Abstract
The cancer risk is unknown for those families in which a microsatellite instable tumour is neither explained by MLH1 promoter methylation nor by a germline mutation in a mismatch repair (MMR) gene. Such information is essential for genetic counselling. Families suspected of Lynch syndrome (n=614) were analysed for microsatellite instability, MLH1 promoter methylation and/or germline mutations in MLH1, MSH2, MSH6, and PMS2. Characteristics of the 76 families with a germline mutation (24 MLH1, 2 PMS2, 32 MSH2, and 18 MSH6) were compared with those of 18 families with an unexplained microsatellite instable tumour. The mean age at diagnosis of the index patients in both groups was comparable at 44 years. Immunohistochemistry confirmed the loss of an MMR protein. Together this suggests germline inactivation of a known gene. The Amsterdam II criteria were fulfilled in 50/75 families (66%) that carried a germline mutation in an MMR gene and in only 2/18 families (11%) with an unexplained microsatellite instable tumour (P<0.0001). Current diagnostic strategies can detect almost all highly penetrant MMR gene mutations. Patients with an as yet unexplained microsatellite instable tumour likely carry a different type of mutation that confers a lower risk of cancer for relatives.
Keywords: colorectal neoplasms, hereditary nonpolyposis, microsatellite instability, DNA mismatch repair, DNA methylation
Lynch syndrome (Hereditary NonPolyposis Colorectal Cancer (HNPCC)) accounts for about 5% of colorectal cancers and is caused by a germline mutation in one of the mismatch repair (MMR) genes (Aaltonen et al, 1998; Cunningham et al, 2001; Lynch and de la Chapelle, 2003; Hampel et al, 2005; Barnetson et al, 2006). Known MMR genes causing Lynch syndrome are MLH1, PMS2, MSH2, and MSH6. Typical Lynch syndrome families show autosomal dominant predisposition to a number of cancers of which colorectal cancer is the most important. Conversely, over 90% of colorectal cancers of Lynch syndrome patients have a defect in the MMR system (Lynch and de la Chapelle, 2003). Failure of the DNA MMR system causes microsatellite instability (MSI) in tumours.
MSI reflects either the presence of a germline mutation in the MMR system or somatic hypermethylation of the promoter region of the MLH1 gene (Cunningham et al, 2001). Patients with a tumour with MSI and somatic hypermethylation of the MLH1 promoter rarely carry a germline mutation in the MMR system, although rare exceptions have been reported. A few families have been described in which Lynch syndrome patients display hypermethylation of the MLH1 promoter in tumour as well as in non-tumour tissue (Gazzoli et al, 2002; Miyakura et al, 2004; Suter et al, 2004; Hitchins et al, 2005; Valle et al, 2007a). In addition, a family was recently described in which the susceptibility to tumours is caused by germline methylation of the MSH2 promoter (Chan et al, 2006). Taken together, hypermethylation of the MLH1 promoter indicates a very low likelihood that Lynch syndrome is the cause of the MSI (Samowitz et al, 2005; Weisenberger et al, 2006). Such patients can be offered less stringent surveillance programs (Lindor et al, 2005; Dove-Edwin et al, 2006; Valle et al, 2007b).
Little is known about the cancer risk in those families in which MSI is detected in a tumour, but where the MSI can be explained neither by hypermethylation of the MLH1 promoter nor by a germline mutation in an MMR gene. Such information is essential for genetic counselling.
In the present study, we examined patients with a tumour that indicated possible Lynch syndrome for germline mutations in the MMR genes MLH1, MSH2, MSH6, and PMS2. In addition, we tested tumour DNA for hypermethylation of the MLH1 promoter. Most of these patients were preselected by MSI analysis. Family characteristics of the patients with an MMR germline mutation were compared with those of patients with an unexplained microsatellite instable tumour.
MATERIALS AND METHODS
Patients
We examined 614 families, who visited the Department of Human Genetics of the Radboud University Nijmegen Medical Centre between 1997 and November 2005 because of possible familial colorectal cancer. Families were included because they either fulfilled the Amsterdam II criteria (Vasen et al, 1999) (n=126), or fulfilled the Bethesda guidelines (Rodriguez-Bigas et al, 1997) (n=333), or had a history very close to the Bethesda guidelines (n=155). Such patients are suspected of Lynch syndrome, which is defined as cancer owing to a germline mutation in one of the MMR genes (Jass, 2006).
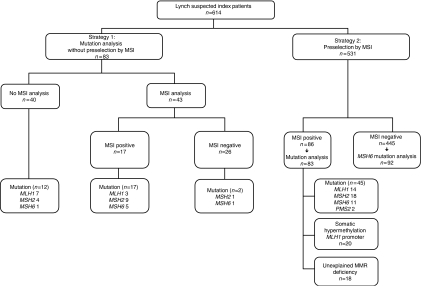
Two diagnostic molecular strategies were used. Mutation analysis of germline DNA was performed as the first test in 83 families, who fulfilled clinical criteria for Lynch syndrome, or in which no tumour material was available. MSI analysis was also performed in 43 of these families. In the other 531 families MSI analysis was used as the initial step to select patients for germline mutation analysis.
The index tumour of a family was defined as the MSI-positive tumour that was diagnosed at the youngest age or, in case MSI analysis was not performed, the first tumour of the patient in whom the germline mutation was detected. The study was performed in accordance with the rules of the Radboud University Nijmegen Medical Centre Medical Ethical Committee.
Germline mutation analysis of the MMR system
Mutation analysis of MLH1, PMS2, MSH2, and MSH6 was performed in DNA from peripheral blood lymphocytes by a combination of either single-strand conformation polymorphism analysis or denaturing gradient gel electrophoresis and direct sequence analysis essentially as described elsewhere (Wu et al, 1997; Hoogerbrugge et al, 2003). Only mutations resulting in a premature termination codon, the recurrent amino-acid deletion in MLH1 c.1852_1854del (p.Lys618del) and the amino-acid deletion c.211_213del (p.Glu71del) in MLH1, which has been shown to abort the function of MLH1 (Raevaara et al, 2002), were considered pathogenic. For the detection of large deletions and duplications in MLH1, MSH2, MSH6, and PMS2, the P003 and P008 Multiplex Ligation-dependent Probe Amplification (MLPA) kits of MRC Holland (Amsterdam, the Netherlands) were used. All deletions and duplications were confirmed by Southern blot analysis essentially as described elsewhere (Wijnen et al, 1998) or with a specific PCR using primers flanking the deletion or one of the breakpoints of a duplicated region.
