Abstract
Increased copy numbers of 17q23 chromosomal region have been shown to occur in different tumour types and to be associated with tumour progression and with poor prognosis. Several genes have earlier been proposed as potential oncogenes at this region largely on the grounds of cell lines studies. In this study, we performed a systematic gene expression survey on 26 primary breast tumours with known 17q23 amplification status by quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR). The 17q23 amplicon is restricted to an ∼5 MB region in breast cancer and contains 29 known genes. Our survey revealed a statistically significant (P<0.01) difference between the high level and no amplification groups in a set of eleven genes whereas no difference between the moderate and the non-amplified tumour groups were observed. Interestingly, these 11 genes were located adjacent to one another within a 1.56 Mb core region in which all except one of the genes were overexpressed. These data suggest that only high-level amplification at the 17q23 amplicon core leads to elevated gene expression in breast cancer. Moreover, our results highlight the fact that 17q23 amplicon carries multiple candidate genes and that this may be a more common event in gene amplification than previously thought.
Keywords: breast cancer, gene amplification, gene expression
Gene amplification is one of the major mechanisms that allow cancer cells to promote expression of genes that are involved in tumour development and progression. Oncogene overexpression as a result of gene amplification has been shown to play a crucial part in the pathogenesis of various human malignancies, especially in solid tumours such as breast, prostate, lung, ovarian, gastric, pancreatic, and colon cancers (Knuutila et al, 1998). The first illustration on the clinical significance of amplified oncogenes in human cancer was the discovery of an association between amplification of the MYC oncogene and more aggressive neuroblastoma variants (Schwab, 1993). To date, one of the clinically most important amplified gene is the ERBB2 oncogene, which is well known for its central role as a prognostic and predictive factor in breast cancer as well as a therapeutic target (Baselga et al, 1999; Cobleigh et al, 1999; Ross and Fletcher, 1999; Vogel et al, 2002). These examples illustrate that genes that are altered by amplification in cancer are likely to have an impact on both disease pathogenesis and the clinical management of cancer patients.
Amplification of the chromosomal region 17q23 was first discovered in breast cancer (Kallioniemi et al, 1994). After that, numerous studies have reported increased copy numbers of 17q23 in tumours of brain, lung, ovary, pancreas, bladder, testis, and liver (Muleris et al, 1994; Ried et al, 1994; Korn et al, 1996; Solinas-Toldo et al, 1996; Brinkschmidt et al, 1997; Marchio et al, 1997; Richter et al, 1997; Schwendel et al, 1997; Sonoda et al, 1997; Weber et al, 1997; Vandesompele et al, 1998; Clark et al, 2002; Willis et al, 2003). We recently performed an extensive survey on the distribution and frequency of the 17q23 copy number increases in 3520 tumours representing 166 different tumour types (Andersen et al, 2002). The results confirmed previous data and indicated that increased 17q23 copy number occurs most commonly in brain, lung, breast, ovarian, urinary bladder, and soft tissue tumours, although high-level amplification was observed exclusively in breast cancer. Importantly, the increased copy number of 17q23 region has also been associated with tumour progression (Andersen et al, 2002) and with poor prognosis in breast cancer (Isola et al, 1995; Barlund et al, 2000a), ovarian clear cell adenocarcinoma (Hirasawa et al, 2003), neuroblastoma (Saito-Ohara et al, 2003), and acute myelogenous leukaemia (Morerio et al, 2001). Taken together, these data suggest that the genes affected by the 17q23 amplification contribute to cancer pathogenesis.
In breast cancer, several studies have been performed to define the limits of the 17q23 amplicon (Barlund et al, 1997, 2000b; Couch et al, 1999; Erson et al, 2001; Monni et al, 2001; Wu et al, 2001). Based on the combination of data from these studies and the current information available in the human genome databases (http://www.ncbi.nlm.nih.gov/mapview/ and http://www.ensembl.org/Homo_sapiens/index.html), the amplicon is considerably large covering an approximately 5 Mb region at 17q23. A number of studies have also aimed to uncover the possible target genes for this amplification, mainly by looking at correlation between amplification and high-level mRNA expression. One of the first genes to be identified as a potential oncogene in this region was the ribosomal protein S6 kinase, RPS6KB1 (Couch et al, 1999; Barlund et al, 2000a). Thereafter several other putative target genes have been proposed including APPBP2 (also known as PAT1), RAD51C, TBX2, TRIM37 (MUL), THRAP1 (TRAP240), PPM1D, and BRIP1 (Barlund et al, 2000b; Wu et al, 2000, 2001; Erson et al, 2001; Monni et al, 2001; Bulavin et al, 2002; Li et al, 2002). However, these studies were carried out at a time when the genome sequence of this region was incomplete and thus all genes within this region had not been identified. In addition, these previous studies mainly concentrated on the analysis of established breast cancer cell lines and the expression levels of only five genes from this region have been evaluated in a small number of primary breast tumours (Couch et al, 1999; Barlund et al, 2000b; Li et al, 2002).
