Abstract
Previous results showed that overexpression of the WTH3 gene in multidrug resistance (MDR) cells reduced MDR1 gene expression and converted their resistance to sensitivity to various anticancer drugs. The WTH3 gene promoter was found to be differentially regulated in paired MDR vs non-MDR MCF7 cells owing to epigenetic modifications and transcription factor modulations. To understand further the mechanisms that govern WTH3's differential expression, we uncovered a p53-binding site in its promoter, which indicated that WTH3 could be regulated by the p53 gene. This hypothesis was then tested by different strategies. The resulting data revealed that (1) the WTH3 promoter was upregulated by the p53 transgene in diverse host cells; (2) there was a correlation between WTH3 expression levels and p53 gene status in a cell line panel; (3) a WTH3 promoter region was directly targeted by the p53 protein in vitro and in vivo. In addition, overexpression of the WTH3 gene promoted the apoptotic phenotype in host cells. On the basis of these findings, we believe that the negative role played by the WTH3 gene in MDR development is through its proapoptotic potential that is regulated by multiple mechanisms at the transcription level, and one of these mechanisms is linked to the p53 gene.
Keywords: WTH3 gene, p53-response element, multidrug resistance, apoptosis
Multidrug resistance (MDR) is a fatal event encountered during cancer chemotherapy (Chen et al, 1986; Gros et al, 1986a, 1986b; Cole et al, 1992; Robinson et al, 1997; Smyth et al, 1998; Johnstone et al, 1999). To understand better MDR development, we employed the methylation sensitive-representational difference analysis technique (Yuan et al, 1999; Shan et al, 2000, 2002a) to study DNA hypermethylation events in a human MDR breast cancer cell line, MCF7/AdrR, as compared to its parental line, MCF7/WT. As a result, the WTH3 gene was discovered. The gene product is homologous to the Rab6 and Rab6c genes that belong to the ras super family and encode small G proteins (Zahraoui et al, 1989; Goud et al, 1990; Echard et al, 1998, 2000; Shan et al, 2000, 2002a). Similar to the Rab6s, WTH3 is a housekeeping gene and its product is capable of binding to GTP molecules (Tian et al, 2005b). However, unlike the Rab6s that reside in the Golgi network, most of WTH3 locates in the cytoplasm and to a less degree in the nuclei. This disparity could be due to WTH3's lack of a cysteine at its C terminus for geranyl–geranylation, a necessary post-translational modification for membrane attachment (Barbacid, 1987). Previous studies found that the WTH3 gene was downregulated in MDR cell lines, MCF7/AdrR and MES-SA/Dx5 (a human uterine sarcoma cell line), and by introducing it back into those lines caused downregulation of MDR1 gene expression and reversed their MDR phenotypes to various anticancer drugs (Shan et al, 2002a; Tian et al, 2005b). In addition, our research also revealed that hypermethylation (an epigenetic modification event in mammals) of the WTH3 promoter and transcription factor modulation were involved in its differential expression in MCF7/AdrR vs MCF7/WT cells (Tian et al, 2005b). Furthermore, the hypermethylation event was also observed in primary drug-resistant breast cancer cells (Tian et al, 2005a). Taken together, our data supported the notion that WTH3 could play an important role in MDR development.
To understand further the mechanisms involved in WTH3's differential expression in MDR cells, the Patch Search Program was utilised to look for consensus sequences in the gene promoter for existing transcription factors. As a result, several candidate motifs were implicated, one of which was a p53-binding domain, although it contained five mismatched base pairs (bp) as compared to the typical p53-binding consensus sequence (5′-RRRCWWGYYY(N=0–13)RRRCWWGYYY-3′) (Lowe, 1995). Consequently, we named it p53M. The p53 gene product is a transcription factor that functions as a tumour suppressor and plays a pivotal role in apoptosis and cell cycle arrest (Lowe, 1995; Aas et al, 1996; Righetti et al, 1996). Various mutations of p53 are associated with human cancers and the onset of MDR in a broad field of solid and haematological malignancies (Ogretmen and Safa, 1997; Schmitt and Lowe, 1999; Smith et al, 2003; Norbury and Zhivotovsky, 2004; Pommier et al, 2004; Kim, 2005; Steele and Lane, 2005). Identification of a potential p53-binding motif in the WTH3 promoter suggested that its activity could be regulated by p53. To test this hypothesis, WTH3 gene promoter function under the influence of the p53 transgene was evaluated by luciferase assays. In addition, the correlation between WTH3 expression levels in 11 cell lines with defined p53 status was examined. Owing to the mismatches in p53M, instead of using the targeted-deletion strategy, we generated serial deletion mutants to determine the p53-response region in the WTH3 promoter. Next, the physical interaction between the defined response region and the p53 protein was examined by the electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP) approach. The resulting data suggested that the WTH3 gene was a direct target of the p53 protein. This information led us to evaluate the possible participation of WTH3 in promoting apoptosis.
MATERIALS AND METHODS
Cell lines and doxorubicin (DOX) treatment
MCF7/AdrR (p53 del 126–132), MCF7/WT (WT p53) (Ramljak et al, 2005), MES-SA/Dx5 and its parental cell line, MES-SA (ATCC.), were grown under the conditions as described (Shan et al, 2002a). Hs578T (p53 V157F), MDA-MB-231 (p53 R280K), MDA-MB-435 (p53 G266E), MDA-MB-436 (mutated p53), MDA-MB-468 (p53 R273H), T47D (p53 L194F) and SKBr3 (p53 R175H) (Camps et al, 1990; Elstner et al, 1995; Nieves-Neira and Pommier, 1999) (gifts from Dr Moll M Ute, Department of Pathology; and Dr Cao Jian Division of Hematology, SUNYSB), were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium medium with 10% foetal calf serum (FCS), 100 μg/ml streptomycin and 100 U/ml penicillin. HEK293 (Human primary embryonic kidney cells, ATCC), and Hela cells were grown at 37°C with 5% CO2 in Roswell Park Memorial Institute 1640 culture medium with 10% FCS, 100 μg/ml streptomycin and 100 U/ml penicillin. To induce p53 expression, MCF7/WT cells were treated with 1 μM of DOX for 20 h.
