Abstract
Homologous antisense constructs were used to down-regulate tobacco cinnamyl-alcohol dehydrogenase (CAD; EC 1.1.1.195) and cinnamoyl-CoA reductase (CCR; EC 1.2.1.44) activities in the lignin monomer biosynthetic pathway. CCR converts activated cinnamic acids (hydroxycinnamoyl–SCoAs) to cinnamaldehydes; cinnamaldehydes are then reduced to cinnamyl alcohols by CAD. The transformations caused the incorporation of nontraditional components into the extractable tobacco lignins, as evidenced by NMR. Isolated lignin of antisense-CAD tobacco contained fewer coniferyl and sinapyl alcohol-derived units that were compensated for by elevated levels of benzaldehydes and cinnamaldehydes. Products from radical coupling of cinnamaldehydes, particularly sinapaldehyde, which were barely discernible in normal tobacco, were major components of the antisense-CAD tobacco lignin. Lignin content was reduced in antisense-CCR tobacco, which displayed a markedly reduced vigor. That lignin contained fewer coniferyl alcohol-derived units and significant levels of tyramine ferulate. Tyramine ferulate is a sink for the anticipated build-up of feruloyl–SCoA, and may be up-regulated in response to a deficit of coniferyl alcohol. Although it is not yet clear whether the modified lignins are true structural components of the cell wall, the findings provide further indications of the metabolic plasticity of plant lignification. An ability to produce lignin from alternative monomers would open new avenues for manipulation of lignin by genetic biotechnologies.
Keywords: monolignol/genetic modification/antisense RNA/coniferyl alcohol/feruloyl-CoA
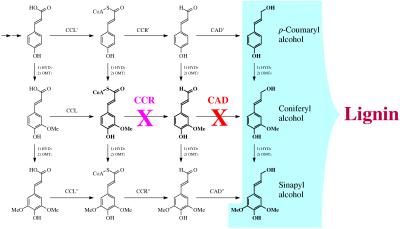
Lignins are phenolic polymers essential for mechanical support, defense, and water transport in vascular terrestrial plants (1–3), but they are a major obstacle to efficient utilization of plants for paper making or animal feed. A recent approach toward improved utilization has been the down-regulation of enzymes involved in the lignin monomer biosynthetic pathway (Fig. 1). Antisense technologies allow selective targeting of single enzymes in the pathway and transgenic plants have been obtained with altered lignin content and/or lignin structure (4–14).
Figure 1.
Lignin biosynthetic pathway, simplified schematic. Not all of the pathways or enzymes are necessarily known or distinct. CAD is a generic term (cinnamyl-alcohol dehydrogenase) for the enzyme class that catalyzes the final reduction to hydroxycinnamyl alcohols—CAD′ may or may not be different from CAD and CAD". Similarly for CCR (cinnamoyl-CoA reductase).
Recently, a lignin isolated from a pine mutant deficient in cinnamyl-alcohol dehydrogenase (CAD; EC 1.1.1.195) was characterized (15). This lignin had significantly lower coniferyl-alcohol-derived units, compensated for by elevated aldehyde levels, and major levels of dihydroconiferyl alcohol units. The structural observations begin to reveal the processes by which substantial lignin levels are maintained in some plants with severely depressed activity of lignification enzymes. The CAD-deficient mutant appeared able to utilize other phenols as substrates for radical coupling reactions to produce polymers that may function similarly to normal lignin (15, 16). Whether this altered lignin is a true structural component of the cell wall remains to be determined.
To explore other plants with enzyme down-regulation, we have selected tobacco plants transformed by CAD and cinnamoyl-CoA reductase (CCR; EC 1.2.1.44) antisense constructs. Tobacco is an excellent model plant with comprehensive data on the genes of the lignification pathway; some data have been published on lignins from CCR (8) and CAD down-regulated tobacco plants (9, 12, 13, 17). For this study, uniformly 13C-enriched lignins were isolated from down-regulated CAD and CCR tobacco plants and compared with control plants through the application of NMR techniques to examine how plants respond to the down-regulation of specific enzymes.
MATERIALS AND METHODS
Transgenic Tobacco Plants (Nicotiana tabacum L. Cv. Samsun).
Antisense CAD plants. We used seeds resulting from self-pollination of primary transformant T37 (17) carrying a 1-kb CAD cDNA in antisense orientation, associated to the 35S cauliflower mosaic virus (CaMV) promoter and the 3′ terminator of the nopaline synthase.
Antisense CCR plants. We used seeds resulting from a test cross on primary transformant B3 (8) carrying a 1.3-kb CCR cDNA in antisense orientation, associated to the 35S CaMV promoter and the 3′ terminator of the nopaline synthase.
Both CAD and CCR antisense constructs also carried the neomycin phosphotransferase gene, which confers resistance to kanamycin.