In the present study, we aimed to further characterise the molecular consequences of the 17q23 amplification on gene expression levels in primary breast tumours. To this end, all known genes localised within the 5 Mb amplicon at 17q23 were obtained from publicly available databases and their expression levels were measured in 26 primary breast tumours using quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR). Our data revealed that high-level amplification at 17q23 in primary breast cancer leads to coordinated overexpression of 11 adjacent genes located at a 1.56 Mb central region of the amplicon.
MATERIALS AND METHODS
Primary tumours and breast cancer cell lines
Freshly frozen primary breast tumour specimens and corresponding paraffin-embedded tissue samples from 26 breast cancer patients were acquired from the Department of Pathology, Tampere University Hospital. These samples represent a subset of a larger material that has been described in detail previously (Rauta et al, 2006). The clinicopathological characteristics of these 26 tumour samples are presented in Table 1. The mean age of the patients at diagnosis was 62 years (range 38–84). The patients had not received any therapy before tumour removal. The use of these specimens in this study was approved by the Ethics Committee of the Pirkanmaa Hospital District and by the National Authority for Medicolegal Affairs in Finland.
Table 1. Clinicopathological characteristics of 26 primary breast tumours.
| Variable | n | % |
|---|---|---|
| Tumour type | ||
| Infiltrating ductal | 22 | 84.6 |
| Infiltrating lobular | 4 | 15.4 |
| Tumour size | ||
| <20 mm | 12 | 46.2 |
| ⩾20 mm | 14 | 53.8 |
| Histological grade a | ||
| I | 0 | 0.0 |
| II | 8 | 36.4 |
| III | 14 | 63.6 |
| Stage | ||
| T1 | 11 | 42.3 |
| T2 | 5 | 19.2 |
| T3 | 4 | 15.4 |
| T4 | 6 | 23.1 |
| Nodal status | ||
| Negative | 12 | 46.2 |
| Positive | 11 | 42.3 |
| Not available | 3 | 11.5 |
| ER | ||
| Positive | 21 | 81.8 |
| Negative | 5 | 19.2 |
| PR | ||
| Positive | 13 | 50.0 |
| Negative | 13 | 50.0 |
| ERBB2 | ||
| Negative | 21 | 80.8 |
| Positive | 5 | 19.2 |
Histological grade was determined only from the infiltrating ductal carcinomas.
Two breast cancer cell lines BT-474 and MCF7 were included in this study. Pancreatic cancer cell lines PANC-1 and HUPT3 as well as prostate cancer cell lines DU145 and PC-3 were used as controls in RT-PCR analyses. HUPT3 cell line came from the German Collection of Micro-organisms and Cell Cultures (Braunschweig, Germany), all other cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cell lines were cultured under recommended conditions. Normal testis and pancreas cDNA (Ambion, Foster City, CA, USA) were also used as controls in RT-PCR analyses. Human Mammary Gland (HMG) cDNA was from BD Biosciences Clontech (Palo Alto, CA, USA).
Copy number analysis by fluorescence in situ hybridisation
Three pairs of bacterial artificial chromosome (BAC) clones corresponding to the 17q23 chromosome region (probe set A: RP11-579A4 and RP11-579O24; probe set B: RP11-634F5 and RP11-1081E4; probe set C: RP11-269G24 and CTD-2501B8) were identified from public databases (http://www.ncbi.nlm.nih.gov/mapview/ and http://www.ensembl.org). BAC DNA was labelled with SpectrumOrange-dUTP (Vysis, Downers Grove, IL, USA) by random priming and a SpectrumGreen labelled chromosome 17 probe (Vysis) was used as a reference. Fluorescence in situ hybridisation (FISH) to normal metaphase chromosomes was performed to confirm that the clone contigs recognised a single copy target at 17q23. Fluorescence in situ hybridisation on paraffin-embedded breast tumour samples was performed using a modified Paraffin Pretreatment Reagent kit protocol (Vysis) as described previously (Rauta et al, 2006). Hybridisation signals were analysed using an Olympus BX50 epifluorescence microscope (Tokyo, Japan) using an × 60 objective (NA 1.4). Specimens containing a three to fivefold increase in the number of gene specific probe signals, as compared with the chromosome 17 centromere signals, were considered to be moderately amplified. In addition, specimens containing a fivefold or higher increase in the number of PPM1D signals or tight clusters of signals were considered to be highly amplified. The results using the RP11-634F5 and RP11-1081E4 BAC contigs were included in a previous publication (Rauta et al, 2006).
RNA isolation and cDNA synthesis
Total RNA was extracted from cancer cell lines using TRIZOL Reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. For primary tumour samples, a representative tumour area was selected from freshly frozen tumour specimen based on haematoxylin–eosin stained tissue section and a core-biopsy (∅2 mm) was obtained for RNA isolation. Tumour tissues were homogenised with a syringe and a needle (20G, ∅0.9 mm) and total RNA was extracted with RNeasy Mini Kit (Qiagen, Valencia, CA, USA). First-strand cDNA synthesis for both cell line and primary tumour RNAs was performed using SuperScript II reverse transcriptase and random hexamer primers (Invitrogen).