Construction of recombinant DNA
Detailed information about generating the pcDNA3.1/WTH3 and pGL/WTH3P constructs containing the coding region and WTH3 promoter, respectively, were described previously (Shan et al, 2002a; Tian et al, 2005b). Six deletion mutants of the WTH3 promoter were created by polymerase chain reaction (PCR) amplification using pGL/WTH3P as the template. Sense primers for deletion 1 (−540 to −1), 2 (−453 to −1), 3 (−396 to −1), 4 (−289 to −1), 5 (−194 to −1) and 6 (−116 to −1) were 5′-AGAG GTACCCACCGCACCATTGTTTTTAGTAC-3′, 5′-AGAGGTACCCGCACTCAGCAGGTTGGGC-3, 5′-AGAGGTACCTGAGAGATCCCGGATACATCTGC-3′, 5′-AGAGGTACCCAAAGCACACCCCTGGCTC-3, 5′-AGAGGTACCGGCGG CTGCCAGTCTGTG-3′ and 5′-AGAGGTACCGGGGCGCAGAGAGCTCGG-3, respectively. The anti-sense primer for all the mutants was 5′-GAAGATCTTCGTGGAACTAGAGGAGCTGTCGCC-3′. Each primer pair contained KpnI and BamHI restriction enzyme sites for cloning the PCR fragment into the pGL3 vector to create pGL/WTH3P-d1, -d2, -d3, -d4, -d5 and -d6. The correct sequence of each construct was verified by sequencing (Genewiz Inc., South Plainfield, NJ, USA). Wild-type p53 in pcDNA/P53 and the mutated p53 gene in pcDNA/P53R249S, which did not contain trans-element activity, were gifts from Dr Moll M Ute.
Transient transfection and luciferase assays
To determine whether the WTH3 promoter was regulated by p53, pGL/WTH3P was cotransfected with pcDNA/P53, pcDNA/P53R249S (negative control) or pcDNA/3.1 (negative control) into MCF7/WT and HEK293 cells. In brief, 0.2 μg of each construct were transfected along with 0.1 μg of pCMV/β-galactosidase when the cells (seeded onto 24-well plates) reached 50–70% confluence. After 24 h, luciferase and β-galactosidase activity was measured using the Luciferase Assay System and Beta-Glo™ Assay System (Promega, Madison, WI, USA) according to the manufacturer's instruction. Luciferase activities of transfectants were compared after normalising their β-galactosidase activities and protein concentrations. To determine p53s influence on endogenous WTH3 gene expression, pcDNA/P53 or the empty vector was transiently transfected into Hela and MCF7/AdrR cells. After 24 h, RNAs were isolated from the cells for semi-quantitative reverse transcriptase (SQRT)-PCR analyses.
SQRT-PCR and Western blot
Total RNAs were isolated from cell lines, transfectants and the corresponding negative controls by the High Pure RNA Isolation Kit (Roche, Indianapolis, IN, USA). Semi-quantitative reverse transcriptase-polymerase chain reaction was performed using the Titan One Tube RT-PCR system based on the manufacturer's protocol (Roche). The sense and anti-sense primers for WTH3 and GAPDH were described previously (Shan et al, 2002a). The PCR and quantification of PCR products were performed as described (Shan et al, 2000, 2002a; Tian et al, 2005a, 2005b). To evaluate WTH3 protein levels in the cell lines, the protein concentrations of cell lysates were determined by absorbance measured at 280 nm and the bicinchoninic acid protein assay reagent kit (BCA Kit, Pierce, Rockford, IL, USA). A total of 100 μg of each cell lysate was loaded onto triplicate 12% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) gels for Western analyses as described previously (Shan et al, 2002b) using the antibodies for WTH3 (1 : 200 dilution) (Research Genetics, Huntsville, AL, USA), Rab6 (1 : 2000 dilution) and MDR1 (1 : 30) (Santa Cruz, Santa Cruz, CA, USA).
EMSA and super-EMSA
Electrophoretic mobility shift assay were performed using the purified p53 protein (Santa Cruz) or and probe (P49) that covered the region from −282 to −330 in the WTH3 promoter (gagccgggtgcggaaggagggaacg[gCCctagcct/TggGaagccA]aagc-3′) and contained the putative p53-response element, p53M (bracketed). The five mismatches in it, comparing to the typical p53-binding site, were capitalised and underlined. Another probe representing the sequence in the albumin gene served as the negative control, which was amplified from genome DNA by PCR using the forward and reverse primers, 5′-GCTGTCATCTCTTGTGGGCTGT-3′ and 5′-ACTCATGGGAGCTGCTGGTTC-3′. The probes were generated by annealing the forward and reverse oligonucleotides, followed by end labelling using T4 polynucleotide kinase in the presence of [γ-32P]dATP. To perform EMSA, extracts were prepared from MCF7/AdrR and MCF7/WT cells as described (Tian et al, 2005a, 2005b). A total of 10 μg nuclear extract and 50 ng of p53 protein were applied to perform EMSAs. The detailed procedure for carrying out EMSAs was described previously (Tian et al, 2005b). Super-EMSA experiments were performed in the presence or absence of 1 μg of monoclonal p53 antibody (p53-(DO)-1, Santa Cruz) for 30 min at room temperature and analysed on a 4% non-denaturing-PAGE gel. The experiments were repeated three times.
ChIP assays
MCF7/WT cells were treated with 1 μM DOX for 20 h to induce p53 expression. Chromatin immunoprecipitation assays were carried out as described previously (Adachi et al, 2004). Briefly, genomic DNA and proteins were crosslinked by the addition of 1% final concentration of formaldehyde directly into the culture medium and incubated for 30 min at 37°C. Cells were lysed and sonicated to 300–1000 bp DNA fragments. After centrifugation, the supernatant was diluted 10 times with the ChIP buffer and incubated with the agarose conjugated with anti-p53 (p53-AC) or -HA (negative control) antibodies (Santa Cruz) at 4°C overnight. Immune complexes were precipitated and washed. The DNA–protein complexes were decrosslinked by heating at 65°C for 5 h with high concentration salt and DNA was purified and resuspended in 50 μl of TE buffer. The input DNA was diluted 100 times before PCR. The bound and input DNAs were analysed by PCR (35 cycles). Specific primers for the p53 response element in the WTH3 promoter were 5′-GCCCTAGCCTTGGGAAGCCAAAG-3′ (forward) and 5′-CGGCAGAGTAGCCGAGCACG-3′ (reverse). The sense and anti-sense primers for p21 (positive control) and albumin (negative control) promoters were 5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAG-3′ as well as 5′-GCTGTCATCTCTTGTGGGCTGT-3′ and 5′-ACTCATGGGAGCTGCTGGTTC-3′.
