation in living cells. We performed spotting assays on galactose-containing agar plates to quantitatively assess the impact of TDP-43 accumulation on cell growth. Expressing TDP-43 or TDP-43-GFP from a 2-μ plasmid (conditions that result in cytoplasmic TDP-43 aggregation) led to dramatic inhibition of growth compared with cells transformed with either an empty plasmid or a plasmid expressing GFP alone (Fig. 2A). TDP-43 also inhibited growth of yeast cells in liquid culture (data not shown). Some TDP-43-expressing yeast cells stained positive for propidium iodide (Fig. S2), and electron microscopy revealed an extensively deranged cellular morphology, with large swollen vacuoles, hypertrophied ER, and other features characteristic of dead and dying cells (data not shown) indicating that TDP-43 expression can cause cellular toxicity and not simply growth inhibition. Thus, the yeast system displays striking cellular toxicity associated with TDP-43 aggregation, raising the intriguing possibility that TDP-43 misfolding and aggregation might be a direct cause of neurodegeneration in human disease.
Fig. 2.
TDP-43 is toxic to yeast cells. Yeast cells were transformed with a galactose-inducible GFP construct, untagged TDP-43, or a TDP-43-GFP fusion. Serial dilutions of transformants were spotted on glucose- or galactose-containing agar plates, and growth was assessed after 2 days. Whereas the transformants grew equally well on the control glucose plates, TDP-43 expression profoundly affected growth.
TDP-43 Inclusions Differ from Other Aggregating Proteins.
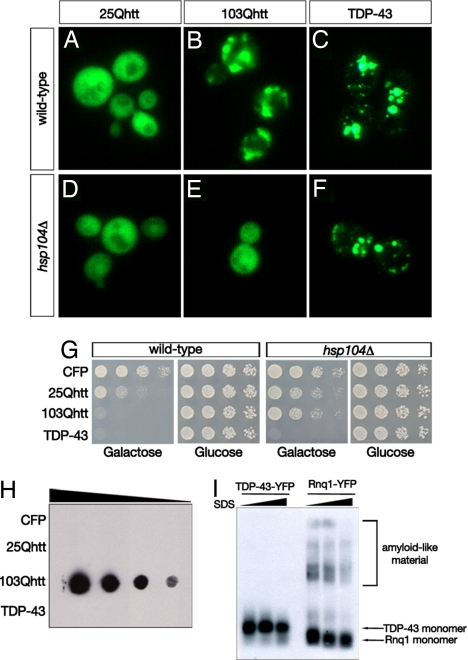
These studies indicate that TDP-43 can be added to the list of human protein-misfolding disease proteins whose overexpression in yeast cells results in intracellular aggregation and toxicity (32). We sought to determine which aspects of the yeast TDP-43 model were similar to and which different from other aggregating disease proteins. Expression of a fragment of the huntingtin (htt) protein harboring pathogenic polyglutamine (polyQ) expansions results in Q-length dependent aggregation and cellular toxicity (26–29) (Fig. 3 A and B). Both polyQ aggregation and toxicity depend on the presence of the yeast prion [RNQ+] (a self-perpetuating amyloid conformation of the Rnq1 protein, which is rich in glutamine and asparagine residues) and the protein-remodeling factor Hsp104 (28, 29). We tested the dependence of TDP-43 aggregation and toxicity on [RNQ+] and Hsp104. Whereas deletion of Hsp104 or Rnq1 (which perforce eliminates the [RNQ+] prion) prevented aggregation and toxicity of htt103Q (Fig. 3 E and G and data not shown), these manipulations had no effect on either the localization or the toxicity of TDP-43 (Fig. 3 F and G and data not shown).
Fig. 3.
TDP-43 inclusions are distinct from polyglutamine aggregates. (A–H) Yeast cells expressing CFP-fusions of htt25Q, htt103Q, or TDP-43 were visualized by fluorescence microscopy. htt25Q-CFP was diffusely distributed (A and D), whereas htt103Q-CFP and TDP-43-CFP formed cytoplasmic aggregates in wild-type cells (strain BY4741) (B and C). (D–G) Deletion of Hsp104 (in cells isogenic to the wild-type BY4741 strain) eliminated the aggregation of htt103Q (E) but not that of TDP-43 (F). Similarly, Hsp104 deletion abolished htt103Q toxicity but not TDP-43 toxicity (G). (H) Filter retardation assay demonstrates SDS-insoluble htt103Q inclusions, unable to pass through cellulose acetate membrane, whereas TDP-43 are fully soluble by this assay. (I) SDD-AGE gel electrophoresis was used to compare TDP-43 aggregates to those of the yeast prion determinant Rnq1. Monomeric and high-molecular-mass amyloid-like forms are detected in yeast lysates expressing Rnq1-YFP, whereas only the monomeric form is detected for TDP-43. Protein samples were incubated in the presence of increasing SDS concentrations (0.1%, 1%, 2%) and detected by using immunoblotting with an anti-GFP antibody (cross-reacts with YFP).