MSI analysis
In total, 667 tumours in 574 families were tested for MSI: 566 colorectal carcinomas, 34 colorectal adenomas, 47 endometrium carcinomas, eight duodenum/small bowel/appendix carcinomas, one sebaceous carcinoma and one other skin tumour (trichoepithelioma or trichoblastoma), one ovarian carcinoma, and nine urothelial cell carcinomas.
MSI analysis was performed using the Bethesda panel of microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26) (Boland et al, 1998). Tumours were scored as MSI positive if at least two of the five Bethesda markers showed instability; they were scored MSI negative if none of the Bethesda markers showed instability. In case of one instable marker, additional markers were included, and immunohistochemistry (IHC) of MMR proteins was performed (Hoogerbrugge et al, 2003). In 273 tumours, the mononucleotide marker BAT40 was added to the standard set of markers and in 558 of 667 tumours IHC of MSH6 was performed irrespective of the MSI status to minimise the chance of missing a tumour that was due to an MSH6 germline mutation.
Immunohistochemistry
IHC was performed on formalin-fixed, paraffin-embedded tissues. Slides were stained with antibodies against MLH1 (Pharmingen code: 51-1327gr), PMS2 (Pharmingen code: 556415), MSH2 (Oncogene Research Products code: NA26), and MSH6 (Transduction Laboratories code: G70220). Staining patterns of MMR proteins were evaluated using normal epithelial, stromal, and inflammatory cells as internal controls. Stained slides were scored as (1) positive, that is showing nuclear staining in at least some tumour cells; (2) negative, that is no staining of the tumour with a positive internal control; or (3) not assessable, that is when the technical quality was insufficient to provide an unambiguous result despite repeated assays (de Jong et al, 2004).
Analysis of hypermethylation of the MLH1 promoter
The DNA methylation status of the MLH1 promoter region was determined after bisulphite treatment of the DNA using the EZ DNA methylation KIT™, ZYMO Research. To avoid conversion of methylated cytosines to uracil, the modification time was optimised. Modification was performed in duplicate for 3 and 6 h, respectively. Methylation of the region of 337–154 bp upstream of the translational start site, which has been shown to correlate with MLH1 expression, was analysed (Deng et al, 2002). FAM-labelled PCR products were generated using primers (5′-TATTTTTGTTTTTATTGGTTGGATA-3′ and 5′-AATACCAATCAAATTTCTCAACTCT-3′) flanking 11 CpG sites and analysed on an ABI PRISM 3730 Genetic Analyzer under denaturing conditions using genemapper software. The products of unmethylated and methylated DNA migrate at 186 and 183 bp, respectively. This was verified by digestion by BstUI (New England Biolabs, Beverly, MA, USA), which cleaves only the CGCG sequence, which is not converted by bisulphite treatment when methylated. To assess the amount of methylation, the peak height of the product at 183 bp was divided by the sum of the peak heights at 183 and 186 bp. The resulting percentage of methylation was corrected for the percentage of tumour cells. Most tumours with methylation of the MLH1 promoter showed percentage of methylation above 80%, whereas in none of the adjacent normal tissues methylation was detected.
Patient characteristics and pedigree analysis
The following information was obtained for all families as part of the genetic counselling procedure: age at diagnosis, type of cancer, number of family members who had cancer, their age at diagnosis, their type of cancer, and their relation with the patient. Pathological and surgical reports were evaluated whenever possible.
Pedigrees were scored as fulfilling the Amsterdam I criteria (Vasen et al, 1991), the Amsterdam II criteria (Vasen et al, 1999), or the criterion described by Rodriguez-Bigas et al (1997) that is: two first degree relatives with a cancer associated with Lynch syndrome, one of them with an age at diagnosis below 50 years. As this study was mainly directed at MSI in tumours, a positive score for fulfilment of one of these three criteria was only given if the index patient was part of the criterion. The occurrence of metachronous or synchronous cancers associated with Lynch syndrome was noted.
Data analysis
Descriptive statistics were used to describe the results of the molecular laboratory tests, germline mutation analysis, and pedigree analysis. Categorical variables were checked for statistically significant differences using either the χ2 test or logistic regression. Continuous variables were checked for statistically significant differences using either the Student's t-test or analysis of variance. The Tukey–Kramer test was used to calculate differences in continuous variables between two groups with adjustment for multiple testing. P-values <0.05 were considered statistically significant. Analyses were performed with the SAS system for Windows V8.2.
RESULTS
Identification and characterisation of families with a germline MMR gene mutation
Our strategy 1 involved germline mutation analysis without prior testing for MSI in those families that fulfilled clinical criteria for Lynch syndrome, and those for whom no tumour DNA was available for MSI analysis. We found a germline mutation in 31 out of these 83 families (Figure 1, Table 1). In total, 43 index patients were tested for MSI within this group. MSI was detected in tumours of 17/43 patients. In all 17 MSI-positive index patients, a germline mutation in MLH1, MSH2, or MSH6 was identified. One MSH2 mutation was detected in a patient with an endometrial tumour diagnosed at age 39 that was tested MSI negative without loss of MLH1, PMS2, MSH2, or MSH6 protein staining. No other tumours from this family were available. One MSH6 mutation was detected in a patient in whom a rectum tumour at an age of 51 years was MSI negative without loss of MLH1, PMS2, MSH2, or MSH6 protein staining. Material from a sigmoid carcinoma that occurred in another family member at the age of 33 years was not available.
Figure 1.
Analytic strategy of the study and number of patients in each analysis and for each result.