Expression screen by RT-PCR
Gene specific primers for 29 genes from the 17q23 region (primer sequences are available on request) were obtained from TIB MolBiol (Berlin, Germany). The PCR contained 1 × PCR Gold buffer (Applied Biosystems, Foster city, CA, USA), 1.5 mM MgCl2, 0.2 mM dNTPs each, 0.2–0.4 mM gene specific primers, 1.5–2.5 units of Amplitaq Gold DNA polymerase (Applied Biosystems), and 1 μl cDNA template (MCF7 or BT474) adjusted to 50 μl with sterile H2O. The PCR programme began with initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 60 s and elongation at 72°C for 60 s, with final elongation at 72°C for 10 min. The PCR products were run on a 1.5% agarose gel. For those genes with no detectable expression in MCF7 and BT-474 cells, the functionality of the PCR primers was verified using cDNA from PANC-1, HUPT3, DU145, PC-3, normal testis or normal pancreas as a template.
Quantitative real-time RT-PCR
DNA Hybridisation Probe Sets for 24 genes at 17q23 and the housekeeping gene TATA-box-binding protein (TBP) were obtained from TIB MolBiol (Berlin, Germany). The PCR reactions were performed in the LightCycler apparatus using the LC FastStart DNA Hybridisation Probes Kit according to manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Briefly, the PCR contained 1 × LightCycler FastStart DNA Master HybProbe mix (Roche Applied Science, Mannheim, Germany), 4 mM MgCl2, 0.2–0.4 μM gene specific probes, 0.8 μM gene specific primers, and 1.5 μl cDNA template adjusted to 20 μl with sterile H2O. After 10 min of initial denaturation at 95°C, the cycling conditions (55 cycles) were as follows: denaturation at 95°C for 10 s, annealing at 55–58°C for 10–20 s, and elongation at 72°C for 6–12 s depending on the gene of interest. Quantitative analysis was performed using the LightCycler software according to the fit-point method as described earlier (Kauraniemi et al, 2003). The expression levels were normalised against the housekeeping gene TBP.
Statistical analysis
For each gene, two-tailed Mann–Whitney U-test was used to examine the possible statistical significance of differences in expression levels between the three tumour groups. Three separate comparisons (high vs moderate, high vs no, and moderate vs no amplification tumour group) were made.
RESULTS
Primary tumour selection and copy number analysis
We had previously determined the DNA copy number levels within the centre of the 17q23 amplicon (at the PPM1D gene locus) in a set of 146 primary breast tumours by using formalin-fixed, paraffin-embedded samples (Rauta et al, 2006). On the basis of these data, a total of 26 tumours, including eight cases with increased copy number and 18 tumours with no copy number increase, were selected for this study owing to the availability of freshly frozen tumour material required for the mRNA expression analyses. To determine further the 17q23 amplification status in this set of samples, we used additional probe pairs from both ends of the 5 Mb amplicon and determined their copy number levels by FISH (Table 2). As might be expected, the 18 non-amplified tumours did not show copy number increase with the additional probe sets either (data not shown). The eight amplified cases demonstrated more or less consistent copy number changes across the entire amplicon and could now be classified into groups with high (n=4) and moderate (n=4) level of amplification (Table 2).
Table 2. Copy number levels at the 17q23 amplicon relative to chromosome 17 centromere in eight primary breast tumours by FISH.
|
Probe setsa
|
|||
|---|---|---|---|
| Tumour number | Ab | Bc | Cd |
| High | |||
| 133 | 1.5 | Ampe | 3 |
| 206 | Ampe | Ampe | 2.5 |
| 209 | Ampe | Ampe | Ampe |
| 236 | 1 | >5 | 1 |
| Moderate | |||
| 249 | 1.5 | 3.5 | 1 |
| 251 | 3 | 3.5 | 3.5 |
| 255 | 2 | 3 | 2 |
| 256 | 3.5 | 3.5 | 3 |
FISH=fluorescence in situ hybridisation.
See Materials and Methods.
Includes RAD51C-PPM1E genes.
Includes PPM1D gene.
Includes CYB561-ACE-KCNH6-WDR68 genes.
Tight cluster of signals indicative of amplification.
Transcript identification
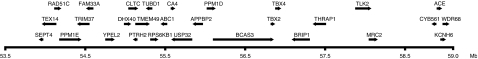
On the basis of previous studies (Barlund et al, 1997, 2000b; Couch et al, 1999; Erson et al, 2001; Monni et al, 2001; Wu et al, 2001) and the current genome sequence information (http://www.ncbi.nlm.nih.gov/mapview/ and http://www.ensembl.org/Homo_sapiens/index.html), the 17q23 amplicon is restricted to an ∼5 Mb region (53.95–59.02 Mb) and contains 29 genes with known function (Figure 1, Table 3). First, the expression levels of these 29 genes were screened by regular RT-PCR in MCF7 and BT-474 breast cancer cell lines that are known to harbour high level 17q23 amplification (Couch et al, 1999; Barlund et al, 2000a; Monni et al, 2001). Five (SEPT4, TEX14, CA4, MRC2, and KCNH6) of the 29 genes had either very low or undetectable expression in both MCF7 and BT-474 cells (data not shown) and were thus excluded from further analyses based on the fact that a potential amplification target gene is expected to show elevated mRNA expression.
