Abstract
In a previous study, we showed that levels of cell-free DNA (cfDNA) were significantly higher in sera of patients with hepatocellular carcinoma (HCC) associated with hepatitis C virus (HCV) than in sera of non-HCC patients with HCV. To confirm this finding, we analysed serum cfDNA levels in a cohort of 96 patients with HCV-related HCC and in 100 HCV carriers without known HCC. Again we found that serum cfDNA levels were significantly higher in HCC patients than in HCV carriers (115.9±98.3 vs 34.4±40.4 ng ml−1 (mean±s.d.), P<0.0001). Of 87 eligible patients who underwent curative hepatectomy, those with a high cfDNA level had a significantly shorter overall survival (OS) time than those in whom the cfDNA level was not high. Cox proportional hazards model showed the cfDNA level to be an independent prognostic factor for OS and cancer recurrence in distant organs. Our results suggest that the serum cfDNA level reflects the metastatic potential of HCV-related HCC and that it can be a useful predictive biomarker for distant metastasis after curative surgery.
Keywords: HCV, HCC, blood testing, cell-free DNA, real-time PCR, GSTP1
Hepatocellular carcinoma is one of the most common cancers, with an estimated 564 000 new cases registered worldwide in 2000 (Parkin et al, 2001), and it represents a major international health problem because its incidence is increasing in many countries (Deuffic et al, 1998; Llovet et al, 2003; El Serag, 2004). Particularly in Europe and North America, the incidences of HCC have increased markedly in the last decade and will increase further in the upcoming two decades due to hepatitis C virus (HCV) infection (Deuffic et al, 1998; El Serag, 2004). Despite much effort in HCC research, the prognosis of HCC remains poor because of both a high frequency of intrahepatic recurrence (IHR; Iizuka et al, 2003, 2004; Llovet et al, 2003) and high mortality of associated with extrahepatic recurrence (EHR) in distant organs (Itoh et al, 2002; Yang et al, 2007). Therefore, new non-invasive prognostic markers are urgently needed to improve the prognosis of HCC, especially of HCV-related HCC.
It may become possible to use serum biomarkers to screen for HCC patients at high risk for recurrence (Mann et al, 2007). To date, many candidate markers have been reported in relation to the clinical outcome of HCC patients (Marrero and Lok, 2004; Kuramitsu and Nakamura, 2006). In addition to these biomarkers, circulating cell-free DNA (cfDNA) has attracted a great deal of attention as an easy-to-use tool for evaluation of the malignant potential of cancer (Gautschi et al, 2004; Umetani et al, 2006). By means of real-time polymerase chain reaction (PCR) assay, we found that cfDNA levels were significantly higher in sera from patients with HCV-related HCC than in sera from HCV carriers without known HCC (Iizuka et al, 2006a). An additional intriguing finding was that cfDNA levels increased in parallel with tumour dedifferentiation and were positively associated with tumour size. Ren et al (2006) showed that levels of circulating cfDNA correlated inversely with the prognosis of HCC, in most cases attributable to hepatitis B virus (HBV) infection, suggesting that cfDNA may be a robust predictive marker for the prognosis of HBV-related HCC. However, a marker specific to HBV-related HCC may not be useful for HCV-related HCC because there are many differences in genetic changes and the clinical course between the two types of HCC (Iizuka et al, 2002; Llovet et al, 2003). We were prompted to examine whether cfDNA levels were predictive of outcomes in a larger cohort of patients with HCV-related HCC. Our present study showed for the first time in a large study group that the cfDNA level in the bloodstream can function as a predictor for overall survival (OS) and EHR in distant organs after curative hepatectomy in patients with HCV-related HCC.
MATERIALS AND METHODS
Patients and samples
Between April 1998 and August 2006, 96 patients underwent surgical treatment of HCC at Yamaguchi University Hospital. All were positive for HCV antibody. Clinical characteristics of these patients based on the TNM classification (Sobin and Wittekind, 2002) of the Union Internationale Contre le Cancer (UICC) are shown in Table 1. To explore the relation between serum cfDNA levels and patient outcomes, we excluded six patients who had any residual tumour, and three patients who died of other disease. As a result, the remaining 87 patients were subjected to the following study. Patients were followed up as described previously (median follow-up time: 39 months) (Iizuka et al, 2003; Matoba et al, 2005). In brief, all 87 patients were followed up at least once every 3 months postoperatively by routine X-ray, ultrasonography (US), computed tomography, or magnetic resonance imaging, levels of serum α-fetoprotein and protein induced by vitamin K absence II (PIVKA-II) were also measured. When tumour recurrence was suspected, CT angiography was included as a follow-up examination.