4′,6-Diamidino-2-phenylindole (DAPI) staining assays
Hela cells were seeded onto glass cover slips placed in 6-well plates. When the cells reached 50% confluence, they were transfected with pcDNA3.1/WTH3 or pcDNA3.1 in parallel using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) as described previously (Shan et al, 2000, 2002a) following the manufacturer's instructions. Nuclear staining with DAPI was performed as described (Kim et al, 2002). After 24 h of transfection, cells were washed with 1 × phosphate-buffered saline (PBS), fixed with 70% ethanol and washed again with PBS. The cells then were treated with DAPI (1 μg/ml) (Sigma) for 12 min, washed with PBS for 5 min, and treated with VectaShield (Vector Laboratories, Burlingame, CA, USA). Stained nuclei were visualised under a fluorescent microscope. Apoptotic cells were morphologically defined by cytoplasmic and nuclear shrinkage and chromatin condensation. The experiments were repeated three times.
Terminal deoxynucleotidyl transferase biotin-dUTP nick end labelling (TUNEL) assays
Hela and HEK293 cells were seeded in 6-well plates. When the cells reached 50% confluence, they were transfected with pcDNA3.1/WTH3 or pcDNA3.1. After 18 (Hela) and 30 (HEK293) hours of transfection, cells were trypsinised and washed with 2 × PBS and transferred onto a glass slide and air dried. The cells were fixed in 3% paraformaldehyde for 30 min and washed in PBS. Terminal deoxynucleotidyl transferase biotin-dUTP nick end labelling assays were performed using the FragEL™ DNA Fragmentation Detection Kit (Calbiochem, San Diego, CA, USA) following the manufacturer's protocol. The apoptotic cells, which exhibited a brown stain, were visualised under a microscope. The experiments were repeated three times.
Flow cytometry
Approximately 5 × 105 cells/well of HEK293 were transfected with zero or equal mole ratio of pcDNA/WTH3 or pcDNA3.1. The pCMV/β-galactosidase was also transfected in parallel as a transfection efficiency control. After 24 h of transfection, the cells were harvested by trypsinisation, washed in PBS, and the DNA was stained with propidium iodide (PI, 50 μg/ml) containing 250 μg/ml of ribonuclease A, followed by flow cytometry analysis as described previously (Shan et al, 2002a). The experiments were repeated three times.
RESULTS
The WTH3 promoter was positively regulated by p53
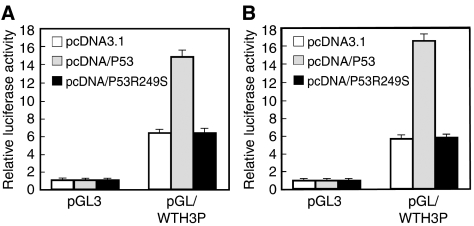
Since results generated by the Patch Search indicated that the p53M sequence in the WTH3 promoter could be a potential binding site for p53, we speculated that the p53 gene could regulate the WTH3 promoter. To test this hypothesis, pGL/WTH3P or the empty vector, pGL3, was cotransfected with pcDNA3.1, pcDNA/P53 or pcDNA/P53R249S, as well as pCMV/β-galactosidase into MCF7/WT and HEK293 cells. The enzyme activities driven by the WTH3 promoter under the influence of wild-type and mutated p53 were measured with justification of protein concentrations and transcription efficiency (Figure 1A and B). We found that the wild-type p53, but not its mutant, increased WTH3 promoter activity approximately 2.5–3 times in both hosts. As the TSP50 gene, which was negatively regulated by p53, was available in our lab, we cotransfected pGL/TSP50P (pGL containing the TSP50 promoter) with pcDNA/P53, in parallel, into MCF7/WT and HEK293 cells. The results clearly showed that p53 downregulated the TSP50 promoter (Xu et al, 2007), whereas it upregulated the WTH3 promoter. Similar results were obtained when MCF7/AdrR and Hela cells were used as hosts (data not shown), which suggested that WTH3 was upregulated by the p53 gene in a cell-type independent manner. This information provided us with a plausible explanation that WTH3's low expression observed in MCF7/AdrR could be the result of p53 dysfunction owing to the mini deletion in its DNA-binding domain, whereas MCF7/WT, which contains the wild-type p53 gene, expressed a relatively high level of WTH3 (Norbury and Zhivotovsky, 2004). To examine further the correlations between these two genes, we measured WTH3 gene expression levels in a cell line panel with defined p53 gene status.
Figure 1.
Results obtained from luciferase assays where pGL3 or pGL/WTH3P was cotransfected with pcDNA3.1, pcDNA/P53 or pcDNA/P53R249S into (A) MCF7/WT and (B) HEK293 cells. Relative luciferase activities driven by the WTH3 promoter under the influence of the empty vector, wild-type and mutated p53 were compared.