Growth and Labeling Conditions.
Seeds were sterilized for 5 min in 10% sodium hypochlorite and rinsed in sterile water. After 15 days in vitro [light/dark regime of 16 h, 20–30 μE⋅m−2⋅sec−1, 27°C; 1 E (einstein) = 1 mol of photons] on a solid MS (Murashige and Skoog) medium supplemented with kanamycin (500 mg/liter) to select the seedlings harboring the transgenes, resistant plantlets were transferred to vermiculite and grown in a culture room (same culture conditions) for 26 days. Six-week-old plants (10 expanded leaves, stem height 7–10 cm) were transferred to Cadarache for labeling in a C23A chamber (18). This strictly closed module (750 liters) derived from glove boxes used for nuclear manipulation, allows accurate regulation of atmospheric gas composition and environmental parameters (14 h photoperiod, photon flux rate 330 μmol⋅m−2⋅s−1, temperature 25°C day/20°C night, relative humidity 70% day/80% night, watering eight times per day with nutrient solution). CO2 concentration into the chamber during the light period was maintained at 350 μl⋅liter−1 by automatic injections of concentrated CO2, to compensate for photosynthetic assimilation. During the first 4 days in the chamber, CO2 with natural 13C abundance (about 1%) was injected. Then it was replaced by 13C-enriched CO2 (10.14%; purchased from Euriso-top, Saint-Aubin, France) until the beginning of flowering (33 days). Plants were harvested and stems were fixed in liquid nitrogen and lyophilized.
Transgenic tobacco plants harboring the antisense-CAD construct were phenotypically normal but displayed a characteristic red-brown coloration of the xylem (17), whereas the transgenic plants carrying the antisense-CCR construct showed a reduction of growth and an orange-brown coloration of the xylem. Significant reductions of CAD and CCR activities were respectively demonstrated in the two types of transformants.
Isolation of Lignins.
Lignins were isolated essentially as previously reported (15). Tobacco stem material was first ground in a Wiley mill (1-mm screen), and soluble components were removed by successive Soxhlet extractions with water, methanol, acetone, and chloroform. Cell wall Klason lignin (19) levels were 17% wt/wt for the normal tobacco, 15% for the antisense-CAD tobacco, and 8% for the antisense-CCR tobacco. The extractive-free cell wall preparation was then ball milled, polysaccharides were reduced by digestion with crude cellulases (Cellulysin, Calbiochem), and the resulting “enzyme lignin” was extracted with 96:4 vol/vol dioxane/water (20). The extracts were lyophilized to yield 12%, 24%, and 18% of the original Klason lignin as isolated lignins. Water-soluble components and metal ions were removed with aqueous EDTA (6 M, pH 8) (21), and the insoluble materials were lyophilized. The final yields of lignin were 7%, 17%, and 8.5%. The 13C-labeled samples were too valuable to determine lignin levels at this time, but NMR indicates <15% saccharide contamination in each lignin. After the series of NMR experiments, the lignins were extracted with methylene chloride to remove possible low molecular weight components. This washing caused neither substantial material loss nor the reduction of the aldehyde or tyramine ferulate signals, which were therefore included in the polymer. Acetylation of small amounts of each of the lignins was by means of acetic anhydride and pyridine. The acetylated lignins were extracted into freshly distilled ethyl acetate and washed with aqueous EDTA to remove trace metal contaminants prior to NMR study.
NMR.
NMR experiments were performed at 360 MHz on a Bruker AMX-360 using a narrow-bore probe with conventional coil geometry and without gradients, or at 750 MHz on a Bruker DMX-750 using a narrow-bore (5 mm) triple resonance probe (1H, 13C, 15N; the 15N was not used) optimized for inverse experiments and with three-axis gradients. Experiments used were standard Bruker implementations of traditional or gradient-selected versions of inverse (1H-detected) heteronuclear multiple quantum coherence (HMQC), heteronuclear single quantum coherence (HSQC), HMQC-total correlation spectroscopy (TOCSY), HSQC-TOCSY, and heteronuclear multiple bond correlation (HMBC) experiments. TOCSY experiments used a 100-ms spin lock period; HMBC experiments used a 100-ms long-range coupling delay.
RESULTS AND DISCUSSION
Definition of Isolated Tobacco Lignin.
All following comments refer to an extracted “lignin” that is not necessarily representative of lignin in situ. Indeed, it has been suggested (49) that the isolated lignins [in particular from the mutant pine previously studied (15)] may represent partially polymerized phenolic extractives, similar to those that occur in heartwood (22, 23). Whether these isolates really derive from the structural plant polymer remains to be elucidated. Nevertheless, isolation of polymeric components from control and transgenic plants by identical standard lignin-isolation procedures ensures that differences noted are indicative of real compositional differences in the original plants and elucidates responses due to down-regulated lignin-biosynthetic-pathway enzymes.