Table 1. Characteristics of patients and HCCs per cfDNA level.
|
cfDNA amount
|
|||
|---|---|---|---|
| Low (<117.8 ng ml−1) | High (⩾117.8 ng ml−1) | P-value | |
| Sex | 0.132 | ||
| Male (n=71) | 42 | 29 | |
| Female (n=25) | 19 | 6 | |
| Age (year) | 0.76 | ||
| <60 (n=18) | 12 | 6 | |
| >60 (n=78) | 49 | 29 | |
| Tumour size | 0.879 | ||
| <5 cm (n=76) | 48 | 28 | |
| >5 cm (n=20) | 13 | 7 | |
| Number of primary lesion | 0.299 | ||
| Single (n=56) | 38 | 18 | |
| Multiple (n=40) | 23 | 17 | |
| Venous invasion | 0.586 | ||
| Negative (n=69) | 45 | 24 | |
| Positive (n=27) | 16 | 11 | |
| Tumour differentiation | 0.155 | ||
| Well (G1) (n=24) | 18 | 6 | |
| Moderately (G2) (n=64) | 40 | 24 | |
| Poorly (G3) (n=8) | 3 | 5 | |
| UICC TNM stage | 0.217 | ||
| I (n=44) | 32 | 12 | |
| II (n=42) | 23 | 19 | |
| IIIA/IV (n=10) | 6 | 4 | |
cfDNA=cell-free DNA; HCV=hepatitis C virus; HCC=hepatocellular carcinoma.
To evaluate the high and low of cfDNA levels, we used a cutoff value of 117.8 ng ml−1, which is equal to the mean+2s.d. of the control value (value in HCV carriers without HCC).
For control, we used 100 serum samples from 100 HCV-positive patients with chronic liver disease who were recruited from outpatient clinics of the Yamaguchi University Graduate School of Medicine between July 2001 and October 2006. These control samples were selected to minimize the difference in age (66.3±7.3 vs 64.9±8.4 years (mean±s.d.), P=0.209) between the HCC group and the control group. Laboratory tests and imaging studies including US and CT did not reveal any HCC in the 100 HCV-positive patients during the median follow-up time of 18 months.
The study protocol was approved by the Institutional Review Board for the Use of Human Subjects at the Yamaguchi University School of Medicine, and written informed consent was obtained from each patient.
Extraction and quantification of DNA in sera
Blood samples were collected as described previously (Iizuka et al, 2006a). After clotting, which occurred within 1 h of collection, blood samples were spun at 3000 r.p.m. (1600 × g) for 10 min at room temperature. Sera were stored at −80°C until use. DNA was extracted from 1 ml of serum with a DNA Extractor SP Kit for Serum and Plasma (Wako Pure Chemical Industries Ltd, Osaka, Japan) according to the manufacturer's instructions. DNA was quantified as described previously (Iizuka et al, 2006a). Briefly, 1 μl of DNA solution was subjected to real-time PCR amplification for quantitative analysis of the GSTP1 gene. Finally, we calculated the amount of DNA on the basis of standard DNAs (leucocyte genomic DNAs) at 16–2000 ng ml−1.
Statistical analysis
Values are shown as mean±s.d. The Mann–Whitney U-test, Student's t-test and χ2 test were used to analyse differences in values between two groups, and analysis of variance was used to analyse differences between three groups. Overall survival and disease-free survival (DFS) were determined by the Kaplan–Meier method and analysed by log-rank test. The effect of eight clinicopathologic factors (sex, age, cfDNA level, tumour size, number of primary lesion, venous invasion, tumour differentiation grade and TNM stage) on OS and EHR in distant organs was assessed by means of the Cox proportional hazards model, hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated. Multivariate analysis was also performed to identify independent factors for early IHR by means of the stepwise logistic regression model. The eight above-mentioned clinicopathologic factors were also entered into a forward stepwise regression model. Each model was tested for goodness of fit by –2 log likelihood and χ2 in each step. All analyses were performed with SPSS 11.0J software (SPSS Inc., Chicago, IL, USA) run on a Windows computer. A P-value of less than 0.05 was considered statistically significant.