There was a correlation between WTH3 expression levels and p53 gene status
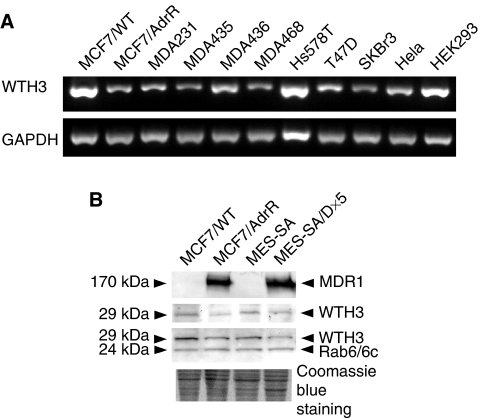
WTH3 expression levels in nine breast carcinoma cell lines, Hs578T, MCF7/AdrR, MCF7/WT, MDA-MB-231, MDA-MB-435, MDA-MB-436, MDA-MB-468, T47D and SKBr3, as well as in Hela and HEK293 were examined by SQRT-PCR. The results showed that MCF7/WT and HEK293, which contained the wild-type p53 gene, produced the highest WTH3 RNA as compared to the remaining cell lines whose p53 gene was either mutated, as in Hs578 T, MCF7/AdrR, MDA-MB-231, −435, −436, −468, T47D and SKBr3, or attenuated, as in Hela cells by the papillomavirus E6 protein that targets p53 for degradation (Li et al, 2004) (Figure 2A). Although each cell line contains its own unique, complicated biological characteristics, the results showed a trend that the cell lines possessing mutated p53 expressed relatively low WTH3 RNA as compared to those containing its wild type. However, we will further confirm this correlation via other approaches in the future. In addition, reduced WTH3 transcripts were also reflected at the protein level when antibodies for WTH3 and Rab6 were used to measure the WTH3 protein in MCF7/AdrR vs MCF/WT and MES-SA/D × 5 vs MES-SA cells. Densitometer analysis showed that the protein amount detected by the WTH3 antibody was significantly higher in MCF7/WT and MES-SA as compared to their MDR counterparts (Figure 2B). Rab6 antibody, which recognised both Rab6c (a housekeeping gene, served as the quantitative control) and WTH3 protein, generated similar results. This was consistent with previous data that found WTH3 transcript levels in MCF7/WT and MES-SA were much higher than those in MCF7/AdrR and MES-SA/Dx5 (Shan et al, 2002a). Since both MDR cell lines expressed extremely high levels of the MDR1 gene relative to their corresponding parental cell line, the MDR1 antibody was also used to detect MDR1 protein levels in those cells, which served as another control.
Figure 2.
(A) Results obtained from SQRT-PCR to estimate endogenous WTH3 gene expressions in 11 cell lines with defined p53-gene status. (B) Western blot analysis of WTH3 proteins in MCF7/AdrR vs MCF7/WT and MES-SA/D × 5 vs MES-SA using WTH3 (1 : 200 dilution) and Rab6 (1 : 2000 dilution) antibodies. In addition, MDR1 detected by the MDR1 antibody (1 : 30 dilution) served as one of the controls. Proteins and their molecular weights are indicated on the right and left. Identical SDS-PAGE gels with the same amount of protein (100 μg/lane) were loaded. Three gels were used for Western blot by three antibodies; the fourth gel was stained with Coomassie blue for protein concentration normalisation.
The p53 transgene elevated endogenous WTH3 gene expression
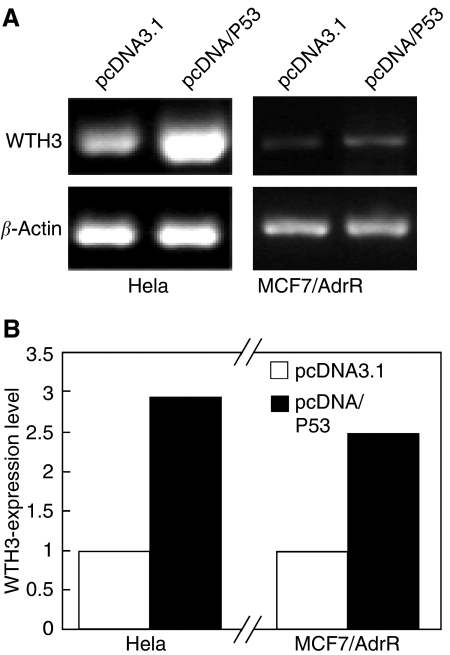
Both Hela and MCF7/AdrR cells were used as the hosts for p53 gene transfection. Briefly, pcDNA/P53 or pcDNA3.1 and pCMV/β-galactosidase were introduced into the cells. After 24 h, total RNA was isolated from the transfectants and SQRT-PCR was performed to evaluate the amount of WTH3 transcripts. As expected, cells expressing the p53 transgene generated approximately 2.5 times higher WTH3 transcripts than the control cells (Figure 3A and B). Taken together, the data gathered from performing different experiments suggested that WTH3 could be a target of the p53 gene. To gain detailed information on how p53 controlled WTH3 gene expression, we next wanted to identify the p53-response element in the gene promoter by creating serial deletion mutants.
Figure 3.
Results obtained from SQRT-PCR to estimate endogenous WTH3 gene expressions in Hela and MCF7/AdrR cells, which were influenced by the p53 transgene. (A) The two cell lines were either transfected with pcDNA3.1 or pcDNA/P53, where β-actin served as the quantitative control. (B) Quantitative comparison of the results presented in (A). The empty and black columns represent WTH3 expression levels under the influence of the empty vector or the p53 transgene, respectively.
The region (−396 to −289) in the WTH3 promoter contained the p53-response element
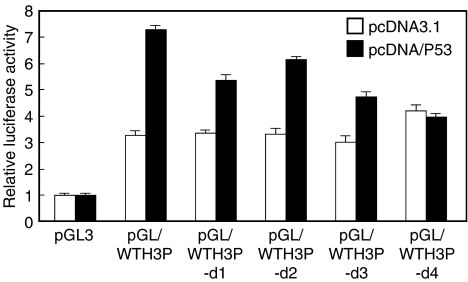
Since p53M contained five mismatches, we were not sure if it was the real p53-binding site. Instead of generating a targeted deletion mutant, we created six serial deletions (each was ∼100 bp shorter than the adjacent one) by PCR amplification using pGL/WTH3P as the template. The resulting PCR products were constructed into pGL3 to obtain pGL/WTH3P-d1 to -d6 constructs. Each of the plasmids was then cotransfected with pcDNA/P53 or pcDNA/3.1 along with pCMV/β-galactosidase. After 24 h, the luciferase activity driven by the wild-type and mutated promoters under the influence of p53 was determined. We found that pGL/WTH3P-d4 no longer responded to the p53 transgene as its enzymatic activity was similar to that in the control cells that were transfected with pGL3 and pcDNA/P53. This finding suggested that the deleted region (from −396 to −289) could contain the p53-response site (Figure 4). As the p53M sequence resided in this region, we believed it could be a direct target of the p53 protein. To test this possibility, EMSA and super-EMSA assays were performed.