We compared the biophysical properties of TDP-43 aggregates to those of polyQ aggregates. Although both appeared similar by fluorescence microscopy (Fig. 3 B and C), their behavior in a filter retardation assay was strikingly different (Fig. 3H). Htt103Q formed SDS-insoluble aggregates that are unable to pass through a 0.2-μm cellulose acetate membrane, whereas TDP-43 aggregates passed freely through the membrane. We also performed a sedimentation assay by differential centrifugation and found that, unlike htt103Q, which completely partitioned into the pellet fraction, TDP-43 remained almost entirely in the soluble supernatant fraction (Fig. S3). Thus, although both TDP-43 and polyQ are toxic, the nature of the aggregating species is very different. ImmunoEM indicated that the protein was localized to foci in the cytoplasm but not to any recognizable cellular feature, suggesting that it is not being sequestered into a membrane-bounded functional compartment or organelle. Some label also appeared in the vacuole, a site where misfolded proteins and protein aggregates are often degraded.
Expressing human neurodegenerative-disease proteins α-synuclein (25), polyglutamine (29), and now TDP-43 in yeast cells results in cellular toxicity. Genetic screens have been used to discover modifiers of toxicity associated with α-synuclein and polyglutamine (24, 37). We found that of 71 genes that modified α-synuclein toxicity (A.D.G. and S.L., unpublished work) and 48 genes that modified polyglutamine toxicity when overexpressed (K. Matlack and S.L., unpublished work), only two polyglutamine modifiers and one α-synuclein modifier also mitigated TDP-43 toxicity (A. Elden, X. Chen, and A.D.G., unpublished observations). This indicates that the toxicity of these proteins is unique to their specific biology and is not simply due to a general effect of aggregated misfolded proteins.
TDP-43 Aggregates Are Not Amyloid-Like in Yeast Cells.
A unifying characteristic of many neurodegenerative-disease proteins is their ability to readily form amyloid-like fibrillar aggregates (38, 39). Some amyloid or amyloid-like protein aggregates can be visualized by semidenaturing detergent-agarose gel electrophoresis (SDD-AGE) (40). We compared the amyloid-forming ability of Rnq1, the protein determinant of the yeast prion [RNQ+], to that of TDP-43. Whereas Rnq1 formed SDS-resistant high-molecular-mass amyloid-like material, we were unable to detect similar formations by TDP-43 (Fig. 3I). TDP-43-YFP inclusions also did not stain positive for the amyloid-binding dye thioflavin T (Fig. S4). These results are concordant with data showing that, unlike other neurodegenerative diseases (e.g., Alzheimer's, Huntington's, Parkinson's) pathologic inclusions containing TDP-43 are not detected by using the amyloid-binding dyes thioflavin T or Congo red (33). Moreover, analysis by electron microscopy did not reveal visible aggregates similar to those formed by the NM portion of the yeast Sup35 prion nor vesicular clusters similar to those formed by the Parkinson's disease protein α-synuclein (ref. 41 and data not shown). Although additional, more sensitive assays are required to definitively exclude amyloid formation, it seems that, similar to human disease, yeast TDP-43 accumulations are non-amyloid-like protein aggregates.
Dissecting Molecular Determinants of TDP-43 Aggregation and Toxicity.