RESULTS
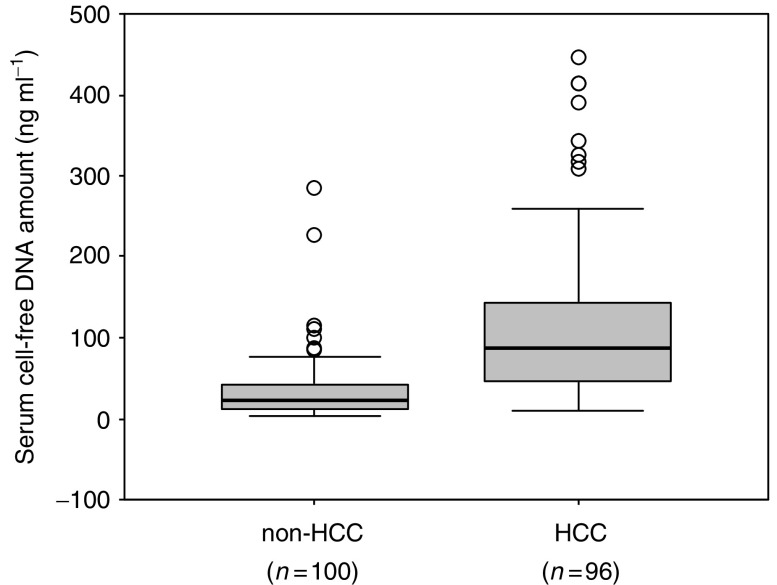
Serum cfDNA levels were significantly higher in HCC patients than in HCV carriers without known HCC (115.9±98.3 vs 34.4±40.4 ng ml−1 (mean±s.d.), P<0.0001 by Mann–Whitney U-test) (Figure 1). There was no significant difference in serum cfDNA levels between the 100 HCV carriers without known HCC and 18 healthy people or patients with benign disease who had no HCV infection (34.4±40.4 vs 45.8±22.6 ng ml−1); (data not shown). Serum cfDNA levels were not associated with any clinicopathologic factors in the total 96 HCC patients (Table 1).
Figure 1.
Box and whisker plot of cell-free DNA (cfDNA) levels in sera as determined by GSTP1-specific PCR assay. Levels of cfDNA in sera were significantly higher in hepatocellular carcinoma (HCC) patients than in hepatitis C virus (HCV) carriers (115.9±98.3 vs 34.4±40.4 ng ml−1 (mean±s.d.), P<0.0001 by Mann–Whitney U-test).
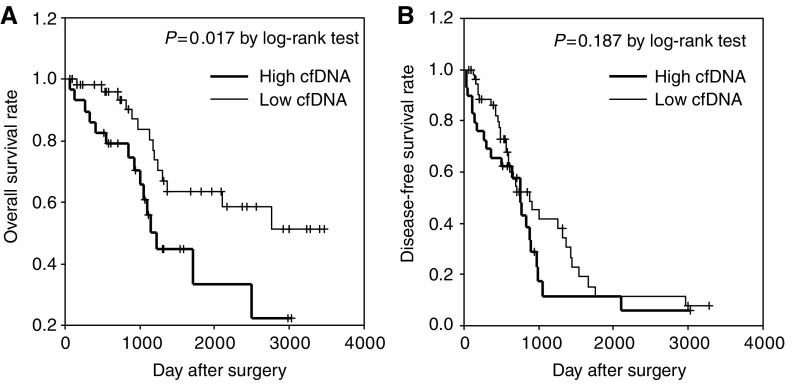
Of the 87 patients who underwent curative hepatectomy, 35 had no recurrence during the follow-up period and the remaining 52 had IHR. Of the 52 patients with IHR, 13 had EHR in distant organs such as lung and bone. Given the finding that cfDNA levels of HCV carrier without HCC are similar to those of controls without HCV infection, to examine the relation between cfDNA and follow-up data, we used a cfDNA cutoff value of 117.8 ng ml−1, which is equal to the mean+2 s.d. of the control value (value in HCV carriers without HCC). Patients with a high cfDNA level (n=29) had significantly shorter OS than those with a low cfDNA level (n=58) (P=0.017 by log-rank test; Figure 2A). By contrast, serum cfDNA levels were not associated with DFS (Figure 2B). According to the multivariate Cox proportional hazards model, cfDNA (HR, 3.4; 95% CI, 1.5–7.6; P=0.004) and tumour size (HR, 3.8; 95% CI, 1.7–8.5; P=0.001) were the only independent prognostic factors for OS (Table 2). In addition, cfDNA (HR, 4.5; 95% CI, 1.3–14.9; P=0.014) was the only independent prognostic factor for EHR in distant organs (Table 3).