Figure 4.
Luciferase assay results for the WTH3 promoter deletion mutants under the influence of the p53 gene in HEK293 cells. The empty and black columns represent relative luciferase activity driven by the empty vector (pGL3), wild-type WTH3 and four deleted promoter mutants (pGL/WTH3P-d1, -d2, -d3 and -d4), which were either influenced by the empty vector (pcDNA3.1) or the p53 transgene.
The p53 protein bound to the WTH3 promoter in the region from −330 to −282
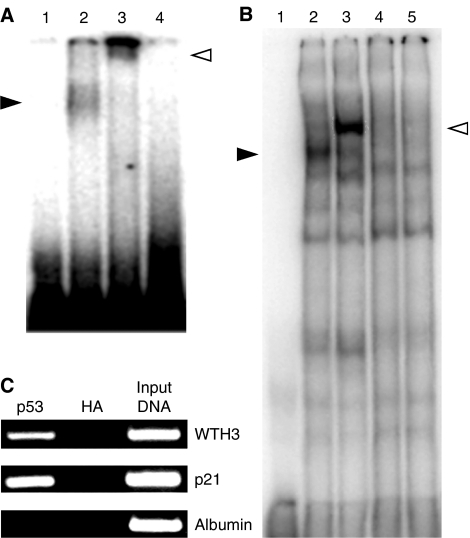
A 49 bp probe (from −330 to −282), P49, which contained the p53M sequence, and an albumin genomic DNA sequence (negative control) were utilised to perform EMSA assays. After isotope labelling, the probes were first incubated with purified p53 protein. The DNA/protein complex was realised on a nondenaturing-PAGE gel. We found that the P49 probe, but not the control probe (data not shown), interacted with the p53 protein. This interaction was specific as the p53 antibody shifted the P49/protein complex into a much higher position and excessive amounts of the cold P49 probe competed away p53 from the complex (Figure 5A). In addition, the same strategy was applied where P49 and nuclear extracts prepared from paired MCF7 cells were used. The results showed that the P49 probe interacted with the p53 proteins generated from MCF7/WT, but not the mutated ones in MCF7/AdrR cells (Figure 5B). However, no forms of the p53 proteins bound to the negative control probe (data not shown) (Figure 5B). Proteins of both sources did not bind to the negative control probe (data not shown). To confirm further that the p53 protein was indeed involved, supershift EMSA was performed utilising p53 antibody. We found that the p53 antibody only shifted the P49-MCF7/WT-protein complex into a much higher position (Figure 5B). To understand further if p53M was the direct target of the p53 protein, three probes were generated, which represented 28 bp of the right (P49R, containing p53M), 36 bp of the left (P49L) and 31 bp of the centre (P49C, from 10 to 40) of the P49 sequence, respectively. The EMSA results showed that the p53 protein did not bind to any of them (data not shown), which suggested that the whole P49 sequence was essential for interacting with p53. To verify further that WTH3 was a direct target of the p53 gene, ChIP assays were carried out.
Figure 5.
(A) Results obtained from EMSA and super-EMSA assays where P49 was used as the probe that was interacted with the purified p53 protein. Lane 1 contains the labelled probe. Lane 2 contains the labelled probe and 50 ng of p53 protein. The solid arrow indicates the DNA/p53 complexes. Lane 3 contains the labelled probe, 50 ng of p53 protein and the anti-p53 antibody. The empty arrow indicates the antibody shifted DNA/p53 complexes. Lane 4 contains the labelled probe, 50 ng of p53 protein and 100 times excess of cold P49. (B) Results obtained from EMSA and super-EMSA assays where the P49 probe and nuclear extracts obtained from MCF7/AdrR and MCF7/WT were used. Lane 1 contains the labelled probe. Lane 2 contains the labelled probe and 10 μg of nuclear extract prepared from MCF7/WT cells. The solid arrow indicates the DNA/p53 complexes. Lane 3 contains the labelled probe, 10 μg of MCF7/WT nuclear extract and the anti-p53 antibody. The empty arrow indicates the antibody shifted DNA/p53 complexes. Lane 4 contains the labelled probe, 10 μg of nuclear extract prepared from MCF7/AdrR cells. Lane 5 contains the labelled probe, 10 μg of MCF7/AdrR nuclear extract and the anti-p53 antibody. (C) Chromatin immunoprecipitation assay using MCF7/WT cells treated with 1 μM DOX. p53–DNA complexes were captured with anti-p53 antibody. Anti-HA antibody was used as the negative control. PCR products representing the WTH3, p21 (positive control) and albumin promoter (negative control) sequences are noted.
The p53 protein bound to the WTH3 promoter in vivo
Usually, under normal condition, p53 gene expression is relatively low in cells. However, when responding to DNA-damaging reagents, p53 transcripts are significantly increased. The elevated gene product either functions as a transcription factor for its targeted genes or directly performs its cellular roles, such as promoting apoptosis. Here, we tested whether p53 could directly target the WTH3 promoter in MCF7/WT cells. To this end, the cells were treated with DOX over different time courses to induce endogenous p53 gene expression. Western blot analysis determined that during 8–24 h treatment period, similar amounts of p53 expression, elevating to approximately 10 times the original level, were exhibited (data not shown). Next, the p53–DNA complexes were immunoprecipitated with anti-p53 or anti-HA antibodies. To see if the WTH3 promoter containing the p53-response site was in the enriched DNA fragments, PCR was performed. The upstream sequences of the p21 and albumin promoters, which were with or without a p53-response element, were also amplified and served as the positive and negative control, respectively. The results showed that both WTH3 and p21, but not albumin, promoter regions were enriched by the p53 antibodies. In addition, none of the promoter regions were enriched by the HA antibodies (Figure 5C). These findings suggested that WTH3 was a direct target of the p53 gene. As past research demonstrated that confirmed p53 target genes either are p53 functional mediators, such as p21, Bax and PUMA, or p53 functional regulators, such as MDM2, COP1 and PML (Liu and Chen, 2006), we then examined if WTH3 and p53 shared some biological functions. To date, several approaches were employed to test if WTH3 played a role in promoting apoptosis.