The Unusual Nature of Isolated Tobacco Lignin.
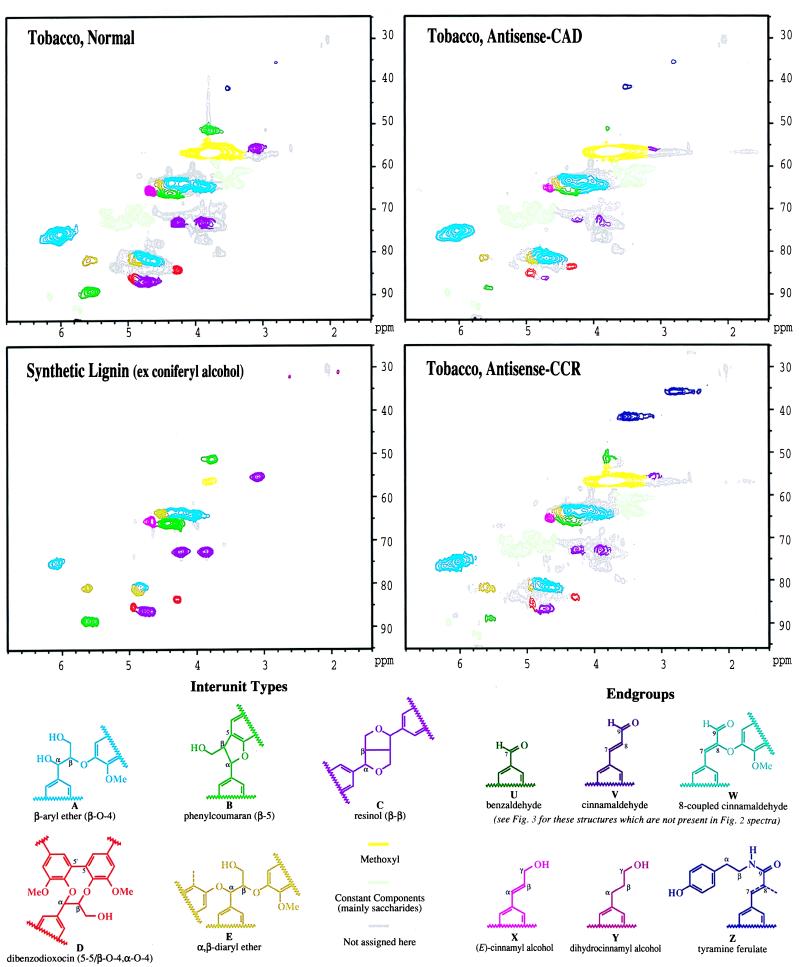
Tobacco lignin isolated from the control had unusually high amounts of α-ethers E (see Fig. 2). Similarly, high amounts of cinnamyl alcohol endgroups X were reminiscent of a lignin from Picea abies suspension cultures (24); such endgroups also dominate synthetic lignins. In contrast, lignins previously isolated from herbaceous or woody plants typically contain low levels of cinnamyl alcohol endgroups. This is because lignification results from slow diffusion of hydroxycinnamyl alcohols into the wall where their radicals encounter radicals of preformed lignin oligomers, so-called endwise polymerization (25). Only rarely do two monomers couple to form a dimer. Lignification thereby produces structures that are in different proportions from those encountered in synthetic lignins obtained from high levels of monomer-coupling products (bulk polymerization) (26–28). Synthetic lignins are usually rich in cinnamyl alcohol endgroups X. They also contain substantial amounts of α–ethers E from addition of phenols to the β-O–4 quinone methide intermediate. Once thought to involve some 6–8% of native lignin units (29), noncyclic α-ethers E have recently been found at extremely low levels in isolated wood lignins (30). Recently discovered dibenzodioxocin structures D (31, 32) probably contributed to erroneous estimates previously.
Figure 2.
(Upper) Partial two-dimensional HSQC NMR (gradient selected) spectra of isolated lignins (acetylated) from normal, antisense-CAD, and antisense-CCR tobacco, and of a synthetic lignin from coniferyl alcohol. Interunit types A−E (structures shown in Lower) are apparent in all spectra. Endgroups are not all visible in the side-chain region shown. These isolated tobacco lignins resemble synthetic lignins more than any lignin characterized by NMR to date; they have significant numbers of β-β units and unsaturated side chains, indicating frequent dimerization reactions of monolignols. Lignins isolated from other plants have low levels of these units and arise from endwise coupling of hydroxycinnamyl alcohol units with growing lignin oligomers. Evidence for coniferyl alcohol depletion is seen in both antisense lignins. Aldehydes (not visible in this range) are the major feature of the antisense-CAD lignin, whereas tyramine units Z are strikingly enriched in the antisense-CCR lignin.