Figure 1.
Physical map of the 17q23 amplicon. The known genes mapping to the ∼5 Mb minimal region of amplification at 17q23 are represented using horizontal lines and their orientation is indicated with arrowheads.
Table 3. List of the 29 known genes located in the 5 Mb minimal region of amplification at 17q23 and their minimum, maximum and median expression levels in 26 primary breast tumours.
| Gene | Description | Min | Max | Median |
|---|---|---|---|---|
| SEPT4 | Peanut-like protein 2 | ND | ND | ND |
| TEX14 | Testis expressed sequence 14 | ND | ND | ND |
| RAD51C | DNA repair protein RAD51 homolog | 0.0 | 1.9 | 0.3 |
| PPM1E | Protein phosphatase 1E | 0.0 | 1.7 | 0.0 |
| TRIM37 | Tripartite motif-containing 37 protein | 0.0 | 1.1 | 0.2 |
| FAM33A | Family with sequence similarity 33, member A | 0.0 | 3.3 | 0.3 |
| YPEL2 | Yippee-like 2 protein | 0.1 | 1.0 | 0.3 |
| DHX40 | DEAH (Asp-Glu-Ala-His) box polypeptide 40 | 0.0 | 4.5 | 0.2 |
| CLTC | Clathrin heavy chain 1 | 0.0 | 3.7 | 0.4 |
| PTRH2 | Peptidyl-tRNA hydrolase 2 | 0.1 | 14.8 | 1.4 |
| TMEM49 | Transmembrane protein 49 | 0.0 | 1.2 | 0.2 |
| TUBD1 | Tubulin δ chain | 0.0 | 2.4 | 0.3 |
| RPS6KB1 | Ribosomal protein S6 kinase | 0.0 | 1.0 | 0.2 |
| ABC1 | Amplified in breast cancer | 0.0 | 1.7 | 0.3 |
| CA4 | Carbonic anhydrase IV precursor | ND | ND | ND |
| USP32 | Ubiquitin C-terminal hydrolase 32 | 0.0 | 1.5 | 0.3 |
| APPBP2 | Amyloid β precursor protein binding protein 2 | 0.0 | 1.6 | 0.1 |
| PPM1D | Protein phosphatase 2C δ isoform, magnesium-dependent | 0.0 | 2.8 | 0.2 |
| BCAS3 | Breast carcinoma amplified sequence 3 | 0.1 | 2.7 | 0.3 |
| TBX2 | T-box transcription factor TBX2 | 0.0 | 0.7 | 0.1 |
| TBX4 | T-box transcription factor TBX4 | 0.0 | 9.0 | 0.3 |
| BRIP1 | BRCA1 interacting protein C-terminal helicase 1 | 0.0 | 2.1 | 0.2 |
| THRAP1 | Thyroid hormone receptor-associated protein | 0.0 | 1.1 | 0.2 |
| TLK2 | Serine/threonine-protein kinase tousled-like 2 | 0.1 | 1.6 | 0.3 |
| MRC2 | Mannose receptor, C type 2 | ND | ND | ND |
| CYB561 | Cytochrome b561 | 0.3 | 9.2 | 1.3 |
| ACE | Angiotensin-converting enzyme, somatic isoform precursor | 4.0 | 233.0 | 32.1 |
| KCNH6 | Potassium voltage-gated channel, subfamily H, member 6 | ND | ND | ND |
| WRD68 | WD-repeat protein 68 | 0.1 | 1.8 | 0.8 |
ND=not determined (low or now expression in breast cancer cell lines MCF7 and BT-474 with high-level amplification).
Expression screen in primary breast tumours
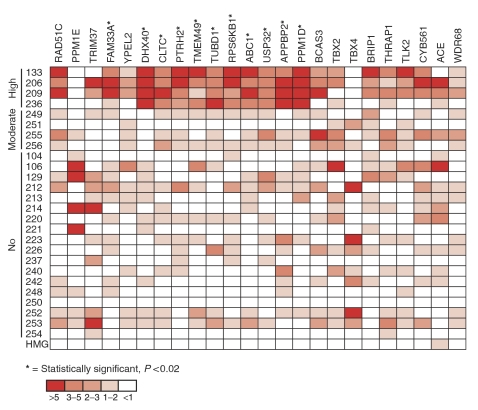
The expression levels of the remaining 24 genes from the 17q23 amplicon were then measured in the 26 primary breast tumours using qRT-PCR. Two genes, PTRH2 and ACE, showed considerably higher expression levels than any other gene within this region (Table 3). However, ACE was also expressed at a similar level in normal HMG, and therefore does not appear to be interesting as putative target gene for 17q23 amplification. The expression levels of PPM1E were very low across all 26 tumour samples and, additionally, both PPM1E and TBX4 showed highest expression levels in non-amplified tumour samples (Table 3, Figure 2).
Figure 2.