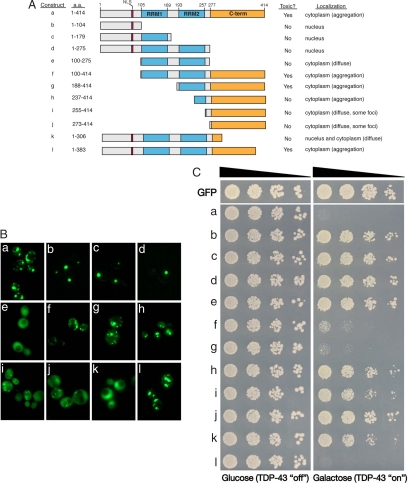
An understanding of the sequence determinants within TDP-43 that govern its nuclear localization and eventual cytoplasmic aggregation will aid the development of therapeutic strategies. The yeast system is well suited for providing a mechanistic framework that can then be validated in mammalian cells and eventually neurons (24). TDP-43 contains two RNA-recognition motifs (RRM1 and RRM2) and a glycine-rich region within the C-terminal region. To determine which regions within TDP-43 are necessary and sufficient for nuclear localization, aggregation, and/or toxicity we generated a panel of TDP-43 truncations starting with the 2-μ plasmid, which produces toxic levels of the full-length protein (Fig. 4A). We expressed each of the truncated TDP-43 constructs as GFP fusions and determined their subcellular localization by fluorescence microscopy (Fig. 4B) and their effect on cell growth by spotting assays (Fig. 4C). By immunoblotting with either GFP- or TDP-43-specfic antibodies, all of the fusion proteins were expressed at comparable levels (Fig. S5 and data not shown).
Fig. 4.
Dissecting the molecular determinants of TDP-43 aggregation and toxicity. (A) A diagram illustrating the domain structure of TDP-43 along with various truncation constructs used in this study (a–l). (B) Structure/function analysis testing the effects of truncations on TDP-43-GFP localization by fluorescence microscopy and toxicity by spotting assays (C). The C-terminal region is required for aggregation and toxicity (compare constructs a and d), but by itself is not sufficient (construct j). A construct harboring the C-terminal region along with RRM2 recapitulates the complete aggregation and toxicity of full-length TDP-43 (construct g). RRM, RNA recognition motif; C-term, C-terminal region; NLS, nuclear localization signal.
Compared with full-length TDP-43, which formed multiple cytoplasmic aggregates, deleting the C terminus had a dramatic effect, resulting in an entirely nuclear localized protein (Fig. 4B, C-terminal deletion, construct d). Subdeletions of the remaining N-terminal region, containing as few as the first 104 residues, which contains a weak predicted nuclear localization sequence (NLS), were also localized to the nucleus. Moreover, a construct containing both RRMs but lacking the predicted NLS was diffusely distributed in the cytoplasm (Fig. 4B, N- and C-terminal deletion, construct e). Most of the C-terminal region was required for aggregation; addition of only a portion of the glycine-rich region immediately after RRM2 caused TDP-43 to now be distributed between both the nucleus and cytoplasm, but not aggregate (Fig. 4B, compare constructs k and l). Thus, sequences within the N-terminal portion of TDP-43 are necessary to direct nuclear localization, whereas the C-terminal region is required for cytoplasmic aggregation.
Having established that the C-terminal region is necessary for aggregate formation, we asked whether it was also sufficient. Surprisingly, a construct containing only the C-terminal portion was almost entirely diffusely localized within the cytoplasm, with occasional small cytoplasmic puncta (Fig. 4B, C-terminal region alone, construct j). Proteins containing larger fragments of RRM2 increased aggregate formation (Fig. 4B, compare constructs g–j); a construct containing the entire RRM2 along with the C-terminal region completely recapitulated the cytoplasmic aggregation pattern of full-length TDP-43 (Fig. 4B, compare constructs a and g). Thus, the TDP-43 C-terminal region alone is not sufficient for aggregation but, rather, requires an intact RRM for robust cytoplasmic foci formation.
We performed spotting assays to determine which of the TDP-43 truncation constructs retained toxic potential (Fig. 4C). In addition to being required for aggregate formation, the C-terminal region was also necessary for cellular toxicity (Fig. 4C, compare constructs a and d). The C-terminal region, however, was not sufficient for toxicity. Rather, the entire RRM2 domain was also needed (Fig. 4C, compare constructs g–j). Nuclear localization was not required for toxicity, because construct f (lacks NLS) formed cytoplasmic aggregates (Fig. 4B) and was toxic (Fig. 4C). Aggregation per se was not sufficient for toxicity because construct h was not toxic, despite significant aggregation. Taken together, these data indicate that the RNA recognition motif and C-terminal region are together required for TDP-43 to form toxic aggregates. Thus, we have defined the sequence requirements for TDP-43 aggregation and cellular toxicity in vivo. It is noteworthy that the particular toxic fragment we have defined (construct g) is nearly a perfect match to one of the recently reported caspase-dependent cleavage fragments of TDP-43 (42), thus providing a mechanistic link between the presence of aggregating TDP-43 cleavage products and cellular pathology.