Table 1. Molecular laboratory tests results and patient characteristics of 76 patients with a pathogenic germline mutation in MLH1, PMS2, MSH2, or MSH6.
| No. | S | Germline mutation | MSI | Meth | IHC | Tumour tested for MSI | Age | Other tumour(s) | AC | 2fam |
|---|---|---|---|---|---|---|---|---|---|---|
| MLH1 | ||||||||||
| 8 | 1 | c.15_28del (p.Gly6fs) | NT | NT | NT | 42 | co42,co47 | + | + | |
| 288 | 2 | c.18_34del (p.Val7fs) | + | − | MLH1/PMS2− | Caecum | 40 | − | − | − |
| 189 | 2 | c.211_213del (p.Glu71del) | + | − | PMS2/MSH6− | Endometrium | 39 | − | − | − |
| 118 | 1 | c.299C>T (p.Arg100X) | NT | NT | NT | 26 | co26,co37 | + | + | |
| 645 | 2 | c.578C>G (p.Ser193X) | + | − | MLH1/PMS2− | Caecum | 26 | co37,co39 | − | + |
| 163 | 1 | c.588+3_588+6del (affects ss) | NT | NT | NT | 50 | co50,co50,co50 | + | + | |
| 198 | 2 | c.677G>A (p.Arg226Gln) (affects ss) | + | − | MLH1/PMS2− | Colon descendens | 41 | − | + | + |
| 318 | 2 | c.806C>G (p.Ser269X) | + | − | MLH1/PMS2− | Colon transversum | 59 | en49,co27 | − | + |
| 19 | 1 | c.806C>G (p.Ser269X) | NT | NT | NT | 45 | co45 | + | + | |
| 345 | 2 | c.1225C>T (p.Gln409X) | + | − | MLH1/PMS2− | Sigmoid | 44 | − | + | + |
| 454 | 2 | c.1354del (p.Thr452fs) | + | − | MLH1/PMS2− | Colon transversum | 49 | − | + | + |
| 335 | 2 | c.1549G>T (p.Gly517X) | + | − | MLH1/PMS2− | Sigmoid | 49 | − | + | + |
| 734 | 2 | c.1852_1854del (p.Lys618del) | + | NT | MLH1/PMS2− | Urothelial cell carcinoma | 60 | co40,en54 | − | + |
| 428 | 2 | c.1852_1854del (p.Lys618del) | + | NT | MLH1/PMS2− | Caecum | 50 | + | + | |
| 15 | 1 | c.1852_1854del (p.Lys618del) | + | NT | MLH1/PMS2− | Endometrium | 52 | ov52,co52 | − | + |
| 451 | 2 | c.1852_1854del (p.Lys618del)a | + | − | MLH1na/PMS2− | Colon ascendens | 27 | − | − | − |
| 168 | 2 | c.1852_1854del (p.Lys618del) | + | NT | NT | Caecum | 42 | − | + | + |
| 42 | 1 | c.1852_1854del (p.Lys618del) | NT | NT | NT | 36 | co36 | + | + | |
| 3 | 1 | c.1852_1854del (p.Lys618del) | + | NT | NT | Colon NOS | 32 | − | + | + |
| 324 | 2 | c.2103+1G>A | + | − | MLH1/PMS2− | Flexura linealis | 50 | co50 | + | + |
| 392 | 2 | c.2103+1G>A | + | − | MLH1/PMS2− | Colon ascendens | 38 | − | + | + |
| 65 | 1 | c.2103+1G>A | + | − | MLH1/PMS2− | Rectum | 42 | − | + | + |
| 222 | 1 | c.2103+1G>A | NT | NT | NT | 38 | co38 | + | + | |
| 260 | 1 | c.2103+1G>A | NT | NT | NT | 55 | co55,en61 | + | + | |
| PMS2 | ||||||||||
| 386 | 2 | entire gene deletion | + | − | PMS2− | Colon ascendens | 36 | − | − | − |
| 641 | 2 | c.989–296_1144+706del | + | NT | PMS2− | Trichoepithelioma/trichoblastoma | 45 | − | − | − |
| MSH2 | ||||||||||
| 322 | 2 | c.1-?_211+?del | + | NT | MSH2/MSH6− | Colon ascendens | 58 | ur49 | + | + |
| 92 | 1 | c.1-?_211+?del | + | NT | MSH2/MSH6− | Endometrium | 41 | − | + | + |
| 5 | 1 | c.1-?_211+?del | + | NT | NT | 38 | co42 | + | + | |
| 614 | 2 | c.1-?_366+?del | + | NT | MSH2/MSH6− | Flexura lienalis | 21 | − | − | − |
| 730 | 2 | c.1-?_1076+?del | + | NT | MSH2/MSH6− | Urothelial cell carcinoma | 57 | co42 | + | + |
| 528 | 2 | c.1-?_1076+?delb | + | NT | none− | Caecum | 32 | en44 | − | + |
| 139 | 1 | c.1-?_1076+?del | + | NT | MSH2/MSH6− | Endometrium | 41 | co42 | + | + |
| 287 | 2 | c.1-?_1276+?del | + | − | MSH2/MSH6− | Caecum | 33 | − | + | + |
| 700 | 2 | c.212–1G>A | + | NT | MSH2-/MSH6na | Caecum | 52 | co61 | − | + |
| 350 | 2 | c.212-?_366+?del | + | − | MSH6− | Caecum | 72 | − | + | + |
| 117 | 1 | c.212-?_366+?del | + | − | MSH2/MSH6− | Ileocecum | 48 | − | + | + |
| 73 | 1 | c.212-?_366+?del | + | NT | NT | Colon ascendens | 44 | en37,ov37 | + | + |
| 237 | 2 | c.255dup (p.Glu86X) | + | NT | MSH2/MSH6− | Sebaceuous gland carcinoma | 39 | co25 | + | + |
| 190 | 2 | c.367-?_645+?del | + | − | MSH2/MSH6− | Caecum | 50 | co50,co50 | − | − |
| 210 | 2 | c.367-?_645+?del | + | − | MSH2/MSH6− | Colon transversum | 43 | − | + | + |
| 366 | 2 | c.642_645del (p.Gln215X) | + | NT | MSH2/MSH6− | Flexura linealis | 46 | − | + | + |
| 18 | 1 | c.793-?_1076+?del | + | − | MSH2/MSH6− | Rectum | 29 | co29 | + | + |
| 25 | 1 | c.836del (p.Leu279fs)c | + | − | MSH2/MSH6− | Flexura linealis | 46 | − | + | + |
| 37 | 1 | c.862C>T (p.Gln288X) | NT | NT | NT | 26 | co26 | + | + | |
| 196 | 2 | c.915_922dup (p.Arg308fs) | + | NT | MSH2/MSH6− | Sigmoid | 46 | co46 | − | − |
| 11 | 1 | c.943-?_1076+?del | + | − | MSH2/MSH6− | Endometrium | 45 | − | + | + |
| 14 | 1 | c.1147C>T (p.Arg383X) | NT | NT | NT | 33 | en33,co50 | + | + | |
| 497 | 2 | c.1165C>T (p.Arg389X) | + | NT | MSH2−/MSH6na | Flexura lienalis | 44 | − | − | − |
| 462 | 2 | c.1203dup (p.Gln402fs) | + | − | MSH2/MSH6− | Sigmoid | 37 | − | + | + |
| 74 | 1 | c.1203dup (p.Gln402fs) | + | − | MSH2−/MSH6na | Rectum | 32 | − | + | + |
