Abstract
This study examines the clinical impact of PTEN genomic deletions using fluorescence in situ hybridisation (FISH) analysis of 107 prostate cancers, with follow-up information covering a period of up to 10 years. Tissue microarray analysis using interphase FISH indicated that hemizygous PTEN losses were present in 42/107 (39%) of prostatic adenocarcinomas, with a homozygous PTEN deletion observed in 5/107 (5%) tumours. FISH analysis using closely linked probes centromeric and telomeric to the PTEN indicated that subband microdeletions accounted for ∼70% genomic losses. Kaplan–Meier survival analysis of PTEN genomic losses (hemizygous and homozygous deletion vs not deleted) identified subgroups with different prognosis based on their time to biochemical relapse after surgery, and demonstrated significant association between PTEN deletion and an earlier onset of disease recurrence (as determined by prostate-specific antigen levels). Homozygous PTEN deletion was associated with a much earlier onset of biochemical recurrence (P=0.002). Furthermore, PTEN loss at the time of prostatectomy correlated with clinical parameters of more advanced disease, such as extraprostatic extension and seminal vesicle invasion. Collectively, our data indicates that haploinsufficiency or PTEN genomic loss is an indicator of more advanced disease at surgery, and is predictive of a shorter time to biochemical recurrence of disease.
Keywords: interphase FISH, PTEN haploinsufficiency, prognostic biomarker, PSA, biochemical recurrence
Prostate cancer is the most commonly diagnosed malignancy in men in the North America and the third leading cause of cancer-related mortality after lung and colorectal cancer in males aged 40 years and older (Jemal et al, 2006). In spite of significant progress in its clinical management, comparatively little is known about the disease aetiology. Widely used biochemical, histopathological and clinical criteria, for example prostate-specific antigen (PSA) level, Gleason score, and the clinical tumour stage, have demonstrated a significant variability in predicting subgroups of prostate cancer patients with distinct clinical outcome (Miller et al, 2001; DeMarzo et al, 2003; Glinsky et al, 2004). As such, there is an absolute necessity to improve the current patient stratification methods using biomarkers identified through studies of prostate cancer genomics.
Previous cytogenetic and genomic profiling analyses have identified several tumour-associated chromosomal rearrangements in the initial stages of sporadic primary prostate cancer, consisting predominantly of losses (Visakorpi et al, 1995; Verhagen et al, 2000; Elo and Visakorpi, 2001). For example, well-recognised changes in early prostatic carcinogenesis include loss of 8p, 6q, 10q, 13q, 16q and 18q, and gain of 8q (Qian et al, 1998; Zitzelsberger et al, 2001; Kasahara et al, 2002; Wolf et al, 2004; van Dekken et al, 2004; Hughes et al, 2006; Ribeiro et al, 2006a, 2006b). The importance of genomic rearrangements in prostate cancer was emphasised by the discovery of recurrent translocations in 40–60% of prostate carcinoma and in 21% of the presumed premalignant lesion, high-grade prostatic intraepithelial neoplasia (HPIN), involving the TMPRSS2 gene with members of the erythroblast transformation-specific (ETS) transcription factor family (Tomlins et al, 2005, 2006; Ahlers and Figg, 2006; Cerveira et al, 2006; Perner et al, 2006; Soller et al, 2006; Wang et al, 2006; Yoshimoto et al, 2006b). Indeed, HPIN lesions reveal similar genetic features to those found in prostate carcinomas, including the loss of 8p, gain of 8q (Hughes et al, 2006; Ribeiro et al, 2006b), and genomic losses of chromosome 10 (Hughes et al, 2006; Ribeiro et al, 2006b). More detailed investigation of the 10q losses using fluorescence in situ hybridisation (FISH) and tissue microarrays (TMAs) showed that PTEN microdeletions were present in 68% of carcinomas and in 23% of HPIN lesions found in radical prostatectomies. The detection of PTEN deletion in HPIN suggested that somatic haploinsufficiency per se might be an early pivotal step in the transition from HPIN to invasive carcinoma (Yoshimoto et al, 2006a).