Discussion
We have established a yeast model to investigate the mechanisms underpinning the conversion of TDP-43 to a toxic species. As in human neurons (17), TDP-43 was nuclear localized in yeast cells but, upon increased expression, formed multiple cytoplasmic inclusions. TDP-43 inclusions lacked amyloid and had properties distinct from polyglutamine aggregates. Our data also link TDP-43 aggregation to cellular toxicity, suggesting a potential causal role for TDP-43 in disease pathogenesis.
What is the mechanism of TDP-43 cellular toxicity? The answer to this question will be vital for devising effective therapeutic strategies. Toxicity might be caused by aggregating TDP-43 being sequestered away from its proposed normal function (for example in regulating transcription, splicing, and mRNA stability). Yeast cells do not have a true TDP-43 homolog, although they do contain RNA-binding proteins with similar domain architectures (A.D.G. and B.S.J., unpublished observations) and TDP-43 might coaggregate and sequester these proteins away from their essential role. Alternatively, the TDP-43 aggregates might have a toxic gain-of-function independent of TDP-43's normal cellular role. If the first scenario were correct, designing therapies aimed at targeting TDP-43 for elimination would not be warranted. However, if the C-terminal TDP-43 fragments themselves were toxic, such an approach might be efficacious. Our in vivo structure/function analyses revealed that only aggregating forms of the protein were toxic, suggesting that TDP-43 causes a toxic gain-of-function phenotype because of protein misfolding. However, aggregation per se is not sufficient for toxicity; it was the addition of an intact RRM that conferred the full toxic potential. Thus, we propose that TDP-43 cellular toxicity, and perhaps TDP-43 pathophysiology, is intimately associated with a function that depends on the RRM and is not simply attributable to general cellular stress associated with accumulating misfolded proteins.
Zhang and colleagues (42) recently described a caspase-dependent pathway leading to the proteolytic cleavage of TDP-43, resulting in the C-terminal fragments observed in the TDP-43 pathologic “signature.” Remarkably, the minimum toxic fragment in yeast is very similar to one of these caspase-cleavage products. Mutations in the secreted factor progranulin have been linked to familial forms of FTD (43, 44), and recent evidence suggests that reduced progranulin function might lead to increased caspase-dependant TDP-43 cleavage (42). Our work showing that these C-terminal fragments are toxic to cells provides a potential link among progranulin dysfunction, TDP-43 cleavage, and cellular degeneration.
During the course of our work, two groups reported the identification of mutations in TDP-43 in familial and sporadic ALS (45, 46), clearly establishing a direct link between TDP-43 and neurodegeneration. Now that TDP-43 has risen in prominence as a neurodegenerative-disease protein (33), intense efforts will be focused on understanding its biology. Because we are able to recapitulate many features of TDP-43 pathology in the genetically tractable yeast model system, including cellular toxicity, genome-wide screens for modifiers of aggregation and toxicity will likely be powerful as one approach in the quest for effective diagnostic and treatment paradigms. Indeed, similar approaches using yeast models of Parkinson's and Huntington's diseases have been fruitful (24, 37, 47). Moreover, the yeast model is a potential platform for preclinical drug discovery aimed at defining molecules that can antagonize TDP-43 aggregation and/or restore disrupted cellular pathways.
Materials and Methods
Yeast Strains, Media, and Plasmids.
Strain and plasmid construction is detailed in SI Materials and Methods.
Yeast Transformation and Spotting Assays.
Yeast procedures were performed according to standard protocols (48). We used the PEG/lithium acetate method to transform yeast with plasmid DNA (49). For spotting assays, yeast cells were grown overnight at 30°C in liquid media containing raffinose (SRaf/-Ura) until they reached log or midlog phase. Cultures were then normalized for OD600, serially diluted, and spotted onto synthetic solid media containing glucose or galactose lacking uracil and were grown at 30°C for 2–3 days.
SDD-AGE.
SDD-AGE is detailed in SI Materials and Methods.
Immunoblotting.
Immunoblotting protocols and antibodies are described in SI Materials and Methods.
Filter Retardation and Sedimentation Assays.
Filter retardation using cellulose acetate membranes and sedimentation assays were performed as described (50) and as detailed in SI Materials and Methods.
Thioflavin T Staining.
Yeast cells were stained with thioflavin T according to a protocol adapted from ref. 48 and as detailed in SI Materials and Methods.
Fluorescence Microscopy.