| 654 | 1 | c.1255C>T (p.Gln419X) | NT | NT | NT | 45 | co45,co63 | + | + | |
| 301 | 2 | c.1277–2A>G | + | NT | MSH2/MSH6− | Caecum | 33 | − | − | + |
| 637 | 2 | c.1386+1G>T | + | − | MSH2/MSH6− | Colon transversum | 47 | co42 | − | + |
| 625 | 2 | c.1387-?_1510+?del | + | − | MSH2/MSH6− | Endometrium | 45 | co53 | + | + |
| 107 | 1 | c.1494dup (p.Ala499fs) | NT | NT | NT | 46 | co46 | + | + | |
| 327 | 2 | c.1861C>T (p.Arg621X) | + | NT | MSH2/MSH6− | Sigmoid | 58 | − | − | − |
| 480 | 1 | c.2005+1G>C | − | NT | none− | Endometrium | 39 | co45 | + | + |
| MSH6 | ||||||||||
| 206 | 1 | c.1-?_457+?deld | + | NT | MSH6− | Urothelial cell carcinoma | 56 | ur57 | − | + |
| 657 | 2 | c.261-?_457+?dupd | + | NT | MSH6− | Colon descendens | 42 | − | + | + |
| 745 | 2 | c.467C>G (p.Ser156X) | + | − | MSH6− | Ileocecum | 63 | − | + | + |
| 138 | 2 | c.651dup (p.Lys218X)d | + | − | MSH6− | Sigmoid | 52 | en37 | + | + |
| 342 | 1 | c.814G>T (p.Glu272X)d,f | + | − | MSH6− | Endometrium | 57 | − | + | + |
| 338 | 2 | c.1135_1139del (p.Arg379X)d | + | − | MSH6− | Endometrium | 38 | ov38 | − | − |
| 137 | 1 | c.1784del (p.Leu595fs) | − | NT | none− | Rectum | 51 | + | + | |
| 515 | 2 | c.2815C>T (p.Gln939X)a,d | + | NT | MSH6− | Iieum | 65 | co38,co51,co58,ur69 | − | + |
| 105 | 1 | c.3261del (p.Phe1088fs)d | + | − | MSH6− | Colon ascendens | 39 | − | + | + |
| 766 | 2 | c.3261del (p.Phe1088fs) | + | − | MSH6− | Endometrium | 41 | − | − | − |
| 446 | 2 | c.3261dup (p.Phe1088fs)d | + | − | MSH2/MSH6− | Endometrium | 43 | − | + | + |
| 450 | 2 | c.3273dup (p.Lys1092X)d | + | − | MSH6− | Colon ascendens | 50 | co46,co50 | + | + |
| 711 | 2 | c.3438+1G>A | + | NT | MSH6na | Rectum | 45 | − | + | + |
| 692 | 2 | c.3438+1G>A | + | NT | MSH6na | Colon transversum | 43 | en53,ov43 | − | − |
| 500 | 2 | c.3514dup (p.Arg1172fs) | + | + | MSH6− | Colon transversum | 70 | ur70 | − | − |
| 434 | 1 | c.3678_3706dup (p.Ala1236fs)d | NT | NT | NT | 38 | en38 | − | + | |
| 128 | 1 | c.3838C>T (p.Gln1280X)d | + | − | MSH6− | Endometrium | 36 | − | + | + |
| 886 | 1 | c.4001G>A (p.Arg1334Gln) affects ss | + | NT | MSH6nae | Colon NOS | 44 | co61: MSI pos,IHC MSH6 NA | − | + |
S, strategy of molecular testing; MSI, microsatellite instability; Meth, methylation analysis of MLH1 promoter; IHC, immunohistochemical analysis of MLH1, PMS2, MSH2, and MSH6; Tumour tested for MSI, tumour origin or exact location of tumour in case of colon cancer of tumour tested for MSI; Age, age at diagnosis of tumour tested for MSI or age at diagnosis of (first) tumour in case MSI analysis was not performed; Other tumour(s), metachronous or synchronous cancer associated with Lynch syndrome of index patient who had MSI analysis or tumour(s) of index patient who did not have MSI analysis and age at diagnosis; AC, Amsterdam II criteria (Vasen et al, 1999); 2fam, 2 first degree relatives (including index) with cancer associated with Lynch syndrome, one of them with an age at diagnose below 50 years (Rodriguez-Bigas et al, 1997); 1, strategy 1 (germline mutation analysis without preselection by MSI analysis); 2, strategy 2 (first MSI analysis); ss, splice site; +, positive; −, negative; NT, not tested; NA, not assessable; NOS, not otherwise specified; co, colon; en, endometrium; ur, urothelial; ov, ovarian.
Carrier status of patient deduced from mutation status of relatives.
Also carrier of variant c.1A>G (p.Met1?) in MSH2 (paper in preparation).
Also carrier of variant c.965G>A(p.Gly322Asp) in MSH2.
Mutations published elsewhere (Kets et al, 2006).
Adenocarcinoma Caecum of sister with same mutation MSI and IHC MSH6− (1co46 MSI IHC MSH6−).
Also carrier of variant c.65G>C (p.Gly22Ala) in MLH1.
Strategy 2 involved 531 families clinically suspected of Lynch syndrome. Here, a positive MSI test result was used to select families for germline mutation analysis. In 86/531 families, at least one MSI-positive tumour was detected. Although their tumours were MSI positive, three patients declined testing for germline mutations and for MLH1 promoter methylation in their tumour. In the remaining 83 families, 45 MMR gene germline mutations were detected (Table 1).
Thus, a pathogenic germline mutation was found in 76 out of 614 families (12%) with a clinical history suggestive of Lynch syndrome. There were 24 mutations in MLH1, two in PMS2, 32 in MSH2, and 18 in MSH6.
Most MSI-positive tumours were also tested by IHC. Tumour cells of MLH1 mutation carriers generally lacked MLH1 and PMS2 protein by IHC staining. Those of MSH2 mutation carriers lacked MSH2 and MSH6. Tumours of MSH6 mutation carriers lacked MSH6, and those of PMS2 mutation carriers lacked PMS2. The IHC pattern correctly pinpointed the mutated gene in 50 of the 53 tumours (94%) where IHC was sufficiently informative.