PTEN plays an important role in the modulation of the phosphatidylinositol-3-kinase (PI3K) pathway by catalysing degradation of phosphatidylinositol-(3,4,5)-trisphosphate generated by PI3K (Besson et al, 1999). Phosphatidylinositol-(3,4,5)-trisphosphate activates the protein kinase Akt and its regulator PDK1 (Goberdhan and Wilson, 2003), which then modulates a number of downstream targets with important roles in apoptosis and the cell-cycle progression, including BAD (Datta et al, 1997), CASP3 and CASP9 (Cardone et al, 1998), MDM2 (Ashcroft et al, 2002), mTOR (Majumder et al, 2004), the forkhead family of transcription factors (FKHR) (Brunet et al, 1999) and p27 (Graff et al, 2000). The lack of inhibition of these pathways by PTEN inactivation is associated with high Gleason score (Koksal et al, 2004) and tumour progression in prostate cancer (Koksal et al, 2004; Bertram et al, 2006).
The occurrence of PTEN mutation in prostate cancer is considered common, with reported frequencies (based on relatively small sample sizes), ranging from 30 to 60% (Whang et al, 1998; Kwabi-Addo et al, 2001). Given the high frequency of PTEN inactivation by genomic deletion in prostate cancer using FISH methods (Yoshimoto et al, 2006a), it has become more practical to conduct much larger correlative studies of patient outcome in situations when the PTEN gene is either lost, or retained in tumours. Such findings would further implicate a role for PTEN haploinsufficiency in poor prognosis and tumour progression. Moreover, knowledge of the PTEN deletion in the primary tumour, in addition to current clinico-pathological features, might be of value when selecting the optimum treatment for a particular patient.
MATERIALS AND METHODS
Tissue specimens
The collection of tissue specimens, clinical and follow-up data was obtained and handled in accordance with the Hospital do Câncer Research Ethics guidelines (São Paulo, Brazil). Archival formalin-fixed, paraffin-embedded tissues were obtained from 107 radical prostatectomies performed between 1997 and 2000 at the Hospital do Câncer, AC Camargo, São Paulo, Brazil. For control purposes, 10 non-neoplastic prostate tissue samples were obtained from patients undergoing surgery solely for benign prostate hyperplasia. The prostate cancer cohort comprising 107 tumour samples and control specimens were sampled using a 0.6 mm diameter tissue core distributed on TMA slide. Adjacent haematoxylin and eosin (H&E)-stained section was reviewed by two pathologists to determine the presence and extent of morphologically representative areas of the original tumours in each tissue core and Gleason grading. The clinico-pathological (TNM) stage and Gleason scores (range from 4 to 9) for each case were obtained from the medical and surgical pathology reports. The size of tumour was based on assessment of total surface area of the gland examined histologically involved by carcinoma. Preoperative PSA level was available for all patients and the PSA non-failure was defined as PSA remaining below 0.2 ng ml−1 after radical prostatectomy. Recurrence-free interval was defined as the time between date of surgery and the date of first PSA increase above 0.2 ng ml−1.
A separate evaluation of PTEN deletion status was also performed using a study group of clinically distinct prostate cancers in which inclusion criteria were the availability of paraffin-embedded formalin-fixed tissue from both an initial primary adenocarcinoma surgical specimen and from metastatic prostate adenocarcinoma in the regional lymph node metastases (Hospital do Câncer, AC Camargo, São Paulo, Brazil). A cohort of 10 such paired tumour samples were identified. Adjacent H&E-stained section was reviewed by two pathologists to determine the presence and extent of morphologically representative areas of the original tumours in each tissue. Blood preoperative PSA levels ranged from 10 to 84 ng ml−1 within this cohort.
Follow-up studies
The 107 patients in the study group were followed up for a period of up to 10 years subsequent to the initial surgery. The median age at diagnosis was 63 years (range from 41 to 76). At the end of the follow-up period for each patient, 58 (54%) had a biochemical recurrence within the period of 0.79–86.28 months after initial surgery. Metastatic progression occurred in 10 patients at a median follow-up of 36.21 months from initial surgery. Survival time for the remaining 49 disease-free cases varied from 13.85 to 127.60 months. The median overall survival for the entire cohort of patients was 80.65 months. All relevant clinico-pathological features of the 107-tumour cohort are summarised in Table 1.