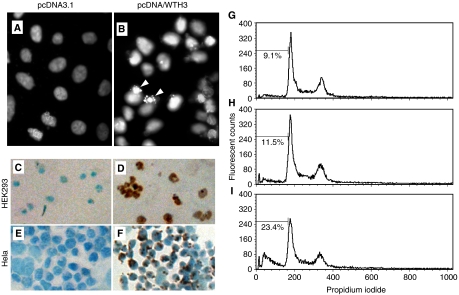
Overexpression of WTH3 induced apoptotic nuclear condensation
To test whether WTH3 could cause cell death, pcDNA/WTH3 was transiently introduced into Hela cells. After 24 h, the transfectants containing the exogenous WTH3 gene or empty vector were treated with DAPI, a fluorescent DNA-binding dye. The cells' nuclear morphology was examined under a fluorescent microscope. The typical morphological features of apoptotic cells were observed in the population of cells transfected with pcDNA/WTH3, but not in the control cells transfected with the empty vector (Figure 6A and B). These findings indicated that the WTH3 gene-induced apoptosis in Hela cells.
Figure 6.
(A and B) results obtained from DAPI staining assays using Hela cells. In (A) and (B) Hela cells contain pcDNA3.1 and pcDNA/WTH3, respectively. Cells were stained with DAPI, and the stained nuclei were visualised under a fluorescent microscope (magnification, × 400). Arrows indicate apoptotic nuclear condensation. (C–F) results obtained from TUNEL assays using Hela and HEK293 cells as hosts. (C and D) HEK293 cells were transfected with pcDNA3.1 and pcDNA/WTH3, respectively, for 30 h. (E and F) Hela cells were transfected with pcDNA3.1 and pcDNA/WTH3, respectively, for 18 h. DNA fragmentation in the apoptotic cells induced by the WTH3 gene were stained with a brown colour as compare to the control cells containing the empty vector, which were blue in colour. (G–I) results generated by flow cytometry assays using HEK293 cells. (G) Cells served as vehicle control. (H and I) cells were transfected with pcDNA/3.1 and pcDNA/WTH3, respectively. The cells were then stained with PI and the intercellular fluorescence was measured by a flow cytometer. The sub-G1 DNA content in each group, which represented apoptotic cells, is indicated by a percentage.
Overexpression of WTH3-induced cell death
To confirm further WTH3's apoptotic potential, TUNEL assays were carried out using both HEK293 and Hela cells as hosts. After receiving pcDNA/WTH3 or pcDNA3.1, TUNEL staining was performed. We found that both host cells transfected with the WTH3 transgene displayed brown colour staining that indicated a typical apoptotic condition, whereas the corresponding controls were stained with blue colour (Figure 6C–F). These findings were consistent with the results of the DAPI staining assays. Thus, WTH3-induced apoptosis in the two cell lines tested.
Overexpression of WTH3 increased sub-G1 cell population
HEK293 cells, which exhibited the highest transfection efficiency, were transfected with pcDNA/WTH3 or pcDNA3.1 to perform flow cytometry assays. By measuring the cells with sub-G1 DNA content, which is believed to represent apoptotic cells, we found that after 24 h, 23.4% of the cell population had under gone apoptosis after receiving the pcDNA/WTH3 construct. However, only 11.5 and 9.1% of the cell population underwent apoptosis after receiving the pcDNA3.1 vector and vehicle control, respectively (P<0.01) (Figure 6G–I). These results further suggested that the WTH3 gene stimulated apoptosis.
DISCUSSION
The WTH3 gene was discovered owing to its hypermethylation in MCF7/AdrR cells. Earlier studies suggested that it was a negative regulator for MDR development (Shan et al, 2002a; Tian et al, 2005a, 2005b), which made it extremely interesting as most MDR-related genes discovered so far exert a positive effect. To understand the mechanisms involved in its downregulation in MDR cells the gene's promoter was identified and analysed (Tian et al, 2005b). We found that it was differentially regulated in MCF7/AdrR and MCF7/WT cells. Several mechanisms could be involved in this differential regulation, which included drug-induced epigenetic modifications and alteration of trans-elements, we believe this is the situation as the WTH3 promoter was found to be hypermethylated in cultured and primary drug resistant cells, and a region targeted by DNA methylation and a repeat sequence in the promoter interacted with diverse transcription complexes prepared from MCF7/AdrR vs MCF7/WT (Tian et al, 2005a, 2005b).