We tested whether MLH1 promoter methylation occurs in the presence of a germline mutation in one of the MMR genes. This was found to be a rare event. We tested a total of 42 microsatellite instable tumours from families with a germline mutation in MLH1 (13), PMS2 (1), MSH2 (14), or MSH6 (14). In only one of these tumours (a tumour with an MSH6 germline mutation and absence of MSH6 protein staining, but presence of MLH1 protein staining), we detected incomplete MLH1 promoter methylation (about 60%). Table 1 presents the results of analysis of MLH1 promoter hypermethylation of index patients.
The majority (66%) of the proven Lynch syndrome families (50/76) fulfilled the Amsterdam II criteria. This was true for both strategy 1 (87%) and for strategy 2 (51%). The mean age at diagnosis of the index patients was 44 years (42 and 46 years in strategy 1 and 2, respectively).
Identification and characterisation of families with hypermethylation of the MLH1 promoter in their tumour
Pathogenic germline mutations in the MMR genes could not be detected in 38 families with at least one MSI-positive tumour (Figure 1). We therefore examined the methylation status of the MLH1 promoter in 42 MSI-positive tumours of these 38 families. Methylation of the MLH1 promoter was detected in 22 tumours (20 families) (Table 2). In the corresponding normal tissues MLH1 promoter methylation was never detected, suggesting that the promoter methylation was not present in the germline.
Table 2. Molecular laboratory tests results and family history of 20 patients with an MSI-positive tumour with somatic MLH1 promoter methylation.
| No. | Meth | IHC | Tumour tested for MSI | Age | Other tumour(s) | AC | 2fam | Tumours of close relatives |
|---|---|---|---|---|---|---|---|---|
| 312 | + | MLH1/PMS2− | Caecum | 63 | − | + | + | 1co53 MSS |
| 408 | + | MLH1/PMS2− | Colon ascendens | 62 | co40 | + | + | |
| 642a | + | MLH1/PMS2− | Colon transversum | 62 | co62: MSI, IHC MLH1−, meth+ | − | − | 1co75 MSI,IHC: MLH1−,meth+ |
| 477 | + | MLH1/PMS2− | Colon transversum | 75 | co75 | − | − | 2co50 MSS |
| 226 | + | MLH1/PMS2− | Colon ascendens | 67 | − | + | + | |
| 755 | + | MLH1/PMS2− | Colorectum NOS | 72 | co72 | − | − | |
| 299 | + | MLH1/PMS2− | Colon ascendens | 50 | co50 | + | − | 1co59 MSS |
| 154 | + | MLH1/PMS2− | Colon ascendens | 71 | − | + | + | 1co38 MSS |
| 771 | + | MLH1/PMS2− | Colon ascendens | 65 | co65 | − | − | |
| 683 | + | PMS2− | Colon ascendens | 69 | − | − | − | 1co58 MSS |
| 785 | + | MLH1/PMS2− | Colon descendens | 54 | co54 | − | − | |
| 142 | + | MLH1/PMS2− | Colon ascendens | 55 | en57 | + | + | 1co22 MSS |
| 86 | + | MLH1/PMS2− | Colon ascendens | 55 | − | + | + | |
| 482b | + | MLH1/PMS2− | Caecum | 65 | co64 | + | − | |
| 748 | + | MLH1/PMS2− | Colorectum NOS | 45 | − | − | − | |
| 57 | + | MLH1/PMS2− | Colon transversum | 49 | − | + | + | |
| 441 | + | MLH1/PMS2− | Caecum | 51 | − | − | − | |
| 316 | + | MLH1/PMS2− | Colon NOS | 71 | en70 MSS,co71 | + | + | 2co43 MSS |
| 331 | + | MLH1/PMS2− | Duodenum | 47 | − | − | − | |
| 655 | + | MLH1/PMS2/MSH2/MSH6− | Colon descendens | 64 | co47 MSS, ur64 MSS | + | + |
Meth, methylation analysis of MLH1 promoter; IHC, immunohistochemical analysis of MLH1, PMS2, MSH2, and MSH6; Tumour tested for MSI, tumour origin or exact location of tumour in case of colon cancer of tumour tested for MSI; Age, age at diagnosis of tumour tested for MSI; Other tumour(s), metachronous or synchronous cancer associated with Lynch syndrome of index patient and age at diagnosis; AC, Amsterdam II criteria (Vasen et al, 1999); 2fam, 2 first degree relatives (including index) with a Lynch syndrome associated cancer, one of them with an age at diagnose below 50 years (Rodriguez-Bigas et al, 1997); Tumours of close relatives, tumours of close relatives tested for MSI and/or IHC, meth; +, positive; −, negative; NOS, not otherwise specified; co, colon; en, endometrium; ur, urothelial; MSI, MSI positive; MSS, MSI negative; 1, first degree relative; 2, second degree relative. (e.g. 1co53 MSS, a first degree relative of index patient had a colon tumour diagnosed at the age of 53 years which was MSI negative).
Carrier of unclassified variant c.3744_3773dup (p.His1248_Ser1257dup) in MSH6.
Carrier of unclassified variant c.663A>C (p.Glu221Asp) in MSH6.
In the group of 20 index patients with tumours with MLH1 promoter methylation, we performed subsequent molecular analyses of other tumours of index patients or family members. Tumours either were MSI negative or showed methylation of the MLH1 promoter in combination with absence of MLH1 protein staining (Table 2).
The Amsterdam II criteria were met in 11 of the 20 families (55%) with an MSI-positive tumour that was due to somatic hypermethylation of the MLH1 promoter. The mean age at diagnosis of these index patients was 61 years.
Characterisation of families with an unexplained MSI-positive tumour
There were 18 families with MSI-positive tumours, where neither a pathogenic MMR gene mutation was present, nor could hypermethylation of the MLH1 promoter be demonstrated in the tumour (Table 3). These tumours were truly MSI positive, as in 17 of these tumours more than 75% of markers were instable, whereas the remaining tumour (no. 421) had three instable mononucleotide and three stable dinucleotide markers.