Table 1. Clinico-pathological parameters from 107 prostatic adenocarcinoma patients.
|
PTEN
|
|||||
|---|---|---|---|---|---|
| Clinico-pathological parameters | Number of cases | Not deleted | Hemizygous | Homozygous | |
| Age median (min–max) | 61.9 years (48–69) | ||||
| Preoperative PSA (ng ml−1)a | P=0.556 | ||||
| 0.9–4.0 | 7 (6.7%) | 3 (42.9%) | 4 (57.1%) | 0 (0%) | |
| 4.1–10.0 | 55 (52.3%) | 32 (58.2%) | 22 (40%) | 1 (1.8%) | |
| 10.1–20.0 | 28 (26.7%) | 19 (67.8%) | 8 (28.6%) | 1 (3.6%) | |
| 20.1–84.0 | 15 (14.3%) | 6 (40%) | 8 (53.3%) | 1 (6.7%) | |
| Median tumour volume (%)a | P=0.158 | ||||
| 0–10.0 | 32 (34%) | 19 (59.4)% | 13 (40.6%) | 0 (0%) | |
| 10.1–20.0 | 19 (20.2%) | 12 (63.2%) | 7 (36.8%) | 0 (0%) | |
| 20.1–85.0 | 43 (45.8%) | 21 (48.9%) | 17 (39.5%) | 5 (11.6%) | |
| Gleason score | P=0.142 | ||||
| Gleason 4–6 | 61 (57%) | 39 (63.9%) | 20 (32.8%) | 2 (3.3%) | |
| Gleason 7 | 33 (30.8%) | 16 (48.48%) | 16 (48.48%) | 1 (3.03%) | |
| Gleason 8–9 | 13 (12.2%) | 5 (38.46%) | 6 (46.15%) | 2 (15.38%) | |
| Pathologic stage | P=0.569 | ||||
| pT2a | 6 (5.6%) | 1 (16.7%) | 5 (83.3%) | 0 (0%) | |
| pT2b | 49 (45.8%) | 30 (61.2%) | 16 (32.7%) | 3 (6.1%) | |
| pT3a | 32 (29.9%) | 18 (56.2%) | 13 (40.6%) | 1 (3.1%) | |
| pT3b | 13 (12.2%) | 4 (57.1%) | 3 (42.9%) | 0 (0%) | |
| pT4 | 7 (6.5%) | 4 (57.1%) | 3 (42.9%) | 0 (0%) | |
| Seminal vesicle invasion | P=0.859 | ||||
| Negative | 94 (87.9%) | 53 (56.4%) | 37 (39.4%) | 4 (4.2%) | |
| Positive | 13 (12.1%) | 7 (53.8%) | 5 (38.5%) | 1 (7.7%) | |
| Perineural infiltration | P=0.517 | ||||
| Negative | 15 (14.0%) | 10 (66.7%) | 5 (33.3%) | 0 (0%) | |
| Positive | 92 (86%) | 50 (54.4%) | 37 (40.2%) | 5 (4.4%) | |
| Angiolymphatic invasiona | P=0.346 | ||||
| Negative | 78 (77.2%) | 46 (59%) | 29 (37.2%) | 3 (3.8%) | |
| Positive | 23 (22.8%) | 10 (43.5%) | 11 (47.8%) | 2 (8.7%) | |
| Capsular invasion | P=0.729 | ||||
| Negative | 46 (43%) | 25 (54.4%) | 18 (39.1%) | 3 (6.5%) | |
| Positive | 61 (57%) | 35 (57.4%) | 24 (39.3%) | 2 (3.3%) | |
| Extraprostatic extensiona | P=0.713 | ||||
| Negative | 83 (81.4%) | 50 (60.2%) | 30 (36.2%) | 3 (3.6%) | |
| Positive | 19 (18.6%) | 9 (47.4%) | 8 (42.1%) | 2 (10.5%) | |
| Lymphonodal invasiona | P=0.812 | ||||
| Negative | 94 (95.9%) | 54 (57.4%) | 35 (37.2%) | 5 (5.3%) | |
| Positive | 4 (4.1%) | 2 (50%) | 2 (50%) | 0 (0%) | |
| Biochemical recurrence | P=0.005 | ||||
| Negative | 49 (46%) | 35 (71.4%) | 14 (28.6%) | 0 (0%) | |
| Positive | 58 (54%) | 25 (43.1%) | 28 (48.3%) | 5 (8.6%) | |
Values not available for all 107 cases.