Schematic representation of expression levels of 24 known genes within the 17q23 chromosomal region in 26 primary breast tumours and normal HMG. Primary tumours are arranged according to their 17q23 amplification status and genes are organised based on their physical order at the 17q23 region from centromere to telomere. Expression levels were determined using qRT-PCR and were normalised against a housekeeping gene TBP. The relative expression values of each gene were median-corrected and displayed using a colour code (shown at the bottom).
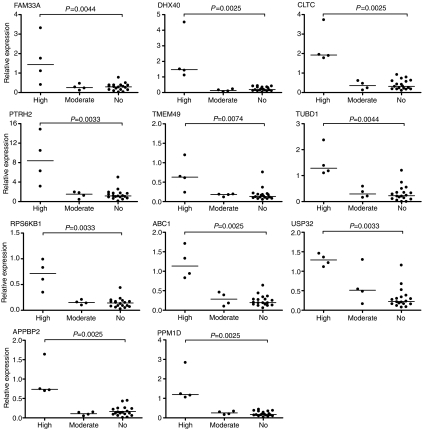
Finally, the expression levels of the 24 genes within the 17q23 amplicon were compared between the three tumour groups. A statistically significant (P<0.01) difference between the high level and no amplification groups were observed for a set of 11 genes (FAM33A, DHX40, CLTC, PTRH2, TMEM49, TUBD1, RPS6KB1, ABC1, USP32, APPBP2, and PPM1D) (Figure 3). All of these, except FAM33A and USP32, also showed significant difference (P<0.05) in expression levels between the high-and moderate-level amplification groups suggesting that these genes are activated by high-level copy number increases (Figure 3). No difference between the moderate and the no amplification tumour groups were observed. Interestingly, these 11 genes are located adjacent to each other within a 1.56 Mb region (54.54–56.10 Mb) at the centre of the 17q23 amplicon (Figures 1 and 2). It was even more intriguing that only a single gene, YPEL2, within this region did not demonstrate an association between amplification and increased mRNA expression.
Figure 3.
Comparison of FAM33A, DHX40, CLTC, PTRH2, TMEM49, TUBD1, RPS6KB1, ABC1, USP32, APPBP2, and PPM1D expression in highly, moderately, and no amplification tumour groups by qRT-PCR. Median value of expression is indicated by a horizontal line. Two-tailed Mann–Whitney U-test was used to assess the statistical significance of differences in expression levels between tumour groups. P-values between highly non-amplified groups are shown.
DISCUSSION
Several studies have illustrated recurrent amplification of 17q23 in various human tumours and its association to poor clinical outcome (Isola et al, 1995; Barlund et al, 2000a; Morerio et al, 2001; Andersen et al, 2002; Hirasawa et al, 2003; Saito-Ohara et al, 2003). These studies have proposed multiple genes from this region to be important for cancer development and progression and thus promising targets for diagnostic, prognostic, and therapeutic approaches. However, no systematic survey of all genes within the 17q23 region has earlier been performed in primary tumour material. Here 29 known genes located within the common amplified segment at 17q23 were defined. Five of these were excluded from further analysis, because they showed very low or undetectable expression in cell lines with high level amplification. The expression levels of the remaining 24 genes were then examined using qRT-PCR in 26 primary breast tumours to assess their role as putative target genes for 17q23 amplification in breast cancer.
Our systematic study revealed eleven genes (FAM33A, DHX40, CLTC, PTRH2, TMEM49, TUBD1, RPS6KB1, ABC1, USP32, APPBP2, PPM1D) that showed significantly (P<0.01) higher expression levels in primary breast tumours with high level 17q23 amplification compared to tumours without amplification. Interestingly, these eleven genes are all located within a 1.56 Mb region at the centre of the 17q23 amplicon. The amplicon core also includes one additional gene, YPEL2, but for unknown reasons it does not demonstrate copy number dependent overexpression. As only very little is known about the function of this gene, it is not possible to speculate on the basis of this phenomenon. Overall, our findings are in good agreement with previous studies that have implicated gene copy number alterations as significant determinants of gene expression patterns (Hyman et al, 2002; Pollack et al, 2002; Wolf et al, 2004; Fridlyand et al, 2006). However, it has to be noted that the tumour group with moderate amplification showed expression levels similar to those seen in the non-amplified tumours, indicating that low level copy number increases at this region did not have a significant effect on gene expression levels. This finding is supported by recent studies demonstrating that the high-level amplifications but not copy number gains are associated with poor prognosis in various tumour types (Sticht et al, 2005; Staebler et al, 2006). In summary, the qRT-PCR screen demonstrated that high level amplification at 17q23 in breast cancer leads to concomitant overexpression of virtually all genes within the amplicon core.