Figure 2.
Relation between serum cell-free DNA (cfDNA) levels and overall and disease-free survival (DFS) in hepatocellular carcinoma (HCC) patients. (A) cfDNA levels and overall survival (OS). (B) cfDNA levels and DFS. Note that patients with a high cfDNA level had significantly shorter OS than did those with a low level of cfDNA (n=58) (P=0.017 by log-rank test). By contrast, serum cfDNA levels were not related to DFS.
Table 2. Independent risk factors for OS.
| Variable | HR (95% CI) | P-value |
|---|---|---|
| cfDNA | 0.004 | |
| Lowa | 1 | |
| Higha | 3.4 (1.5–7.6) | |
| Tumour size | 0.001 | |
| Less than 5 cm | 1 | |
| More than or equal to 5 cm | 3.8 (1.7–8.5) |
cfDNA=cell-free DNA; CI=confidence interval; HCC=hepatocellular carcinoma; HR=hazard ratio; OS=overall survival.
Low, less than 117.8 ng ml−1; high, more than or equal to 117.8 ng ml−1.
Table 3. Independent risk factors for cancer recurrence in distant organs.
| Variable | HR (95% CI) | P-value |
|---|---|---|
| cfDNA | 0.014 | |
| Lowa | 1 | |
| Higha | 4.5 (1.3–14.9) | |
| Tumour differentiation grade | 0.069 | |
| G1b | 1 | |
| G2+G3b | 2.5 (0.9–7.0) |
cfDNA=cell-free DNA; CI=confidence interval; HCC=hepatocellular carcinoma; HR=hazard ratio.
Low, less than 117.8 ng ml−1; high, more than or equal to 117.8 ng ml−1.
G1, well-differentiated HCC; G2, moderately differentiated HCC; G3, poorly differentiated HCC.
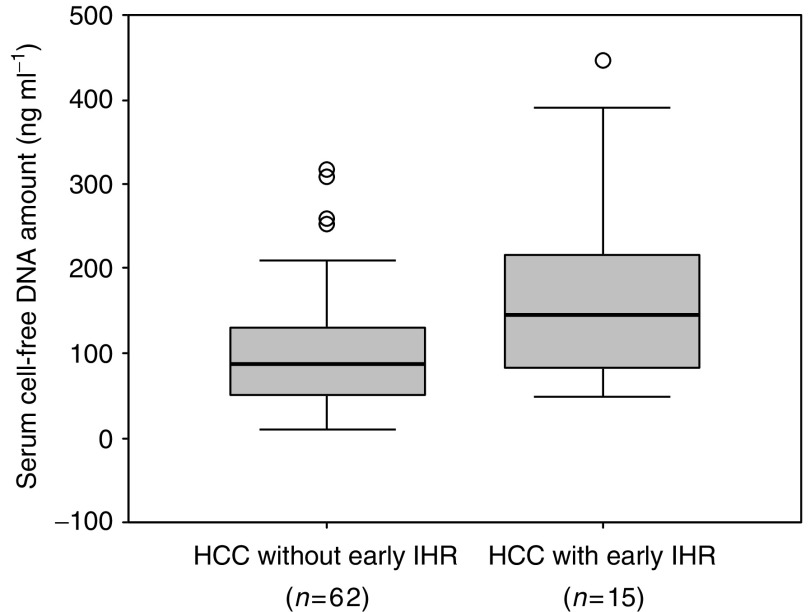
To investigate the relation between cfDNA level and early IHR due to intrahepatic metastasis of HCC, we excluded 10 patients who had the follow-up periods of less than 1 year after surgery. Among the eligible 77 patients, 15 (19.5%) had early IHR within 1 year of surgery and the remaining 62 (80.5%) did not have early IHR. Among the 62 patients, 37 had IHR 1 year or more after surgery and 25 had no IHR during follow-up periods. The serum cfDNA level was significantly higher in patients (n=15) with early IHR than in those (n=62) without early IHR (176.3±124.8 vs 108.7±87.9 ng ml−1, P=0.017 by Student's t-test; Figure 3). The logistic regression model showed that tumour size (relative risk, 7.3; 95% CI, 1.9–27.7; P=0.004) and number of primary lesions (relative risk, 4.5; 95% CI, 1.1–17.7; P=0.033), but not cfDNA level, were independent risk factors for early IHR (data not shown).