Abbreviation: PSA, prostate-specific antigen.
Median overall survival was 80.65 months.
P-value=χ2 analysis.
FISH
Dual-colour FISH on paraffin-embedded tumour tissue was performed using commercially available DNA probes for cytoband 10q23 (Spectrum Orange PTEN locus-specific probe) and region 10p11.1-q11.1 (Spectrum Green centromere of chromosome 10 probe) (LSI PTEN/CEP 10 – Vysis Inc., Downers Grove, IL, USA). The PTEN genomic probe spans 368 kb and starts 166 kb from 5′ end of the gene and extends 98 kb past the 3′ end of the gene. Histologic tissue sections of 5 μm were deparaffinised with a series of xylene before immersion in 100% ethanol. The slide was placed in a 2 × SSC solution at 75°C for 20 min, following by treatment in 0.25 mg ml−1 Proteinase K (Roche Diagnostics, Indianapolis, IN, USA) at 45°C for 20 min. The sections were dehydrated in a graded series of ethanol, and washed in 2 × SSC. Dual-colour probes and target DNA were co-denatured at 80°C for 10 min. Post-hybridisation procedures were performed by 1.5 M urea/0.1 × SSC solution at 45°C for 30 min, 2 × SSC and a graded ethanol series.
Sequential tri-colour FISH method was applied to the prostate cancer TMA to map the flanking regions of genomic loss associated with PTEN deletions in tumours. The following bacterial artificial chromosome (BAC) clones were used. BACs located at: (a) 10q23.2 (88.2–88.7 Mb, 900 kb upstream from the 5′ PTEN gene locus): RP11-141D8, RP11-52G13 and RP11-420K10; (b) 10q23.2 (89.4–89.5 Mb, 100 kb on the centromeric side of the 5′ PTEN): RP11-11O21; (c) 10q23.31 (89.6–89.7 Mb, PTEN gene locus): RP11-383D9; (d) 10q23.33 (96.2–96.6 Mb, 6.6 Mb on the telomeric side of the 3′ PTEN gene locus): RP11-119K6, RP11-90J1 and RP11-466J14; (e) 10q25.1 (108.9–109.3 Mb, 19.3 Mb downstream from the 3′ PTEN gene locus): RP11-246B13, RP11-49H17 and RP11-432B10. The position and name of the BAC clones were taken from the Human March 2006 assembly of the UCSC Genome Browser 1. DNA was extracted and labelled with either Spectrum Green-dUTP, SpectrumOrange-dUTP (Vysis) or DEAC-dUTP (PerkinElmer Life and Analytical Sciences, Boston, MA, USA) using the Vysis nick-translation kit (Vysis). Labelling of probes was done as described previously (Merscher et al, 1997; Sirvent et al, 2000). The chromosome localisation of all BAC clones was confirmed by both normal metaphase and tri-colour FISH analysis.
Data analysis
PTEN copy number was evaluated for each probe by counting spots in a range from 50 to 100 non-overlapped, intact interphase nuclei per tumour tissue core. 4′,6-Diamidino-2-phenylindole, dihydrochloride staining of nuclei with reference to the corresponding H&E-stained tissue identified the areas of adenocarcinoma. Based on hybridisation in 10 control cores (data not shown), hemizygous deletion of PTEN were defined as >20% (mean+3 s.d. in non-neoplastic controls) of tumour nuclei containing one PTEN locus signal and by the presence of CEP 10 signals. Homozygous deletion of PTEN was exhibited by the simultaneous lack of the both PTEN locus signals and by the presence of control signals (Mezzelani et al, 1999; Kawai et al, 2004; Korshunov et al, 2005; Ventura et al, 2006) in >30% of cells (Korshunov et al, 2005).