Fluorescence microscopy was performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Andrea Stout for expert assistance with microscopy; Randal Halfmann and Jijun Dong for advice on semidenaturing detergent-agarose gel electrophoresis; Martin Duennwald (Boston Biomedical Research Institute, Watertown, MA) for huntingtin constructs and for advice on the filter retardation and sedimentation assays; Jens Tyedmers for advice on thioflavin T staining; Alex Chavez (University of Pennsylvania, Philadelphia) for the hsp104Δ yeast strain; Zongtian Tong for advice on immunocytochemistry; and Dan Kessler, Nancy Bonini, Jim Shorter, Paul Taylor, Virginia Lee, and John Trojanowski for helpful discussions. Many thanks to Jim Shorter, Nancy Bonini, Dan Kessler and members of the A.D.G. laboratory for critical comments and suggestions on the manuscript. This work was supported in part by a Pilot grant from the University of Pennsylvania Institute on Aging (A.D.G.). S.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: S.L. is a cofounder of and owns stock in FoldRx Pharmaceuticals, a company developing therapies for diseases of protein misfolding. S.L. and A.D.G. are inventors on patents and patent applications that have been licensed to FoldRx Pharmaceuticals.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802082105/DCSupplemental.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: A decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 3.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. A68: A major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 4.Glenner GG, Wong CW. Alzheimer's disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 5.Spillantini MG, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 6.McKinley MP, Bolton DC, Prusiner SB. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- 7.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 9.Hutton M, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 10.McKhann GM, et al. Clinical and pathological diagnosis of frontotemporal dementia: Report of the Work Group on Frontotemporal Dementia and Pick's Disease. Arch Neurol. 2001;58:1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 11.Neary D, et al. Frontotemporal lobar degeneration: A consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 12.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 13.Hodges JR, et al. Clinicopathological correlates in frontotemporal dementia. Ann Neurol. 2004;56:399–406. doi: 10.1002/ana.20203. [DOI] [PubMed] [Google Scholar]
- 14.Forman MS, et al. Frontotemporal dementia: Clinicopathological correlations. Ann Neurol. 2006;59:952–962. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josephs KA, Jones AG, Dickson DW. Hippocampal sclerosis and ubiquitin-positive inclusions in dementia lacking distinctive histopathology. Dementia Geriatr Cognit Disorders. 2004;17:342–345. doi: 10.1159/000077168. [DOI] [PubMed] [Google Scholar]
- 16.Sampathu DM, et al. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol. 2006;169:1343–1352. doi: 10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 18.Ou SH, et al. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buratti E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strong MJ, et al. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Ayala YM, Pagani F, Baralle FE. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580:1339–1344. doi: 10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 22.Wang IF, Reddy NM, Shen CK. Higher-order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci USA. 2002;99:13583–13588. doi: 10.1073/pnas.212483099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothstein JD. TDP-43 in amyotrophic lateral sclerosis: Pathophysiology or patho-babel? Ann Neurol. 2007;61:382–384. doi: 10.1002/ana.21155. [DOI] [PubMed] [Google Scholar]
- 24.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models, Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duennwald ML, et al. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci USA. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meriin AB, et al. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagriantsev S, Liebman S. Modulation of Abeta42 low-n oligomerization using a novel yeast reporter system. BMC Biol. 2006;4:32. doi: 10.1186/1741-7007-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J, Lindquist S. De novo generation of a PrPSc-like conformation in living cells. Nat Cell Biol. 1999;1:358–361. doi: 10.1038/14053. [DOI] [PubMed] [Google Scholar]
- 32.Gitler AD. Beer and bread to brains and beyond: Can yeast cells teach us about neurodegenerative disease? NeuroSignals. 2008;16:52–62. doi: 10.1159/000109759. [DOI] [PubMed] [Google Scholar]
- 33.Forman MS, Trojanowski JQ, Lee VM. TDP-43: A novel neurodegenerative proteinopathy. Curr Opin Neurobiol. 2007;17:548–555. doi: 10.1016/j.conb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann M, et al. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol Exp Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- 35.Sampathu DM, et al. Ubiquitination of alpha-synuclein is not required for formation of pathological inclusions in alpha-synucleinopathies. Am J Pathol. 2003;163:91–100. doi: 10.1016/s0002-9440(10)63633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 37.Willingham S, et al. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 38.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 39.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 40.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- 41.Gitler AD, et al. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci USA. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YJ, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 44.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 45.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gitcho MA, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008 doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giorgini F, et al. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet. 2005;37:526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guthrie C, Fink GR. Methods in Ezymology: Guide to Yeast Genetics and Molecular Biology. New York: Academic; 2002. p. 169. [Google Scholar]
- 49.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muchowski PJ, et al. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.