Figure 3.
Box and whisker plot of cell-free DNA (cfDNA) levels in sera from patients with early intrahepatic recurrence (IHR) and patients without. Levels of cfDNA in sera were significantly higher in patients with early IHR than in patients without early IHR (176.3±124.8 vs 108.7±87.9 ng ml−1 (mean±s.d.), P=0.017 by Student's t-test).
DISCUSSION
The possibility of detecting and measuring tumour-derived cfDNA has opened a new avenue in predictive oncology (Leon et al, 1977; Anker et al, 1999; Ziegler et al, 2002). This method provides a non-invasive and easy-to-use tool for screening for malignancy and predicting cancer outcomes. By means of real-time PCR, we showed previously that cfDNA levels were significantly higher in sera from patients with HCV-related HCC than in sera from HCV carriers without known HCC, and the diagnostic performance of cfDNA was superior to that of two representative HCC markers, α-fetoprotein and PIVKA-II (Iizuka et al, 2006a). Our present study confirmed the significantly increased levels of serum cfDNA in a large group of patients with HCV-related HCC, suggesting that the increase is related to progression of the disease caused by HCV infection. Indeed, our present study identified two subgroups of patients with HCV-related HCC: those with a high cfDNA level who had an unfavourable outcome after curative surgery, and those with a low cfDNA level, who had a favourable outcome. Notably, cfDNA was the only independent prognostic factor for EHR in distant organs in case of HCV-related HCC treated surgically.
Recurrence of HCC is quite complicated. There are at least three representative modes of postoperative recurrence, early and late IHRs appearing in the remnant liver and EHR appearing in distant organs such as bone and lung. Among the three types of recurrence, late IHR is a de novo primary tumour rather than a metastatic tumour, and it accounts for the majority of HCC recurrences 3 years or more after surgery (Kumada et al, 1997), suggesting that it has less influence on patient survival. The finding that our cohort included many patients with late IHR might account for the lack of association between cfDNA levels and DFS in patients with HCV-related HCC. In contrast, our univariate analysis showed increased levels of cfDNA in HCC patients with early IHR. Most early IHRs can be attributed to intrahepatic metastasis of cancer cells and are detected in 30–50% of patients within 1 or 2 years after surgery, limiting the potential for surgical cure of HCC (Tung-Ping Poon et al, 2000; Iizuka et al, 2003; Llovet et al, 2003; Matoba et al, 2005; Portolani et al, 2006). Thus, the increased levels of serum cfDNA are related to the high metastatic potential, but not to the appearance of de novo tumour, of this type HCC.