Statistical analysis of PTEN deletion in 107-prostate cancer TMA
Fluorescence in situ hybridisation findings for PTEN deletions were correlated in a univariate and multivariate approaches with clinical and pathologic features of disease aggressiveness. Initially, presence and absence of deletion by FISH was correlated with determinants of disease mortality and morbidity including PSA, Gleason score and extraprostatic extension, as well as clinically relevant end points such as time to biochemical relapse, and the development time of metastases following the definitive treatment. For prediction of 5-year biochemical risk failure, PTEN status (not deleted, hemi- or homozygous deletion) was compared with all relevant clinico-pathological features (Table 1). Univariate and multivariate analyses of biochemical risk failure were studied by Cox Proportional Hazard model (Tables 2 and 3, respectively). A significant correlation between two parameters was taken at the 95% confidence interval. P-values <0.05 were considered significant. In addition, the survival rate was estimated by applying the Kaplan–Meier method. The end point for calculating the survival time was defined by the time from radical prostatectomy until the occurrence of metastasis or PSA determined by the biochemical recurrence. Correlation of PTEN copy number changes with Gleason score was tested using Pearson's χ2 statistic. All calculations were performed using Stata 9.1.
Table 2. Univariate Cox proportional hazard analysis of biochemical failure risks (each variable predictor analysed separately).
|
BRFS
|
|||||
|---|---|---|---|---|---|
| Variables | Category | (5 years) | P-value | HR | 95% CI |
| Perineural invasion | Negative | 86.67 | 0.0038 | 1 | Reference |
| Positive | 42.77 | 6.22 | 1.51–25.60 | ||
| Extraprostatic | Negative | 56.71 | 0.0001 | 1 | Reference |
| extension | Positive | 27.78 | 3.2 | 1.72–5.94 | |
| Seminal vesicle | Negative | 54.23 | 0.0001 | 1 | Reference |
| invasion | Positive | 21.43 | 3.41 | 1.74–6.69 | |
| Gleason score | 2–6 | 65.57 | <0.0001 | 1 | Reference |
| 7 | 25.74 | 3.04 | 1.72–5.37 | ||
| 8–9 | 30.77 | 3.4 | 1.60–7.22 | ||
| Preoperative PSA | 0.9–4.0 | 67.69 | <0.0001 | 1 | Reference |
| 4.1–10 | 58.36 | 1.98 | 0.47–8.41 | ||
| 10.1–20 | 51.74 | 2.38 | 0.54–10.42 | ||
| 20.1–84 | 6.67 | 10.27 | 2.30–45.87 | ||
| Lymphonodal | Negative | 49.37 | 0.0068 | 1 | Reference |
| invasion | Positive | 25.00 | 4.43 | 1.36–14.43 | |
| PTEN deletion | Negative | 58.68 | 0.0003 | 1 | Reference |
| Hemizygous | 40.3 | 1.78 | 1.03–3.05 | ||
| Homozygous | 0 | 6.15 | 2.26–16.73 | ||
Abbreviations: BRFS, biochemical recurrence free survival; CI, confidence interval; HR. hazard ratio; PSA, prostate-specific antigen.
Table 3. Multivariate model to biochemical failure risks by Cox logistic regression analysis.
| Variables | Category | HR | P-value | 95% CI |
|---|---|---|---|---|
| Extracapsular extension | Negative | 1 | 0.004 | Reference |
| Positive | 2.81 | 1.38–5.74 | ||
| Seminal vesicle invasion | Negative | 1 | 0.019 | Reference |
| Positive | 2.8 | 1.18–6.65 | ||
| PTEN deletion | Negative | 1 | Reference | |
| Hemizygous | 1.96 | 0.022 | 1.10–3.49 | |
| Homozygous | 11.26 | 0.002 | 2.44–51.93 |
Abbreviations: CI, confidence interval; HR, hazard ratio.
Likelihood-ratio test P=0.0078.
RESULTS
PTEN deletion analysis in 107-prostate cancer TMA
To investigate whether loss of PTEN as determined by interphase FISH indicated a greater prevalence in poor prognosis patients, the frequency of PTEN deletion was investigated in the cohort of 107 tumour samples using a TMA in which anonymous annotation codes allowed interrogation of clinical outcome parameters. Hemizygous PTEN deletion was found in 42 of the 107 (39%) adenocarcinomas samples. As shown in Table 4, homozygous PTEN deletion was found in 5 of the 107 (5%) prostate adenocarcinomas. Representative images of undeleted, hemizygous and homozygous deletions are shown in Figure 1. Comprehensive FISH raw data sets for all samples is summarised in Supplementary Table.