To gain more detailed information regarding the WTH3 gene's differential regulation, we performed the Patch Search, which led to the discovery of a p53-binding motif, p53M. The p53 gene is an important tumour suppressor and is involved in apoptosis and cell cycle arrest, whereas mutations of p53 is associated with human cancers and the onset of MDR in a broad range of malignancies (Schmitt and Lowe, 1999; Norbury and Zhivotovsky, 2004; Pommier et al, 2004; Kim, 2005). Discovery of a putative p53-binding site in the WTH3 promoter led us to explore whether p53-regulated WTH3 expression. By performing luciferase assays we found that the p53 transgene significantly elevated promoter activity in all the host cell lines tested. In addition, evaluating WTH3's expression levels in 11 cancer cell lines with defined p53 status supported this positive influence as the cell lines with wild-type p53 produced much higher levels of WTH3 transcript than those containing mutated or attenuated p53. We also noticed that the observed correlation between these two genes in breast cancer cell lines was not related to their oestrogen receptor positive or negative condition. However, whether WTH3 gene activity coincides with the degree of malignancy remains to be determined. Further observations revealed that the p53 transgene was able to considerably increase endogenous WTH3 gene expression in both Hela and MCF7/AdrR cells, which supported the possibility that WTH3 could be a target of p53. This hypothesis turned out to be true as EMSA and super EMSA assays demonstrated that the P49 probe containing p53M in the WTH3 promoter was specifically targeted by endogenous and purified p53. However, p53 with mini deletion in MCF7/AdrR cells, which lost its DNA-binding capability, did not bind to the probe. In addition, ChIP assays confirmed that the p53 protein physically interacted with the WTH3 promoter region that contained the P49 sequence. In addition, it is worth to mention that EMSA results showed that the p53 protein was not able to bind to the probe that only included the GC rich region, the p53M site, or the sequence containing part of the GC and p53M regions, which suggested that both GC rich and p53M sequences were required for p53 gene targeting. We also noticed that the GC rich region included three CpG sites that were differentially methylated in MDR vs non-MDR cells (Tian et al, 2005a, 2005b). The DNA methylation is one of the epigenetic modifications that is symbolised by reversing traits of gene expression without DNA sequence change (Doerfler, 1983; Bird, 1986; Antequera and Bird, 1993a, 1993b; Kass et al, 1997; Siegfried and Cedar, 1997). At present, little is known about how the epigenetic network interacts with other transcriptional machineries to regulate gene expression in mammalian cells. In the past, p53-binding motifs with GC rich features were observed in several gene promoters including EGFR, Killer/DR4, RB and TGF-α (Qian et al, 2002). However, there are fundamental questions that need to be answered. For example, are those GC-rich regions epigenetically modified? If they are, do they exert a negative impact on p53-transactivity? As the p53-response element in the WTH3 gene promoter was involved in differential methylation, we have been provided with a unique working model system that can possibly answer those questions. Currently, we are designing experiments to explore if there is any interplay between DNA methylation and the p53 transcription factor in regulating WTH3 gene expression.
Prior research demonstrated that confirmed p53 target genes are either p53 functional mediators or regulators (Liu and Chen, 2006). Considering that WTH3 is a target of the p53 gene, we examined if they shared some biological functions. By employing several strategies, we found that WTH3 played a role in promoting apoptosis. It is possible that this proapoptotic potential is the driving force behind WTH3's participation in MDR development. In addition, as WTH3 is a G protein and most likely involved in cellular signalling transduction, we cannot rule out another prospect that it also acts as a p53 functional regulator. Testing this hypothesis is one of the subjects of our future research.
Acknowledgments
We thank for Dr UM Moll and Dr J Cao for the cell lines and DNA constructs. We thank JC Duffy for preparation of the paper. This work was supported in part by the National Cancer Institute (Grant# 1R01CA090443-01A2) and American Cancer Society (Grant# RSG-03-137-01-CDD) awards.
Footnotes
Financial Support: This work was supported in part by the National Cancer Institute (Grant #1R01CA090443-01A2) and American Cancer Society (Grant #RSG-03-137-01-CDD) awards.
References
- Aas T, Borresen AL, Geisler S, Smith-Sorensen B, Johnsen H, Varhaug JE, Akslen LA, Lonning PE (1996) Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 2: 811–814 [DOI] [PubMed] [Google Scholar]
- Adachi K, Toyota M, Sasaki Y, Yamashita T, Ishida S, Ohe-Toyota M, Maruyama R, Hinoda Y, Saito T, Imai K, Kudo R, Tokino T (2004) Identification of SCN3B as a novel p53-inducible proapoptotic gene. Oncogene 23: 7791–7798 [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A (1993a) CpG islands. Exs 64: 169–185 [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A (1993b) Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA 90: 11995–11999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M (1987) ras genes. Annu Rev Biochem 56: 779–827 [DOI] [PubMed] [Google Scholar]
- Bird AP (1986) CpG-rich islands and the function of DNA methylation. Nature 321: 209–213 [DOI] [PubMed] [Google Scholar]
- Camps JL, Chang SM, Hsu TC, Freeman MR, Hong SJ, Zhau HE, von Eschenbach AC, Chung LW (1990) Fibroblast-mediated acceleration of human epithelial tumor growth in vivo. Proc Natl Acad Sci USA 87: 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB (1986) Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 47: 381–389 [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG (1992) Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258: 1650–1654 [DOI] [PubMed] [Google Scholar]
- Doerfler W (1983) DNA methylation and gene activity. Annu Rev Biochem 52: 93–124 [DOI] [PubMed] [Google Scholar]
- Echard A, Jollivet F, Martinez O, Lacapere JJ, Rousselet A, Janoueix-Lerosey I, Goud B (1998) Interaction of a Golgi-associated kinesin-like protein with Rab6. Science 279: 580–585 [DOI] [PubMed] [Google Scholar]