Much effort has been devoted to developing predictive makers for early IHR. Some recent molecular profiling studies (Iizuka et al, 2003, 2004; Kurokawa et al, 2004) raised the possibility of accurately predicting early IHR in a cohort of patients with HCC, most of which were HCV-related HCCs. Predictive systems that are developed will enable to accurately detect patients at high risk for early IHR, but there might be many problems to solve before these markers can be applied to daily clinical practice (Iizuka et al, 2004). Thus far, there have been few reports on a predictor for EHR after surgery, although several studies have identified several key genes or gene products related to distant metastasis (Schimanski et al, 2006; Iizuka et al, 2006b). It was reported that HCC recurred in distant organs in only 3 (7%) of 42 patients who underwent liver transplantation, a radical curative treatment strategies (Nart et al, 2003). A recent large study by Yang et al (2007) showed that, among 348 HCC patients who underwent hepatectomy, 47 (13.5%) had EHR during the follow-up period of 4.8±3.7 years. Thus, the frequency of EHR is lower than that of early IHR; however, once HCC progresses to EHR, it is difficult to control the lesions in most cases because of the limited resectability. Indeed, the frequency of death due to respiratory failure resulting from metastasis of HCC to the lung has increased over the last 30 years in the Japanese population (Itoh et al, 2002). In this regard, our present finding that a patient with a high level of cfDNA has 4.5-fold increased risk for EHR in distant organs may be clinically useful. Such a robust predictive system is urgently needed to screen for patients who will develop EHR.
Taken together, our data suggest that serum cfDNA levels could serve as a useful tool for prediction of EHR after curative surgery in patients with HCV-related HCC. However, it is unlikely that cfDNA alone will be effective in predicting EHR in more global cases. A large training-validation study is needed to construct a robust predictor made up of multiple factors in which cfDNA might play a central role. In addition, identifying and quantifying genetic changes in circulating cfDNAs specific to HCV-related HCC will allow us to determine metastatic potential preoperatively on an individual basis.
Acknowledgments
Grant sponsors: The Ministry of Education, Culture, Sports, Science and Technology (No. 18390366, No. 17591406 and Knowledge Cluster Initiative); the Venture Business Laboratory of Yamaguchi University; the New Energy and Industrial Technology Development Organization (Grant number: 03A02018a).
References
- Anker P, Mulcahy H, Chen XQ, Stroun M (1999) Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 18: 65–73 [DOI] [PubMed] [Google Scholar]
- Deuffic S, Poynard T, Buffat L, Valleron AJ (1998) Trends in primary liver cancer. Lancet 351: 214–215 [DOI] [PubMed] [Google Scholar]
- El Serag HB (2004) Hepatocellular carcinoma: recent trends in the United States. Gastroenterology 127: S27–S34 [DOI] [PubMed] [Google Scholar]
- Gautschi O, Bigosch C, Huegli B, Jermann M, Marx A, Chasse E, Ratschiller D, Weder W, Joerger M, Betticher DC, Stahel RA, Ziegler A (2004) Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol 22: 4157–4164 [DOI] [PubMed] [Google Scholar]
- Iizuka N, Hamamoto Y, Oka M (2004) Predicting individual outcomes in hepatocellular carcinoma. Lancet 364: 1837–1839 [DOI] [PubMed] [Google Scholar]
- Iizuka N, Oka M, Okabe H, Nishida M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, Hamada K, Nakayama H, Ishitsuka H, Miyamoto T, Hirabayashi A, Uchimura S, Hamamoto Y (2003) Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet 361: 923–929 [DOI] [PubMed] [Google Scholar]
- Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, Takemoto N, Tangoku T, Hamada K, Nakayama H, Miyamoto T, Uchimura S, Hamamoto Y (2002) Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method. Cancer Res 62: 3939–3944 [PubMed] [Google Scholar]
- Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, Stark M, Tamatsukuri S, Ishitsuka H, Uchida K, Terai S, Sakamoto K, Tamesa T, Oka M (2006a) Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res 26: 4713–4719 [PubMed] [Google Scholar]