NMR data suggest that tobacco differs from other plants in producing a bulk lignin component. This may be because of rapid monomer supply or to monomer delivery prior to activation of other factors required for peroxidase-catalyzed polymerization. Alternatively, we may have extracted an extracellular “lignin” that is not present in other plants studied to date. The cinnamyl alcohol endgroups were entirely trans. As cis-coniferyl alcohol retains the cis double bond in radical dimerization (33), we may conclude that tobacco lignin does not arise from any significant contribution of cis-coniferyl alcohol.
The control tobacco isolated lignin contained a high proportion of β-ethers A, consistent with its syringyl-guaiacyl nature. NMR signals from phenylcoumarans B and resinols C were also prominent. As noted above, the level of α,β-diethers E and cinnamyl alcohol endgroups X was striking. In addition, the involvement of 5,5-diguaiacyl structures in dibenzodioxocins D makes them stand out in two-dimensional 13C–1H-correlated NMR. The levels in the normal tobacco are quite high, similar to those seen in synthetic guaiacyl lignins, providing further evidence of the bulk nature of this tobacco lignin isolate. Traces of tyramine units Z (see below) are noted in the normal tobacco.
Antisense-CAD Tobacco Lignin.
The Klason lignin contents of control and antisense-CAD tobacco stems were similar (17% and 15%). The yield of isolated lignin from the CAD-deficient plant (24% of the Klason lignin) was twice that of the control (12%), which may reflect different lignin functionality and/or organization of wall polymers. Guaiacyl (G) components from coniferyl alcohol were reduced as evidenced by the decrease in β-5 phenylcoumaran structures which can arise only from G coupling at C-5 (Fig. 2). Syringyl (S) structures arising from coupling of sinapyl alcohol also declined. This was evidenced by reduced S-2/6 carbon signals at ≈105 ppm (Fig. 3). A reduced S:G ratio was found in both pyrolysis-GC-MS and thioacidolysis products (9, 17). In the antisense-CAD lignin, β-ethers A were dominant, whereas phenylcoumarans B were significantly lower, as were resinols C (see Fig. 2 Lower for structures). There was still a significant (although lower) level of dibenzodioxocins D, α,β-diethers E, and cinnamyl alcohol endgroups X. Tyramine ferulate Z levels were modestly higher.
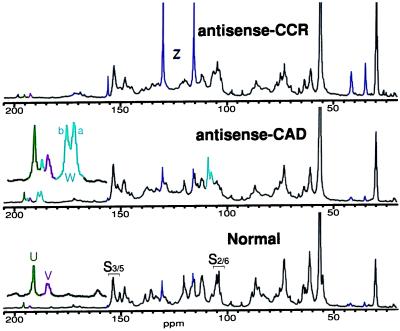
Figure 3.
13C NMR spectra of normal, antisense-CAD, and antisense-CCR tobacco isolated lignins (unacetylated). The normal lignin is a syringyl/guaiacyl lignin, with guaiacyl units predominating. Aldehydes, including newly found aldehydes W (mainly from sinapaldehyde coupling), are elevated in the antisense-CAD sample; tyramine units Z are markedly elevated in the antisense-CCR lignin. Colors define structures as in Fig. 2.
The aldehyde region of the antisense-CAD lignin displayed striking changes (Fig. 3). Both cinnamaldehyde V and benzaldehyde U levels were higher and new aldehydes W became predominant. Cinnamaldehydes are lignin components responsible for the purple staining of lignified tissues by phloroglucinol⋅HCl (34). Vanillin, coniferaldehyde, and the corresponding S analogues are found in extracts of lignified plants. Benzaldehydes U derive from cinnamaldehydes (3). Elevated cinnamaldehyde levels have been reported in the lignins from CAD-deficient plants (9, 12, 15, 17, 35, 36). The red-brown coloration of such plants has been ascribed to the presence of these aldehydes (37), although the molecular basis for the coloration has not been elucidated.
NMR indicated that the benzaldehyde U and cinnamaldehyde V levels in the antisense-CAD isolated tobacco lignin were roughly doubled (Fig. 3). These signals were unaffected by methylene chloride extraction of the lignin—the aldehydes were an integral part of the polymer. Direct incorporation of aldehyde monomers into free-radical polymerization reactions with normal monomers would explain their relatively high contents, but other possibilities are not ruled out. Far more striking was the appearance of new aldehydes W. They were at best barely discernible in the normal lignin but became major contributors to the antisense-CAD lignin (Fig. 3).