Several genes have earlier been proposed as putative target genes of the 17q23 amplification. These include RPS6KB1, APPBP2, and PPM1D (Couch et al, 1999; Barlund et al, 2000a, 2000b; Wu et al, 2001; Bulavin et al, 2002; Li et al, 2002) that also showed strong association between amplification and increased expression in our primary tumour material. Earlier studies have also implicated RAD51C, TRIM37, TBX2, THRAP1, and BRIP1 as genes whose expression correlates with amplification (Barlund et al, 2000b; Wu et al, 2000, 2001; Erson et al, 2001; Monni et al, 2001). In our tumour series, these genes did also show elevated expression in some samples with high level amplification, but the difference in expression levels between the high and no amplification tumour groups, did not reach statistical significance. This finding might reflect the fact that we focussed solely on the analysis of primary tumour material instead of established breast cancer cell lines that were mainly used in these previous studies. Finally, our data revealed seven other protein coding genes (FAM33A, DHX40, CLTC, PTRH2, TMEM49, TUBD1, ABC1, and USP32) that have not been implicated previously in 17q23 studies, but whose expression was clearly associated with copy number status in primary breast tumours. Taken together, our results highlight a set of new genes that are overexpressed because of the 17q23 amplification in breast cancer and thus might contribute to the tumour phenotype.
Similar observations on overexpression of multiple genes within an amplicon have been reported previously in several occasions. For example, recent studies identified a 1 Mb segment of common amplification at 8p11–12 in breast cancer and demonstrated that it contains at least 14 candidate genes showing a strong correlation between amplification and overexpression (Garcia et al, 2005; Gelsi-Boyer et al, 2005). At 11q13, which is amplified in a wide variety of tumours (Saunders et al, 2000; Gollin, 2001, 2004), Huang et al (2006) constructed a detailed 3.6 Mb map of the amplicon core and showed that most of the genes in that region are overexpressed in amplified tumours and cell lines. Similarly, we have demonstrated that amplification of a 280 kb minimal common region at 17q12 in breast cancer leads to simultaneous increase of expression levels of all genes within that genomic segment (Kauraniemi et al, 2001). In general, these data implicate that despite the size of the affected region, the vast majority of the genes within amplicons are overexpressed and hence amplification events typically lead to overexpression of multiple genes. Owing to this tight association between amplification and elevated gene expression, it is difficult to determine which of the genes within particular amplicon actually contribute to tumour pathogenesis. It is likely that all of the overexpressed genes are not critical for cancer progression but are overexpressed merely because of their location within the amplicon. However, it is also possible that, instead of a single target gene, amplicons are driven by a set of genes whose simultaneous overexpression provides growth advantage to cancer cells (Huang et al, 2006; Kao and Pollack, 2006).
Traditionally, the search for amplification target genes has been based on two primary schemes; namely location within the maximal amplitude of the amplicon and overexpression of the putative target gene. Our results demonstrate that a whole set of genes, 11 of the 12 known genes within the 1.56 Mb core region, is overexpressed in primary breast tumours with high level amplification at 17q23. Owing to such coordinated overexpression of genes in the amplification core, overexpression alone is not sufficient to highlight putative target genes but functional studies are required to uncover the possible biological and clinical significance of candidate genes.
Acknowledgments
We thank Ms Kati Rouhento for excellent technical assistance. This study was supported in part by a grant from the Academy of Finland (grant no. 207003).
References
- Andersen CL, Monni O, Wagner U, Kononen J, Barlund M, Bucher C, Haas P, Nocito A, Bissig H, Sauter G, Kallioniemi A (2002) High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am J Pathol 161: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlund M, Forozan F, Kononen J, Bubendorf L, Chen Y, Bittner ML, Torhorst J, Haas P, Bucher C, Sauter G, Kallioniemi OP, Kallioniemi A (2000a) Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J Natl Cancer Inst 92: 1252–1259 [DOI] [PubMed] [Google Scholar]
- Barlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, Kallioniemi O, Kallioniemi A (2000b) Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res 60: 5340–5344 [PubMed] [Google Scholar]
- Barlund M, Tirkkonen M, Forozan F, Tanner MM, Kallioniemi O, Kallioniemi A (1997) Increased copy number at 17q22–q24 by CGH in breast cancer is due to high-level amplification of two separate regions. Genes Chromosomes Cancer 20: 372–376 [DOI] [PubMed] [Google Scholar]
- Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz CC, Dantis L, Sklarin NT, Seidman AD, Hudis CA, Moore J, Rosen PP, Twaddell T, Henderson IC, Norton L (1999) Phase II study of weekly intravenous trastuzumab (Herceptin) in patients with HER2/neu-overexpressing metastatic breast cancer. Semin Oncol 26: 78–83 [PubMed] [Google Scholar]