- Iizuka N, Tamesa T, Sakamoto K, Miyamoto T, Hamamoto Y, Oka M (2006b) Different molecular pathways determining extrahepatic and intrahepatic recurrences of hepatocellular carcinoma. Oncol Rep 16: 1137–1142 [PubMed] [Google Scholar]
- Itoh Y, Ohkubo K, Iuchi H, Michitaka K, Horiike N, Onji M (2002) Chronological changes of causes of death and distant metastasis in hepatocellular carcinoma. Oncol Rep 9: 331–335 [PubMed] [Google Scholar]
- Kumada T, Nakano S, Takeda I, Sugiyama K, Osada T, Kiriyama S, Sone Y, Toyoda H, Shimada S, Takahashi M, Sassa T (1997) Patterns of recurrence after initial treatment in patients with small hepatocellular carcinoma. Hepatology 25: 87–92 [DOI] [PubMed] [Google Scholar]
- Kuramitsu Y, Nakamura K (2006) Proteomic analysis of cancer tissues: shedding light on carcinogenesis and possible biomarkers. Proteomics 6: 5650–5661 [DOI] [PubMed] [Google Scholar]
- Kurokawa Y, Matoba R, Takemasa I, Nagano H, Dono K, Nakamori S, Umeshita K, Sakon M, Ueno N, Oba S, Ishii S, Kato K, Monden M (2004) Molecular-based prediction of early recurrence in hepatocellular carcinoma. J Hepatol 41: 284–291 [DOI] [PubMed] [Google Scholar]
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ (1977) Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 37: 646–650 [PubMed] [Google Scholar]
- Llovet JM, Burroughs A, Bruix J (2003) Hepatocellular carcinoma. Lancet 362: 1907–1917 [DOI] [PubMed] [Google Scholar]
- Mann CD, Neal CP, Garcea G, Manson MM, Dennison AR, Berry DP (2007) Prognostic molecular markers in hepatocellular carcinoma: a systematic review. Eur J Cancer 43: 979–992 [DOI] [PubMed] [Google Scholar]
- Marrero JA, Lok AS (2004) Newer markers for hepatocellular carcinoma. Gastroenterology 127: S113–S119 [DOI] [PubMed] [Google Scholar]
- Matoba K, Iizuka N, Gondo T, Ishihara T, Yamada-Okabe H, Tamesa T, Takemoto N, Hashimoto K, Sakamoto K, Miyamoto T, Uchimura S, Hamamoto Y, Oka M (2005) Tumor HLA-DR expression linked to early intrahepatic recurrence of hepatocellular carcinoma. Int J Cancer 115: 231–240 [DOI] [PubMed] [Google Scholar]
- Nart D, Arikan C, Akyildiz M, Yuce G, Demirpolat G, Zeytunlu M, Karasu Z, Aydogdu S, Killi R, Yuzer Y, Tokat Y, Kilic M (2003) Hepatocellular carcinoma in liver transplant era: a clinicopathologic analysis. Transplant Proc 35: 2986–2990 [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P (2001) Estimating the world cancer burden: Globocan 2000. Int J Cancer 94: 153–156 [DOI] [PubMed] [Google Scholar]
- Portolani N, Coniglio A, Ghidoni S, Giovanelli M, Benetti A, Tiberio GA, Giulini SM (2006) Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg 243: 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren N, Qin LX, Tu H, Liu YK, Zhang BH, Tang ZY (2006) The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol 132: 399–407 [DOI] [PubMed] [Google Scholar]
- Schimanski CC, Bahre R, Gockel I, Muller A, Frerichs K, Horner V, Teufel A, Simiantonaki N, Biesterfeld S, Wehler T, Schuler M, Achenbach T, Junginger T, Galle PR, Moehler M (2006) Dissemination of hepatocellular carcinoma is mediated via chemokine receptor CXCR4. Br J Cancer 95: 210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin LH, Wittekind C (2002) TNM classification of Malignant Tumours, 6th edn. UICC: Wiley-Liss. 81–83 [Google Scholar]
- Tung-Ping Poon R, Fan ST, Wong J (2000) Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg 232: 10–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, Hoon DS (2006) Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol 24: 4270–4276 [DOI] [PubMed] [Google Scholar]
- Yang Y, Nagano H, Ota H, Morimoto O, Nakamura M, Wada H, Noda T, Damdinsuren B, Marubashi S, Miyamoto A, Takeda Y, Dono K, Umeshita K, Nakamori S, Wakasa K, Sakon M, Monden M (2007) Patterns and clinicopathologic features of extrahepatic recurrence of hepatocellular carcinoma after curative resection. Surgery 141: 196–202 [DOI] [PubMed] [Google Scholar]
- Ziegler A, Zangemeister-Wittke U, Stahel RA (2002) Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev 28: 255–271 [DOI] [PubMed] [Google Scholar]