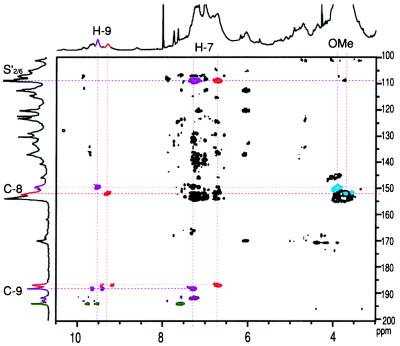
Nature of new aldehydes W. The new aldehydes W showed only one HMQC-TOCSY correlation with the aldehyde proton/carbon, suggesting that they were benzaldehydes. Their carbon chemical shifts (Fig. 3) were, however, lower than normal benzaldehydes. NMR of the acetylated lignins provided misleading correlations between aldehydic carbons and methoxyl protons suggesting that they were 2-methoxybenzaldehydes, for which the low carbon chemical shifts were reasonable. In fact these correlations were simply coincident; the new aldehydes are not 2-methoxybenzaldehydes as originally assigned (15). They arise instead from coniferaldehyde and sinapaldehyde. The upfield peak Wa (186.8 ppm) (Fig. 3) matches a dimer from 8–O–4-coupling of coniferaldehyde (38). The analogous lower-field peak Wb (188.1 ppm) appears to be from heterocoupling (of a cinnamaldehyde with an unconjugated G or S unit). No evidence for 8–8- or 8–5 coupling of cinnamaldehydes could be discerned in these spectra. The HMBC experiment correlated each W aldehyde carbon with side-chain H–7 protons at 7.3 and 6.7 ppm, respectively (Fig. 4). These H–7 protons correlated strongly with S carbons at ≈108 ppm, which suggests that sinapaldehyde is predominantly incorporated. By contrast, a coniferaldehyde-coniferyl alcohol synthetic lignin, which shows similar aldehyde resonances and H–7 correlations, has its H–7 protons correlating with G carbons at >110 ppm (not shown). A detailed comparison of this tobacco lignin with lignin from other CAD-deficient plants, with synthetic lignins containing coniferaldehyde, and with model compounds, will appear elsewhere.
Figure 4.
Partial two-dimensional HMBC NMR spectrum (gradient-selected) of antisense-CAD acetylated lignin, providing evidence of sinapyl aldehydes W, involved in two resolvable structures (colored red and violet here to differentiate). The red component is almost certainly an 8–O–4-coupled sinapaldehyde product; the analogous violet one appears to be a sinapaldehyde cross-coupled product. The diagnostic correlations (colored) are not present in the equivalent spectrum of normal tobacco lignin. The apparent correlation to methoxyl protons (blue) is a coincidence that erroneously suggested 2-methoxybenzaldehydes. H–7 in each structure correlates with the aldehyde carbon (C–9) and also new syringyl resonances at ≈108 ppm, implicating sinapaldehyde. [Colors do not relate to structure assignments in Fig. 2.]
Comparison with lignin from a CAD-deficient pine mutant (15).
An analogous pair of hydroxycinnamaldehyde coupling products W were seen in lignin isolated from a naturally CAD-deficient pine mutant (15). The levels in that lignin were not elevated to the extent they were in this tobacco. These comparisons support the hypothesis that aldehydes in tobacco mainly result from hydroxycinnamaldehyde coupling to structures that are possible from either coniferaldehyde or sinapaldehyde (e.g., 8–O–4). The CAD-deficient pine lignin also had large quantities of another unusual unit, dihydroconiferyl alcohol (15). These units were not detectable in tobacco lignins (Fig. 2). Clearly the mutant pine and the transgenic tobacco display both common and specific responses to a CAD enzyme deficit.
Antisense-CCR Tobacco Lignin.
The lignin content of the antisense-CCR tobacco stem material was about half that of control, but the extractable lignin was at about the same relative level. The stem material was quite flexible and plastic, compared with more brittle control and antisense-CAD tissues. In agreement with a previous study (8), the CCR-down-regulated tobacco showed abnormal development with reduced growth, abnormal leaf morphology, and collapsed vessels. The lignin showed clear evidence of reduced G components, again by the lower levels of β–5 phenylcoumaran structures (Fig. 2). In this case, the S units were relatively retained, as evidenced by the strong S–2/6 carbons (≈105 ppm) in the one-dimensional 13C NMR spectrum (Fig. 3). Similar to the control, the β-ethers A were dominant. Resinols C, dibenzodioxocins D, α,β-diethers E, and cinnamyl alcohols X remained significant. Aldehyde levels were not changed.
In the antisense-CCR tobacco, thioacidolysis revealed unusually high amounts of tyramine ferulates (C.L., unpublished data), about 3 times that of the control. In agreement with this finding, the most striking change in the 13C NMR spectra (Fig. 3) was the dominance of signals from p-hydroxyphenyl units with free phenolic groups, as shown by shifts after acetylation.