- Echard A, Opdam FJ, de Leeuw HJ, Jollivet F, Savelkoul P, Hendriks W, Voorberg J, Goud B, Fransen JA (2000) Alternative splicing of the human Rab6A gene generates two close but functionally different isoforms. Mol Biol Cell 11: 3819–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstner E, Linker-Israeli M, Said J, Umiel T, de Vos S, Shintaku IP, Heber D, Binderup L, Uskokovic M, Koeffler HP (1995) 20-epi-vitamin D3 analogues: a novel class of potent inhibitors of proliferation and inducers of differentiation of human breast cancer cell lines. Cancer Res 55: 2822–2830 [PubMed] [Google Scholar]
- Goud B, Zahraoui A, Tavitian A, Saraste J (1990) Small GTP-binding protein associated with Golgi cisternae. Nature 345: 553–556 [DOI] [PubMed] [Google Scholar]
- Gros P, Ben Neriah YB, Croop JM, Housman DE (1986a) Isolation and expression of a complementary DNA that confers multidrug resistance. Nature 323: 728–731 [DOI] [PubMed] [Google Scholar]
- Gros P, Croop J, Housman D (1986b) Mammalian multidrug resistance gene: complete cDNA sequence indicates strong homology to bacterial transport proteins. Cell 47: 371–380 [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Cretney E, Smyth MJ (1999) P-glycoprotein protects leukemia cells against caspase-dependent, but not caspase-independent, cell death. Blood 93: 1075–1085 [PubMed] [Google Scholar]
- Kass SU, Pruss D, Wolffe AP (1997) How does DNA methylation repress transcription? Trends Genet 13: 444–449 [DOI] [PubMed] [Google Scholar]
- Kim EJ, Park JS, Um SJ (2002) Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo. J Biol Chem 277: 32020–32028 [DOI] [PubMed] [Google Scholar]
- Kim R (2005) Recent advances in understanding the cell death pathways activated by anticancer therapy. Cancer 103: 1551–1560 [DOI] [PubMed] [Google Scholar]
- Li C, Lin M, Liu J (2004) Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis. Oncogene 23: 9336–9347 [DOI] [PubMed] [Google Scholar]
- Liu G, Chen X (2006) DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol 26: 1398–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW (1995) Cancer therapy and p53. Curr Opin Oncol 7: 547–553 [DOI] [PubMed] [Google Scholar]
- Nieves-Neira W, Pommier Y (1999) Apoptotic response to camptothecin and 7-hydroxystaurosporine (UCN-01) in the 8 human breast cancer cell lines of the NCI Anticancer Drug Screen: multifactorial relationships with topoisomerase I, protein kinase C, Bcl-2, p53, MDM-2 and caspase pathways. Int J Cancer 82: 396–404 [DOI] [PubMed] [Google Scholar]
- Norbury CJ, Zhivotovsky B (2004) DNA damage-induced apoptosis. Oncogene 23: 2797–2808 [DOI] [PubMed] [Google Scholar]
- Ogretmen B, Safa AR (1997) Expression of the mutated p53 tumor suppressor protein and its molecular and biochemical characterization in multidrug resistant MCF-7/Adr human breast cancer cells. Oncogene 14: 499–506 [DOI] [PubMed] [Google Scholar]
- Pommier Y, Sordet O, Antony S, Hayward RL, Kohn KW (2004) Apoptosis defects and chemotherapy resistance: molecular interaction maps and networks. Oncogene 23: 2934–2949 [DOI] [PubMed] [Google Scholar]
- Qian H, Wang T, Naumovski L, Lopez CD, Brachmann RK (2002) Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene 21: 7901–7911 [DOI] [PubMed] [Google Scholar]
- Ramljak D, Romanczyk LJ, Metheny-Barlow LJ, Thompson N, Knezevic V, Galperin M, Ramesh A, Dickson RB (2005) Pentameric procyanidin from Theobroma cacao selectively inhibits growth of human breast cancer cells. Mol Cancer Ther 4: 537–546 [DOI] [PubMed] [Google Scholar]
- Righetti SC, Della Torre G, Pilotti S, Menard S, Ottone F, Colnaghi MI, Pierotti MA, Lavarino C, Cornarotti M, Oriana S, Bohm S, Bresciani GL, Spatti G, Zunino F (1996) A comparative study of p53 gene mutations, protein accumulation, and response to cisplatin-based chemotherapy in advanced ovarian carcinoma. Cancer Res 56: 689–693 [PubMed] [Google Scholar]
- Robinson LJ, Roberts WK, Ling TT, Lamming D, Sternberg SS, Roepe PD (1997) Human MDR 1 protein overexpression delays the apoptotic cascade in Chinese hamster ovary fibroblasts. Biochemistry 36: 11169–11178 [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Lowe SW (1999) Apoptosis and therapy. J Pathol 187: 127–137 [DOI] [PubMed] [Google Scholar]
- Shan J, Mason JM, Yuan L, Barcia M, Porti D, Calabro A, Budman D, Vinciguerra V, Xu H (2000) Rab6c, a new member of the rab gene family, is involved in drug resistance in MCF7/AdrR cells. Gene 257: 67–75 [DOI] [PubMed] [Google Scholar]
- Shan J, Yuan L, Budman DR, Xu HP (2002a) WTH3, a new member of the Rab6 gene family, and multidrug resistance. Biochim Biophys Acta 1589: 112–123 [DOI] [PubMed] [Google Scholar]
- Shan J, Yuan L, Xiao Q, Chiorazzi N, Budman D, Teichberg S, Xu HP (2002b) TSP50, a possible protease in human testes, is activated in breast cancer epithelial cells. Cancer Res 62: 290–294 [PubMed] [Google Scholar]
- Siegfried Z, Cedar H (1997) DNA methylation: a molecular lock. Curr Biol 7: R305–R307 [DOI] [PubMed] [Google Scholar]
- Smith ND, Rubenstein JN, Eggener SE, Kozlowski JM (2003) The p53 tumor suppressor gene and nuclear protein: basic science review and relevance in the management of bladder cancer. J Urol 169: 1219–1228 [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Krasovskis E, Sutton VR, Johnstone RW (1998) The drug efflux protein, P-glycoprotein, additionally protects drug-resistant tumor cells from multiple forms of caspase-dependent apoptosis. Proc Natl Acad Sci USA 95: 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele RJ, Lane DP (2005) P53 in cancer: a paradigm for modern management of cancer. Surgeon 3: 197–205 [DOI] [PubMed] [Google Scholar]
- Tian K, Jurukovski V, Wang XP, Kaplan MH, Xu H (2005a) Epigenetic regulation of WTH3 in primary and cultured drug-resistant breast cancer cells. Cancer Res 65: 10024–10031 [DOI] [PubMed] [Google Scholar]
- Tian K, Jurukovski V, Yuan L, Shan J, Xu H (2005b) WTH3, which encodes a small G protein, is differentially regulated in multidrug-resistant and sensitive MCF7 cells. Cancer Res 65: 7421–7428 [DOI] [PubMed] [Google Scholar]
- Xu H, Shan J, Jurukovski V, Yuan L, Li J, Tian K (2007) TSP50 encodes a testis-specific protease and is negatively regulated by p53. Cancer Res 67: 1239–1245 [DOI] [PubMed] [Google Scholar]
- Yuan L, Shan J, De Risi D, Broome J, Lovecchio J, Gal D, Vinciguerra V, Xu HP (1999) Isolation of a novel gene, TSP50, by a hypomethylated DNA fragment in human breast cancer. Cancer Res 59: 3215–3221 [PubMed] [Google Scholar]
- Zahraoui A, Touchot N, Chardin P, Tavitian A (1989) The human Rab genes encode a family of GTP-binding proteins related to yeast YPT1 and SEC4 products involved in secretion. J Biol Chem 264: 12394–12401 [PubMed] [Google Scholar]