- Brinkschmidt C, Christiansen H, Terpe HJ, Simon R, Boecker W, Lampert F, Stoerkel S (1997) Comparative genomic hybridization (CGH) analysis of neuroblastomas–an important methodological approach in paediatric tumour pathology. J Pathol 181: 394–400, doi:10.1002/SICI)1096-9896(199704)181:4〈394∷AID-PATH800〉3.0.CO;2-1 [DOI] [PubMed] [Google Scholar]
- Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, Kallioniemi A, Fornace Jr AJ, Appella E (2002) Amplification of PPM1D in human tumours abrogates p53 tumour-suppressor activity. Nat Genet 31: 210–215, doi:10.1038/ng894 [DOI] [PubMed] [Google Scholar]
- Clark J, Edwards S, John M, Flohr P, Gordon T, Maillard K, Giddings I, Brown C, Bagherzadeh A, Campbell C, Shipley J, Wooster R, Cooper CS (2002) Identification of amplified and expressed genes in breast cancer by comparative hybridization onto microarrays of randomly selected cDNA clones. Genes Chromosomes Cancer 34: 104–114, 10.1002/gcc.10039 [DOI] [PubMed] [Google Scholar]
- Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ (1999) Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 17: 2639–2648 [DOI] [PubMed] [Google Scholar]
- Couch FJ, Wang XY, Wu GJ, Qian J, Jenkins RB, James CD (1999) Localization of PS6K to chromosomal region 17q23 and determination of its amplification in breast cancer. Cancer Res 59: 1408–1411 [PubMed] [Google Scholar]
- Erson AE, Niell BL, DeMers SK, Rouillard JM, Hanash SM, Petty EM (2001) Overexpressed genes/ESTs and characterization of distinct amplicons on 17q23 in breast cancer cells. Neoplasia 3: 521–526, doi:10.1038/sj/neo/7900187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, McLennan J, Ziegler J, Chin K, Devries S, Feiler H, Gray JW, Waldman F, Pinkel D, Albertson DG (2006) Breast tumour copy number aberration phenotypes and genomic instability. BMC Cancer 6: 96, doi:10.1186/1471-2407-6-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, Vias M, Kranjac T, Subkhankulova T, Paish C, Ellis I, Brenton JD, Edwards PA, Caldas C (2005) A 1 Mb minimal amplicon at 8p11–12 in breast cancer identifies new candidate oncogenes. Oncogene 24: 5235–5245, doi:10.1038/sj.onc.1208741 [DOI] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, Lasorsa L, Letessier A, Ginestier C, Monville F, Esteyries S, Adelaide J, Esterni B, Henry C, Ethier SP, Bibeau F, Mozziconacci MJ, Charafe-Jauffret E, Jacquemier J, Bertucci F, Birnbaum D, Theillet C, Chaffanet M (2005) Comprehensive profiling of 8p11–12 amplification in breast cancer. Mol Cancer Res 3: 655–667, doi:10.1158/1541-7786.MCR-05-0128 [DOI] [PubMed] [Google Scholar]
- Gollin SM (2001) Chromosomal alterations in squamous cell carcinomas of the head and neck: window to the biology of disease. Head Neck 23: 238–253 [DOI] [PubMed] [Google Scholar]
- Gollin SM (2004) Chromosomal instability. Curr Opin Oncol 16: 25–31 [DOI] [PubMed] [Google Scholar]
- Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I (2003) Association of 17q21–q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res 9: 1995–2004 [PubMed] [Google Scholar]
- Huang X, Godfrey TE, Gooding WE, McCarty Jr KS, Gollin SM (2006) Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosomes Cancer 45: 1058–1069, doi:10.1002/gcc.20371 [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, Kallioniemi OP, Kallioniemi A (2002) Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 62: 6240–6245 [PubMed] [Google Scholar]
- Isola JJ, Kallioniemi OP, Chu LW, Fuqua SA, Hilsenbeck SG, Osborne CK, Waldman FM (1995) Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol 147: 905–911 [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM (1994) Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 91: 2156–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao J, Pollack JR (2006) RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosomes Cancer 45: 761–769, doi:10.1002/gcc.20339 [DOI] [PubMed] [Google Scholar]
- Kauraniemi P, Barlund M, Monni O, Kallioniemi A (2001) New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res 61: 8235–8240 [PubMed] [Google Scholar]
- Kauraniemi P, Kuukasjarvi T, Sauter G, Kallioniemi A (2003) Amplification of a 280-kilobase core region at the ERBB2 locus leads to activation of two hypothetical proteins in breast cancer. Am J Pathol 163: 1979–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius VM, Vidgren V, Zhu Y (1998) DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 152: 1107–1123 [PMC free article] [PubMed] [Google Scholar]
- Korn WM, Oide Weghuis DE, Suijkerbuijk RF, Schmidt U, Otto T, du Manoir S, Geurts van Kessel A, Harstrick A, Seeber S, Becher R (1996) Detection of chromosomal DNA gains and losses in testicular germ cell tumours by comparative genomic hybridization. Genes Chromosomes Cancer 17: 78–87, doi:10.1002/(SICI)1098-2264(199610)17:2〈78∷AID-GCC2〉3.0.CO;2-Y [DOI] [PubMed] [Google Scholar]
- Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, Gabriele T, McCurrach ME, Marks JR, Hoey T, Lowe SW, Powers S (2002) Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet 31: 133–134, doi:10.1038/ng888 [DOI] [PubMed] [Google Scholar]
- Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A (1997) Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer 18: 59–65 [PubMed] [Google Scholar]