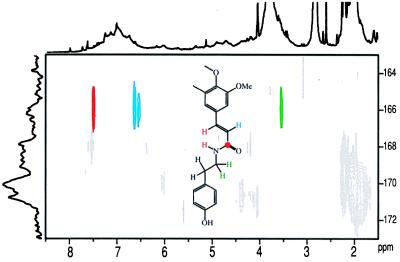
Proof of tyramine ferulate units and biochemical implication. NMR was able to provide elegant evidence for tyramine ferulate units Z (Fig. 2). HMQC-TOCSY spectra correlated the α- and β-carbons (35.1 and 41.6 ppm) with side-chain protons, including the amide N–H (≈7.4 ppm). Synthetic tyramine ferulate shifts fell directly under prominent contours in the antisense-CCR lignin (Fig. 5). As has been fully elucidated for ferulate esters (39, 40), feruloyl amides are expected to incorporate intimately into lignins by the full complement of radical coupling reactions. Without data from synthetic lignins incorporating tyramine ferulate, it is not possible to identify the range of structures involved in these copolymer lignins. However, NMR can readily identify the structure in which the tyramine ferulate has radically coupled at the feruloyl 4–O and/or 5 position, leaving the double bond intact. Fig. 6 shows the region of the HMBC spectrum correlating the feruloyl amide carbonyl carbon with all protons within three bonds, namely the (unresolved) N–H and the β-Hs of the tyramine moiety, and both unsaturated protons in the ferulate side chain (H–8 and H–7). These correlations are diagnostic of the involvement of tyramine ferulate in tobacco lignin and match those of authentic tyramine ferulate. However, these experiments do not ascertain whether ferulate is the only hydroxycinnamate component involved.
Figure 5.
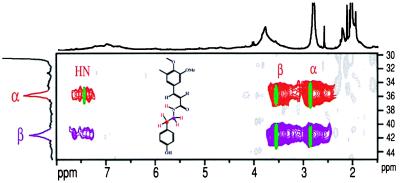
Partial two-dimensional HMQC-TOCSY NMR spectrum of antisense-CCR tobacco isolated lignin (acetylated) proving tyramine units are involved as amides. Correlation between the tyramine side-chain aliphatic carbons (C–α and C–β) and the three side-chain protons (H–α, H–β, and the amide N–H) matches well with data from tyramine ferulate itself (superimposed data in green). [Colors do not relate to structure assignments in Fig. 2.]
Figure 6.
Proof that feruloyl units are amides with tyramine. HMBC spectra show diagnostic correlations between the amide carbonyl to ferulate H–7 and H–8 side-chain protons (as well as the amide N–H, close to H–8) and, across the amide bond, to the H–βs on tyramine. Determination of the full array of feruloyl unit linkages awaits preparation of synthetic lignins containing strategically labeled tyramine ferulate. [Colors do not relate to structure assignments in Fig. 2.]
Tyramine ferulates have been previously reported in tobacco walls (41–43). The required transferase, hydroxycinnamoyl-CoA:tyramine hydroxycinnamoyltransferase, has been found in many plants (44–47). With the caveat that such units increase in stressed tobacco (48), tyramine ferulates are a logical sink for feruloyl–SCoA units that might build up when the CCR enzyme is down-regulated (Fig. 1). It is obviously a leap to infer that the plant up-regulates production of tyramine specifically to provide a sink for feruloyl–SCoA and produce a derivative that can be incorporated into lignin to offset the deficit in coniferyl alcohol. Nevertheless, the antisense-CCR tobacco is making a copolymer that may functionally mimic normal lignin. If the tyramine ferulate is intimately incorporated into the hydroxycinnamyl alcohol polymer, we have a further example of the metabolic plasticity of plants to utilize other phenols to produce “lignin” when normal monomers become limited. That the same units are used in certain stress and wounding responses may indicate that the extractable lignins are polymerized extractives or that these pathways can be up-regulated to provide a functional lignin-like polymer. However, the abnormal development of the antisense-CCR tobacco shows that the normal functions required of lignin were not fully met by the reduced levels of its modified lignin.
CONCLUSIONS
Down-regulating enzymes in the lignin monomer biosynthetic pathway may or may not reduce lignification. In the case of CCR, reduction was substantial. In down-regulated plants that have been structurally examined to date, the plants appear to compensate for the shortage in hydroxycinnamyl alcohol monomers by utilizing other phenols through up-regulation or redirection of other pathways. The isolated lignins from CAD and CCR down-regulated tobacco plants contained substantial levels of unusual components. While both degradative analyses on native lignins and NMR studies on isolated lignins have provided evidence regarding the structural claims for these modified lignins, their functional roles are not clear. The abnormality of heavily CCR down-regulated plants does not clarify whether the plant’s vigor is affected by lignin’s quantity or its functional quality. Viability of transgenic or mutant plants might be because of their ability to produce lignin-like polymers from unconventional components. Such a capability would appear to be evolutionarily wise and may be at least partly responsible for plants’ survival despite mutations that reduce the flux through the normal lignification pathway. The challenge for future studies is to address the features described here on the whole lignin fraction.