Table 3. Molecular laboratory tests results and family history of 18 patients with an MSI-positive tumour with unexplained etiology.
| No. | Meth | IHC | Tumour tested for MSI | Age | Other tumour(s) | AC | 2fam | Tumours of close relatives |
|---|---|---|---|---|---|---|---|---|
| 149a | − | MLH1/PMS2− | Colon ascendens | 43 | − | − | − | 3co45 MSS |
| 499 | − | MLH1/PMS2− | Colon ascendens | 58 | − | − | − | |
| 445 | − | MLH1/PMS2− | Colon transversum | 49 | − | − | + | |
| 498 | − | MLH1/PMS2− | Sigmoid | 58 | co49 | − | − | |
| 172 | − | MLH1/PMS2− | Colon ascendens | 51 | − | − | + | |
| 373a | − | PMS2− | Caecum | 36 | − | − | − | |
| 554 | − | PMS2− | Colon transversum | 55 | co55 | − | − | 1co56 MSS |
| 582 | − | MSH2/MSH6− | Sigmoid | 33 | − | + | + | 1co47 MSI,IHC:MSH2−,meth− |
| 396 | − | MSH2/MSH6− | Appendix | 34 | − | − | − | |
| 224 | − | MSH2/MSH6− | Ileocecum | 53 | co34,co50 | + | + | |
| 580 | − | MSH2/MSH6− | Endometrium | 45 | − | − | − | |
| 718 | − | MSH2/MSH6− | Rectum | 18 | − | − | + | 1co47 MSI,HC:MSH2−,meth− |
| 736 | − | MSH2/MSH6− | Rectum | 58 | co58 MSS | − | − | |
| 421b | − | MSH6− | Colon ascendens | 53 | − | − | − | 1en62 MSI/IHC NA |
| 135 | − | PMS2/MSH2/MSH6−c | Colon ascendens | 27 | − | − | − | |
| 243 | − | PMS2/MSH2/MSH6−c | Colon transversum | 30 | − | − | + | |
| 127 | − | None− | Rectum | 54 | − | − | − | |
| 375d | − | None− | Caecum | 36 | − | − | + |
Meth, methylation analysis of MLH1 promoter; IHC, immunohistochemical analysis of MLH1, PMS2, MSH2, and MSH6; Tumour tested for MSI, tumour origin or exact location of tumour in case of colon cancer of tumour tested for MSI; Age, age at diagnosis of tumour tested for MSI; Other tumour(s), metachronous or synchronous cancer associated with Lynch syndrome of index patient and age at diagnosis; AC, Amsterdam II criteria (Vasen et al, 1999). 2fam, 2 first degree relatives (including index) with Lynch syndrome associated cancer, one of them with an age at diagnose below 50 years (Rodriguez-Bigas et al, 1997); Tumours of close relatives, tumours of close relatives tested for MSI and/or IHC, meth; +, positive; −, negative; NA, not assessable; co, colon; MSI, MSI positive; MSS, MSI negative; 1, first degree relative; 2, second degree relative; 3 third degree relative. (e.g. 3co45 MSS, a third degree relative of index patient had a colon tumour diagnosed at the age of 45 years which was MSI negative).
Carrier of unclassified variant c.1852_1853delinsGC (p.Lys618Ala) in MLH1.
Carrier of unclassified variant c.2117T>C (p.Phe706Ser) in MSH6 (Kets et al, 2006).
IHC difficult to interpret.
Carrier of unclassified variant c.250A>G (p.Lys84Glu) in MLH1 and c.984C>T (silent) in MSH2.
In 16 of the 18 index patients without a detectable germline mutation in an MMR gene and without hypermethylation of the MLH1 promoter, the staining of at least one MMR protein was absent. The IHC patterns were in line with inactivation of MLH1 (five families), PMS2 (two families), MSH2 (six families), and MSH6 (one family (no. 421)). In the last family, an unclassified MSH6 variant was detected for which the pathogenic nature could not be established (Kets et al, 2006). In two families, there was simultaneous loss of IHC protein staining of MSH2 and MSH6, and of PMS2.
Subsequent molecular analyses of tumours of family members of these index patients are presented in Table 3. Two index patients with absent MSH2 staining had a family member with a tumour with an IHC pattern that also matched with MSH2 inactivation, that is absence of MSH2 and MSH6.
Only two of the 18 families (11%) with an unexplained MSI-positive tumour fulfilled the Amsterdam II criteria. The mean age at diagnosis of these index patients was 44 years.
Sensitivity of current mutation detection techniques
Mutation analysis was performed in 100 families with at least one tumour with MSI. In 20 of these families, MLH1 promoter methylation was present. A pathogenic germline mutation was detected in 62 of the remaining 80 families (78%). Mostly, their clinical characteristics matched those of classical Lynch syndrome: a germline mutation was detected in 36 of 38 (95%) of those families that fulfilled the Amsterdam II criteria. Likewise, an MMR gene mutation was found in 47 of 54 (87%) of the families that met the following criterion of the Bethesda guidelines: two first degree relatives (including the index patient) with a tumour associated with Lynch syndrome, of which at least one was diagnosed below the age of 50 years.
Comparison of characteristics of families with and without a detectable MMR gene mutation
The majority of families with an MMR gene mutation fulfilled the Amsterdam II criteria. In the 83 families selected by MSI only (strategy 2), we found that the clinical Amsterdam II criteria were met in 51% of the families in which an MMR gene mutation was eventually found compared with 11% of those in whom no such mutation was detected (P<0.009). The mean age at onset of the index patients was comparable in both groups (46 and 44 years, respectively). The mean age at diagnosis of index patients with a tumour with somatic hypermethylation of the MLH1 promoter (61 years) was significantly higher than that of mutation positive index patients (Table 4).
Table 4. Family history and patient characteristics of index patients with an MSI-positive tumour who were preselected by MSI analysis (strategy 2): a comparison between patients with a germline mutation, with somatic methylation of the MLH1 promoter and with a tumour with unexplained MSI.
| Germline mutation MLH1, PMS2, MSH2, or MSH6 N=45 | Somatic methylation of MLH1 promoter N=20 | Unexplained MMR deficiency N=18 | Germline mutation vs somatic methylation (P-value) | Germline mutation vs unexplained MMR deficiency (P-value) | |
|---|---|---|---|---|---|
| Amsterdam I criteria positivea | 16 (36%) | 6 (30%) | 2 (11%) | 0.66 | 0.07 |
| Amsterdam II criteria positivea | 23 (51%) | 11 (55%) | 2 (11%) | 0.77 | 0.009 |
| Two first degree relatives with Lynch syndrome associated cancer, one below 50 yearsa | 31 (69%) | 9 (45%) | 7 (39%) | 0.07 | 0.03 |
| Metachronous or synchronous Lynch syndrome associated cancers of index patient | 17 (38%) | 10 (50%) | 4 (22%) | 0.20 | 0.24 |
| Age at diagnosis of MSI-positive index tumour | 45.9 [42.6–49.1] | 60.6 [55.7–65.5] | 43.9 [38.8–49.1] | <0.0001 | 0.80 |
| Age at diagnosis of first Lynch syndrome associated cancer of index patient | 43.1 [39.9–46.3] | 58.6 [53.8–63.4] | 42.4 [37.3–47.5] | <0.0001 | 0.97 |
| Mean age of two youngest relatives with Lynch syndrome associated cancerb | 45.7 [40.3–51.1] | 54.0 [46.3–61.7] | 49.5 [31.5–67.5] | 0.19 | 0.91 |
A positive score for fulfilment was only given if the index patient was part of the criterion.