- Monni O, Barlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, Kallioniemi A (2001) Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA 98: 5711–5716, doi:10.1073/pnas.091582298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morerio C, Russo I, Rosanda C, Rapella A, Leszl A, Basso G, Maserati E, Pasquali F, Panarello C (2001) 17q21-qter trisomy is an indicator of poor prognosis in acute myelogenous leukemia. Cancer Genet Cytogenet 124: 12–15 [DOI] [PubMed] [Google Scholar]
- Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B (1994) Detection of DNA amplification in 17 primary breast carcinomas with homogeneously staining regions by a modified comparative genomic hybridization technique. Genes Chromosomes Cancer 10: 160–170 [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO (2002) Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumours. Proc Natl Acad Sci USA 99: 12963–12968, doi:10.1073/pnas.162471999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauta J, Alarmo EL, Kauraniemi P, Karhu R, Kuukasjarvi T, Kallioniemi A (2006) The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours. Breast Cancer Res Treat 95: 257–263, doi:10.1007/s10549-005-9017-7 [DOI] [PubMed] [Google Scholar]
- Richter J, Jiang F, Gorog JP, Sartorius G, Egenter C, Gasser TC, Moch H, Mihatsch MJ, Sauter G (1997) Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res 57: 2860–2864 [PubMed] [Google Scholar]
- Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schrock E, du Manoir S, Cremer T (1994) Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res 54: 1801–1806 [PubMed] [Google Scholar]
- Ross JS, Fletcher JA (1999) The HER-2/neu oncogene: prognostic factor, predictive factor and target for therapy. Semin Cancer Biol 9: 125–138, doi:10.1006/scbi.1998.0083 [DOI] [PubMed] [Google Scholar]
- Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J (2003) PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res 63: 1876–1883 [PubMed] [Google Scholar]
- Saunders WS, Shuster M, Huang X, Gharaibeh B, Enyenihi AH, Petersen I, Gollin SM (2000) Chromosomal instability and cytoskeletal defects in oral cancer cells. Proc Natl Acad Sci USA 97: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M (1993) Amplification of N-myc as a prognostic marker for patients with neuroblastoma. Semin Cancer Biol 4: 13–18 [PubMed] [Google Scholar]
- Schwendel A, Langreck H, Reichel M, Schrock E, Ried T, Dietel M, Petersen I (1997) Primary small-cell lung carcinomas and their metastases are characterized by a recurrent pattern of genetic alterations. Int J Cancer 74: 86–93 [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Wallrapp C, Muller-Pillasch F, Bentz M, Gress T, Lichter P (1996) Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res 56: 3803–3807 [PubMed] [Google Scholar]
- Sonoda G, Palazzo J, du Manoir S, Godwin AK, Feder M, Yakushiji M, Testa JR (1997) Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24, and 20q13 in human ovarian carcinomas. Genes Chromosomes Cancer 20: 320–328 [PubMed] [Google Scholar]
- Staebler A, Karberg B, Behm J, Kuhlmann P, Neubert U, Schmidt H, Korsching E, Burger H, Lelle R, Kiesel L, Bocker W, Shih IEM, Buchweitz O (2006) Chromosomal losses of regions on 5q and lack of high-level amplifications at 8q24 are associated with favorable prognosis for ovarian serous carcinoma. Genes Chromosomes Cancer 45: 905–917, doi:10.1002/gcc.20356 [DOI] [PubMed] [Google Scholar]
- Sticht C, Hofele C, Flechtenmacher C, Bosch FX, Freier K, Lichter P, Joos S (2005) Amplification of Cyclin L1 is associated with lymph node metastases in head and neck squamous cell carcinoma (HNSCC). Br J Cancer 92: 770–774, doi:10.1038/sj.bjc.6602400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, Van Roy N, Van Gele M, Laureys G, Ambros P, Heimann P, Devalck C, Schuuring E, Brock P, Otten J, Gyselinck J, De Paepe A, Speleman F (1998) Genetic heterogeneity of neuroblastoma studied by comparative genomic hybridization. Genes Chromosomes Cancer 23: 141–152 [DOI] [PubMed] [Google Scholar]
- Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, Shak S, Stewart SJ, Press M (2002) Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 20: 719–726 [DOI] [PubMed] [Google Scholar]
- Weber RG, Bostrom J, Wolter M, Baudis M, Collins VP, Reifenberger G, Lichter P (1997) Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA 94: 14719–14724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis S, Hutchins AM, Hammet F, Ciciulla J, Soo WK, White D, van der Spek P, Henderson MA, Gish K, Venter DJ, Armes JE (2003) Detailed gene copy number and RNA expression analysis of the 17q12–23 region in primary breast cancers. Genes Chromosomes Cancer 36: 382–392, doi:10.1002/gcc.10138 [DOI] [PubMed] [Google Scholar]
- Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M, Elkahloun A, Monni O, Chen Y, Kallioniemi A, Kallioniemi OP (2004) High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia 6: 240–247, doi:10.1593/neo.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Sinclair C, Hinson S, Ingle JN, Roche PC, Couch FJ (2001) Structural analysis of the 17q22–23 amplicon identifies several independent targets of amplification in breast cancer cell lines and tumours. Cancer Res 61: 4951–4955 [PubMed] [Google Scholar]
- Wu GJ, Sinclair CS, Paape J, Ingle JN, Roche PC, James CD, Couch FJ (2000) 17q23 amplifications in breast cancer involve the PAT1, RAD51C, PS6K, and SIGma1B genes. Cancer Res 60: 5371–5375 [PubMed] [Google Scholar]