Although the plants’ apparent abilities to circumvent genetic obstacles by utilization of other components to make lignin may interfere with the current objectives of genetic biotechnologies, it opens up new potential for manipulation of lignin’s composition and properties. Studies on lignin-biosynthetic-pathway mutants and transgenic plants will provide a rich source of insight into the processes of lignification. In several instances now, down-regulation of the monolignol pathway has resulted in amplification of unusual units already present in low amounts in normal lignins or in lignin-like polymeric extractives.
Acknowledgments
The authors are grateful to Fachuang Lu for making the tyramine ferulate. The support of the European Community (“Optimization of lignin in crop industrial plants through genetic engineering” and “Tree improvement based on lignin engineering” projects) is gratefully acknowledged, as is partial funding through the U.S. Department of Agriculture National Research Initiatives, no. 96-35304 in the Plant Growth and Development Section.
ABBREVIATIONS
- CAD
cinnamyl-alcohol dehydrogenase
- CCR
cinnamoyl-CoA reductase
- HMQC
heteronuclear multiple quantum coherence
- HSQC
heteronuclear single quantum coherence
- TOCSY
total correlation spectroscopy
- HMBC
heteronuclear multiple bond correlation
- G
guaiacyl
- S
syringyl
Footnotes
A Commentary on this article begins on page 12742.
References
- 1. Harkin J M. In: Chemistry and Biochemistry of Herbage. Butler G W, editor. Vol. 1. London: Academic; 1973. pp. 323–373. [Google Scholar]
- 2.Freudenberg K. Nature (London) 1959;183:1152–1155. doi: 10.1038/1831152a0. [DOI] [PubMed] [Google Scholar]
- 3.Sarkanen K V, Ludwig C H, editors. Lignins: Occurrence, Formation, Structure and Reactions. New York: Wiley-Interscience; 1971. [Google Scholar]
- 4.Sewalt V J H, Ni W, Blount J W, Jung H G, Masoud S A, Howles P A, Lamb C, Dixon R A. Plant Physiol. 1997;115:41–50. doi: 10.1104/pp.115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doorsselaere J V, Baucher M, Chognot E, Chabbert B, Tollier M-T, Petit-Conil M, Leplé J-C, Pilate G, Cornu D, Monties B, et al. Plant J. 1995;8:855–864. [Google Scholar]
- 6.Meyer K, Shirley A M, Cusumano J C, Bell-Lelong D A, Chapple C. Proc Natl Acad Sci USA. 1998;95:6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kajita S, Hishiyama S, Tomimura Y, Katayama Y, Omori S. Plant Physiol. 1997;114:871–879. doi: 10.1104/pp.114.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piquemal J, Lapierre C, Myton K, O’Connell A, Schuch W, Grima-Pettenati J, Boudet A-M. Plant J. 1998;13:71–83. [Google Scholar]
- 9.Halpin C, Knight M E, Foxon G A, Campbell M M, Boudet A-M, Boon J J, Chabbert B, Tollier M-T, Schuch W. Plant J. 1994;6:339–350. [Google Scholar]
- 10.Ni W, Paiva N L, Dixon R A. Transgenic Res. 1994;3:120–126. [Google Scholar]
- 11.Atanassova R, Favet N, Martz F, Chabbert B, Tollier M-T, Monties B, Fritig B, Legrand M. Plant J. 1995;8:465–477. [Google Scholar]
- 12.Hibino T, Takabe K, Kawazu T, Shibata D, Higuchi T. Biosci Biotechnol Biochem. 1995;59:929–931. [Google Scholar]
- 13.Stewart D, Yahiaoui N, McDougall G J, Myton K, Marque C, Boudet A M, Haigh J. Planta. 1997;201:311–318. doi: 10.1007/s004250050072. [DOI] [PubMed] [Google Scholar]
- 14.MacKay J J, O’Malley D M, Presnell T, Booker F L, Campbell M M, Whetten R W, Sederoff R R. Proc Natl Acad Sci USA. 1997;94:8255–8260. doi: 10.1073/pnas.94.15.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ralph J, MacKay J J, Hatfield R D, O’Malley D M, Whetten R W, Sederoff R R. Science. 1997;277:235–239. doi: 10.1126/science.277.5323.235. [DOI] [PubMed] [Google Scholar]