Table 4. Analysis of PTEN gene copy number by FISH.
| PTEN locus | Number of cases (%) |
|---|---|
| Hemizygous deletiona | 42 (39%) |
| Homozygous deletionb | 5 (5%) |
| Not deleted | 60 (56%) |
Abbreviation: FISH, fluorescence in situ hybridisation.
Hemizygous deletion of PTEN was defined as >20% of tumour nuclei containing one PTEN locus signal (mean + 3 s.d. in non-neoplastic controls) (Korshunov et al, 2005).
Homozygous deletions of PTEN was identified by the simultaneous lack of both of the PTEN locus signals and by the presence of CEP 10 signals in >30% of cells (Korshunov et al, 2005).
Figure 1.
PTEN probe enumeration and representative dual-colour FISH images are shown for prostate cancer TMA. (A) Digital images of H&E-stained tissue microarray from 107 prostate cancer samples show areas of prostatic adenocarcinoma at low magnification and at × 20 magnification (rectangle) (ScanScope® System, Aperio Technologies, Inc., Vista, CA, USA). (B–D) A magnified H&E area is displayed in a tissue core with accompanying FISH for the PTEN locus. The rectangles show the main FISH image high magnification. (B) The FISH image shows two signals of both red signals (10q23/PTEN locus) and green signals (CEP 10) in most of the nuclei indicating no deletion of PTEN in tumour cells. (C) The FISH image shows tumour cells with single red signal for 10q23/PTEN locus in most of the nuclei and paired green signals for CEP 10 indicating hemizygous deletion of 10q23/PTEN locus in prostate cancer. (D) Representative FISH image of homozygous deletion in prostate cancer shows absence of red signal for 10q23/PTEN locus in most of the nuclei and retained green signals for CEP 10.
Among the hemizygous PTEN-deleted tumours detected from the cohort of 107 adenocarcinomas, there were 42 patients with hemizygous PTEN deletion that were classified as Gleason score 4–6 (20 tumours), 7 (16 tumours) and 8–9 (6 tumours). A median tumour volume of >20% was found in 17 of the 42 adenocarcinomas. Biochemical recurrence based on PSA level was present in 28 of 42 samples. A median tumour volume of >20% was found in all five adenocarcinomas with homozygous PTEN deletion. In addition, early biochemical recurrence was detected in all five of these samples. A comprehensive description of the clinical parameters associated with the adenocarcinomas having hemizygous or homozygous PTEN deletion is summarised in Table 1.
Statistical analysis
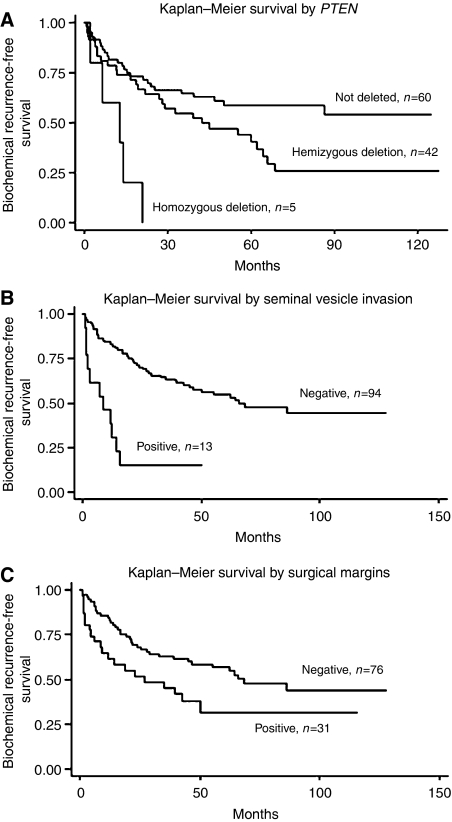
After acquisition of FISH data, the 107 cases were reviewed to search for potential associations between genomic loss of PTEN, clinical variables of disease progression and tumour histology. Univariate analysis of biochemical risk failure was significant for perineural invasion, extraprostatic extension, seminal vesicle invasion, Gleason score, preoperative PSA, lymph nodal invasion and PTEN deletion (Table 2). For prediction of 5-year biochemical risk failure by χ2 analysis, PTEN status (not deleted, hemi- or homozygous deletion) was significantly associated with disease recurrence based on PSA levels (Table 1). Other clinical parameters of aggressive disease such as extraprostatic extension, seminal vesicle and perineural invasion were also significantly associated with biochemical recurrence. By multivariate analysis (Table 3), PTEN deletion, extraprostatic extension and seminal vesicle invasion were observed at an independent level to explain biochemical failure. For comparison purpose, Kaplan–Meier survival analysis applying established clinical markers, such as the preoperative PSA, seminal vesicle invasion and surgical margins status, was considered to identify subgroups with different prognosis with respect to time of relapse after surgery. The estimated disease-free survival curves demonstrated significant association between PTEN deletion and short time based on PSA recurrence intervals. Significantly homozygous PTEN deletion in five tumours was associated with a much earlier onset of biochemical recurrence based on PSA values (Figure 2).