Mean age of the two youngest affected relatives of the index patient. This was only calculated for families that fulfilled the Amsterdam II criteria.
Bold values signify P-values <0.05.
DISCUSSION
A disease causing germline mutation was identified in 78% of patients suspected of Lynch syndrome with an MSI-positive tumour and absence of hypermethylation of the MLH1 promoter. Interestingly, the remaining 22% of patients with an unexplained MSI-positive tumour had a less pronounced family history of cancer, but were diagnosed at an age comparable with that of proven Lynch syndrome patients. Assessment bias is not likely to cause this difference, as the criteria for family history of cancer only include data from close relatives that should be known by the index patients. The large majority of the tumours in this group without gene mutations or promoter methylation, nonetheless showed loss of IHC staining of at least one of the MMR proteins. This is highly suggestive for the presence of either a germline mutation or of a somatic inactivation of the gene involved. These families had a much less prominent history of cancer, which might suggest the presence of a different type of mutation with a lower risk of cancer for relatives. Our current mutation detection protocol, which was highly effective in the classical Lynch syndrome families, did not pick up such putative mutations in this cohort.
The putative mutations might include (1) point mutations in the PMS2 gene that are known to have a lower cancer risk (Truninger et al, 2005; Worthley et al, 2005; Hendriks et al, 2006); (2) germline methylation of the MSH2 promoter as was recently described (Chan et al, 2006); (3) mutations in regulatory sequences or missence variants that might lead to a lower risk of tumour development in relatives because the complete inactivation of the affected MMR gene might be more dependent on modifier genes; and (4) a type of mutation that frequently arises de novo.
To our knowledge, this is the first study in which the difference in family history between patients with unexplained MSI-positive tumours and patients with a recognised germline mutation is addressed. The underlying mechanism for the familial occurrence of a tumour with MSI in such families remains unknown. Irrespective of this, confirmation for these findings in future studies might suggest that the clinical management in these families needs to be modified. For the time being, our surveillance advice for patients with an unexplained MSI-positive tumour and their close relatives remains identical to that of patients with Lynch syndrome including the start of surveillance at an early age.
The present study shows that the currently used techniques to detect a germline mutation have a sensitivity of 78% for patients with an MSI-positive tumour without hypermethylation of the MLH1 promoter. A similar percentage can be calculated from data published by Hampel et al (2005): after exclusion of MSI-positive tumours with methylation of the MLH1 promoter, they found a germline mutation in 23 out of 29 patients (79%) with an MSI-positive tumour. Their methods to analyse germline mutations are comparable with those used in our study. The sensitivity of germline mutation detection could not be deduced from other studies, as they did not include analysis of hypermethylation of the MLH1 promoter and/or performed less comprehensive germline mutation analyses (Mangold et al, 2005; Barnetson et al, 2006; Niessen et al, 2006). Wagner et al (2003) also found a difference in prevalence of pathogenic MLH1, MSH2, or MSH6 mutations between families that fulfilled the Amsterdam criteria (39 mutations in 49 families (80%)) and those not fulfilling these criteria (five mutations in 10 families (50%)) using methods that detect a similar type of mutations as the methods used in the present study. However, in this study data about the MMR deficiency of the tumours are missing.
In conclusion, this is the first study showing that almost all highly penetrant MMR gene mutations are identified with the currently used germline mutation detection techniques. The sensitivity of the currently used germline mutation detection techniques is at least 78% and probably near 100% in Lynch syndrome families with a highly penetrant mutation. A minority of MSI-positive tumours may be due to germline mutations in MMR genes that cannot yet be detected. Such putative mutations may confer a lower risk of cancers associated with Lynch syndrome for relatives.
Acknowledgments
We thank M Ariaans, E Bongers, J de Bruin, I van der Burgt, F van Erp, M Jongmans, T Kleefstra, M Niermeijer, I Reimerink, A van Remortele, M Ruiter, L Schaap, S Vermeer, and M Voorendt for clinical genetic counselling of the patients, M van Asseldonk, E Bosgoed, M Schliekelmann, and S Wezenberg for technical assistance, and R Akkermans for statistical support.
References
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A (1998) Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 338: 1481–1487 [DOI] [PubMed] [Google Scholar]
- Barnetson RA, Tenesa A, Farrington SM, Nicholl ID, Cetnarskyj R, Porteous ME, Campbell H, Dunlop MG (2006) Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med 354: 2751–2763 [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–5257 [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, Leung SY (2006) Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet 38: 1178–1183 [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, Halling KC, McDonnell SK, Schaid DJ, Walsh VC, Kubly V, Nelson H, Michels VV, Thibodeau SN (2001) The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet 69: 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong AE, van Puijenbroek M, Hendriks Y, Tops C, Wijnen J, Ausems MG, Meijers-Heijboer H, Wagner A, van Os TA, Brocker-Vriends AH, Vasen HF, Morreau H (2004) Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res 10: 972–980 [DOI] [PubMed] [Google Scholar]
- Deng G, Peng E, Gum J, Terdiman J, Sleisenger M, Kim YS (2002) Methylation of hMLH1 promoter correlates with the gene silencing with a region-specific manner in colorectal cancer. Br J Cancer 86: 574–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove-Edwin I, de Jong AE, Adams J, Mesher D, Lipton L, Sasieni P, Vasen HF, Thomas HJ (2006) Prospective results of surveillance colonoscopy in dominant familial colorectal cancer with and without Lynch syndrome. Gastroenterology 130: 1995–2000 [DOI] [PubMed] [Google Scholar]
- Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD (2002) A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res 62: 3925–3928 [PubMed] [Google Scholar]
- Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A (2005) Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352: 1851–1860 [DOI] [PubMed] [Google Scholar]
- Hendriks YM, Jagmohan-Changur S, van der Klift H, Morreau H, van Puijenbroek M, Tops C, van Os T, Wagner A, Ausems MG, Gomez E, Breuning MH, Brocker-Vriends AH, Vasen HF, Wijnen JT (2006) Heterozygous mutations in PMS2 cause hereditary nonpolyposis colorectal carcinoma (Lynch syndrome). Gastroenterology 130: 312–322 [DOI] [PubMed] [Google Scholar]
- Hitchins M, Williams R, Cheong K, Halani N, Lin VA, Packham D, Ku S, Buckle A, Hawkins N, Burn J, Gallinger S, Goldblatt J, Kirk J, Tomlinson I, Scott R, Spigelman A, Suter C, Martin D, Suthers G, Ward R (2005) MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology 129: 1392–1399 [DOI] [PubMed] [Google Scholar]
- Hoogerbrugge N, Willems R, van Krieken JH, Kiemeney LA, Weijans M, Nagengast FM, Arts N, Brunner HG, Ligtenberg MJ (2003) Very low incidence of microsatellite instability in rectal cancers from families at risk for HNPCC. Clin Genet 63: 64–70 [DOI] [PubMed] [Google Scholar]
- Jass JR (2006) Hereditary Non-Polyposis Colorectal Cancer: the rise and fall of a confusing term. World J Gastroenterol 12: 4943–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kets CM, van Krieken JH, Hebeda KM, Wezenberg SJ, Goossens M, Brunner HG, Ligtenberg MJ, Hoogerbrugge N (2006) Very low prevalence of germline MSH6 mutations in hereditary non-polyposis colorectal cancer suspected patients with colorectal cancer without microsatellite instability. Br J Cancer 95: 1678–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindor NM, Rabe K, Petersen GM, Haile R, Casey G, Baron J, Gallinger S, Bapat B, Aronson M, Hopper J, Jass J, LeMarchand L, Grove J, Potter J, Newcomb P, Terdiman JP, Conrad P, Moslein G, Goldberg R, Ziogas A, Anton-Culver H, de AM, Siegmund K, Thibodeau SN, Boardman LA, Seminara D (2005) Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA 293: 1979–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A (2003) Hereditary colorectal cancer. N Engl J Med 348: 919–932 [DOI] [PubMed] [Google Scholar]
- Mangold E, Pagenstecher C, Friedl W, Mathiak M, Buettner R, Engel C, Loeffler M, Holinski-Feder E, Muller-Koch Y, Keller G, Schackert HK, Kruger S, Goecke T, Moeslein G, Kloor M, Gebert J, Kunstmann E, Schulmann K, Ruschoff J, Propping P (2005) Spectrum and frequencies of mutations in MSH2 and MLH1 identified in 1,721 German families suspected of hereditary nonpolyposis colorectal cancer. Int J Cancer 116: 692–702 [DOI] [PubMed] [Google Scholar]
- Miyakura Y, Sugano K, Akasu T, Yoshida T, Maekawa M, Saitoh S, Sasaki H, Nomizu T, Konishi F, Fujita S, Moriya Y, Nagai H (2004) Extensive but hemiallelic methylation of the hMLH1 promoter region in early-onset sporadic colon cancers with microsatellite instability. Clin Gastroenterol Hepatol 2: 147–156 [DOI] [PubMed] [Google Scholar]
- Niessen RC, Berends MJ, Wu Y, Sijmons RH, Hollema H, Ligtenberg MJ, de Walle HE, de Vries EG, Karrenbeld A, Buys CH, van der Zee AG, Hofstra RM, Kleibeuker JH (2006) Identification of mismatch repair gene mutations in young colorectal cancer patients and patients with multiple HNPCC-associated tumours. Gut 55: 1781–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raevaara TE, Timoharju T, Lonnqvist KE, Kariola R, Steinhoff M, Hofstra RM, Mangold E, Vos YJ, Nystrom-Lahti M (2002) Description and functional analysis of a novel in frame mutation linked to hereditary non-polyposis colorectal cancer. J Med Genet 39: 747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S (1997) A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst 89: 1758–1762 [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML (2005) Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 129: 837–845 [DOI] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL (2004) Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet 36: 497–501 [DOI] [PubMed] [Google Scholar]
- Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, Bannwart F, Yurtsever H, Neuweiler J, Riehle HM, Cattaruzza MS, Heinimann K, Schar P, Jiricny J, Marra G (2005) Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology 128: 1160–1171 [DOI] [PubMed] [Google Scholar]
- Valle L, Carbonell P, Fernandez V, Dotor A, Sanz M, Benitez J, Urioste M (2007a) MLH1 germline epimutations in selected patients with early-onset non-polyposis colorectal cancer. Clin Genet 71: 232–237 [DOI] [PubMed] [Google Scholar]
- Valle L, Perea J, Carbonell P, Fernandez V, Dotor AM, Benitez J, Urioste M (2007b) Clinicopathologic and pedigree differences in Amsterdam I-Positive Hereditary Nonpolyposis Colorectal Cancer families according to tumor microsatellite instability status. J Clin Oncol 25: 781–786 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Mecklin JP, Khan PM, Lynch HT (1991) The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 34: 424–425 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin JP, Lynch HT (1999) New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116: 1453–1456 [DOI] [PubMed] [Google Scholar]
- Wagner A, Barrows A, Wijnen JT, van der Klift H, Franken PF, Verkuijlen P, Nakagawa H, Geugien M, Jaghmohan-Changur S, Breukel C, Meijers-Heijboer H, Morreau H, van Puijenbroek M, Burn J, Coronel S, Kinarski Y, Okimoto R, Watson P, Lynch JF, de la Chapelle A, Lynch HT, Fodde R (2003) Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet 72: 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38: 787–793 [DOI] [PubMed] [Google Scholar]
- Wijnen J, van der Klift H, Vasen H, Khan PM, Menko F, Tops C, Meijers-Heijboer H, Lindhout D, Moller P, Fodde R (1998) MSH2 genomic deletions are a frequent cause of HNPCC. Nat Genet 20: 326–328 [DOI] [PubMed] [Google Scholar]
- Worthley DL, Walsh MD, Barker M, Ruszkiewicz A, Bennett G, Phillips K, Suthers G (2005) Familial mutations in PMS2 can cause autosomal dominant hereditary nonpolyposis colorectal cancer. Gastroenterology 128: 1431–1436 [DOI] [PubMed] [Google Scholar]
- Wu Y, Nystrom-Lahti M, Osinga J, Looman MW, Peltomaki P, Aaltonen LA, de la Chapelle A, Hofstra RM, Buys CH (1997) MSH2 and MLH1 mutations in sporadic replication error-positive colorectal carcinoma as assessed by two-dimensional DNA electrophoresis. Genes Chromosomes Cancer 18: 269–278 [PubMed] [Google Scholar]