- 16.Boudet A-M. Trends Plant Sci. 1998;3:67–71. [Google Scholar]
- 17.Yahiaoui N, Marque C, Myton K E, Negrel J, Boudet A-M. Planta. 1998;204:8–15. [Google Scholar]
- 18.André M, Du Cloux H. Plant Physiol Biochem. 1993;31:103–112. [Google Scholar]
- 19.Hatfield R D, Jung H G, Ralph J, Buxton D R, Weimer P J. J Sci Food Agric. 1994;65:51–58. [Google Scholar]
- 20.Björkman A. Nature (London) 1954;174:1057–1058. [Google Scholar]
- 21.Ralph J, Hatfield R D, Quideau S, Helm R F, Grabber J H, Jung H-J G. J Am Chem Soc. 1994;116:9448–9456. [Google Scholar]
- 22.Sarkanen K V, Hergert H L. In: Lignins: Occurrence, Formation, Structure and Reactions. Sarkanen K V, Ludwig C H, editors. New York: Wiley-Interscience; 1971. pp. 43–94. [Google Scholar]
- 23.Hergert H L. In: Cellulose Chemistry and Technology, American Chemical Society Symposium Series. Arthur J C, editor. Vol. 48. Washington, DC: Am. Chem. Soc.; 1977. pp. 227–243. [Google Scholar]
- 24.Brunow G, Ede R M, Simola L K, Lemmetyinen J. Phytochemistry. 1990;29:2535–2538. [Google Scholar]
- 25.Sarkanen K V. In: Lignins: Occurrence, Formation, Structure and Reactions. Sarkanen K V, Ludwig C H, editors. New York: Wiley-Interscience; 1971. pp. 95–163. [Google Scholar]
- 26.Freudenberg K. Angew Chem. 1956;68:508–512. [Google Scholar]
- 27.Nimz H H, Lüdemann H-D. Holzforschung. 1976;30:33–40. [Google Scholar]
- 28.Faix O. Holzforschung. 1986;40:273–280. [Google Scholar]
- 29.Adler E. Wood Sci Technol. 1977;11:169–218. [Google Scholar]
- 30.Ede R M, Kilpeläinen I. Res Chem Intermediates. 1995;21:313–328. [Google Scholar]
- 31.Karhunen P, Rummakko P, Sipilä J, Brunow G, Kilpeläinen I. Tetrahedron Lett. 1995;36:169–170. [Google Scholar]
- 32.Karhunen P, Rummakko P, Sipilä J, Brunow G, Kilpeläinen I. Tetrahedron Lett. 1995;36:4501–4504. [Google Scholar]
- 33.Ralph J, Zhang Y. Tetrahedron. 1998;54:1349–1354. [Google Scholar]
- 34.Adler E, Björkquist K J, Häggroth S. Acta Chem Scand. 1948;2:93–94. [Google Scholar]
- 35.Baucher M, Chabbert B, Pilate G, van Doorsselaere J, Tollier M-T, Petit-Conil M, Cornu D, Monties B, van Montagu M, Inz D, et al. Plant Physiol. 1996;112:1479–1490. doi: 10.1104/pp.112.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillonel C, Mulder M M, Boon J J, Forster B, Binder A. Planta. 1991;185:538–544. doi: 10.1007/BF00202964. [DOI] [PubMed] [Google Scholar]
- 37.Higuchi T, Ito T, Umezawa T, Hibino T, Shibata D. J Biotechnol. 1994;37:151–158. [Google Scholar]
- 38.Connors W J, Chen C-L, Pew J C. J Org Chem. 1970;35:1920–1924. [Google Scholar]
- 39.Ralph J, Grabber J H, Hatfield R D. Carbohydr Res. 1995;275:167–178. [Google Scholar]
- 40.Ralph J, Helm R F, Quideau S, Hatfield R D. J Chem Soc Perkin Trans. 1992;1:2961–2969. [Google Scholar]
- 41.Negrel J, Martin C. Phytochemistry. 1984;23:2797–2801. [Google Scholar]
- 42.Negrel J, Jeandet P. Phytochemistry. 1987;26:2185–2190. [Google Scholar]
- 43.Negrel J, Javelle F. Physiol Plant. 1995;95:569–574. [Google Scholar]
- 44.Villegas M, Brodelius E. Physiol Plant. 1990;78:414–420. [Google Scholar]
- 45.Fleurence J, Negrel J. Phytochemistry. 1989;28:733–736. [Google Scholar]
- 46.Hohlfeld H, Scheel D, Strack D. Planta. 1996;199:166–168. [Google Scholar]
- 47.Louis V, Negrel J. Phytochemistry. 1991;30:2519–2522. [Google Scholar]
- 48.Negrel J, Pollet B, Lapierre C. Phytochemistry. 1996;43:1195–1199. [Google Scholar]
- 49.Gang D R, Fujita M, Davin L D, Lewis N G. In: Lignin and Lignan Biosynthesis, ACS Symposium Series 697. Lewis N G, Sarkanen S, editors. Washington, DC: Am. Chem. Soc.; 1998. pp. 389–421. [Google Scholar]