Figure 2.
Kaplan–Meier curves illustrating biochemical recurrence-free survival among 107 prostate cancer patients defined by the status of selected clinico-pathological parameters and PTEN copy number changes. (A) PSA recurrence-free survival curve stratified by the PTEN locus copy number changes on prostate cancer patients. (B) PSA recurrence-free survival analysis stratified by seminal vesicle invasion on prostate cancer patients. (C) PSA recurrence-free survival analysis stratified by surgical margins on prostate cancer patients.
Analysis of PTEN deletion in cohort of 10-paired primary adenocarcinomas and metastatic prostate adenocarcinoma in the regional lymph nodes
PTEN deletion frequency was determined by dual-colour FISH using paired primary prostate adenocarcinomas and metastatic adenocarcinoma in the regional lymph nodes derived from 10 patients. Overall, the presence of PTEN deletion was found at high frequency (9 of 10) in both paired primary and metastatic lymph nodal prostate adenocarcinoma samples in this study group. Only one patient of the 10 retained both copies of the PTEN locus in his primary tumour and in his metastatic lymph nodal biopsy. Hemizygous PTEN deletion was found in both the primary and the metastatic nodal tumour samples in four of 10 patients. Homozygous PTEN deletion was found in both the primary tumour and their metastatic lymph nodes in three of the 10 patients. Significantly, two of the 10 patients with a hemizygous PTEN deletion in their primary adenocarcinomas, had positive lymph node biopsies that had acquired a homozygous PTEN deletion. These findings suggest that loss of the remaining PTEN locus may be associated with metastasis.
Mapping the adjacent genomic regions deleted when PTEN is lost
Tri-colour FISH using the BAC probes spanning the band 10q23.2, 5′ flanking PTEN probe on the centromeric side of the locus, PTEN locus (10pq23.31), and more telomeric probes mapping to 10q23.33 and 10q25.1, was performed on 10 of the 107 adenocarcinomas with hemizygous PTEN deletion and three of the 107 adenocarcinomas with homozygous PTEN deletion (Table 5). Sequential tri-colour FISH analyses from 7 of the 10 PTEN hemizygously deleted adenocarcinomas revealed BAC probes either side of PTEN were retained as two copies, indicating that a hemizygous PTEN deletion was usually accompanied by an interstitial microdeletion within band 10q23.2–q23.31. Among the three homozygous PTEN deletions, sequential tri-colour FISH often revealed the hemizygous loss of the 10q23.2, 10q23.33 and 10q25.1 signals, and homozygous loss of the 5′ flanking PTEN genomic region and the PTEN locus signals.
Table 5. Mapping the adjacent genomic regions deleted when PTEN is lost using BAC DNA probes spanning the predicted deletion interval on chromosome 10q.
| TMA | Centromeric to PTENa | 5′ flanking PTENb | PTEN c | Telomeric to PTENd | Telomeric to PTENe | Predicted deletion |
|---|---|---|---|---|---|---|
| 19 | +/+ | +/− | +/− | +/+ | +/+ | Microdeletion |
| 29 | +/− | +/− | +/− | +/+ | +/+ | Small |
| 35 | +/− | +/− | +/− | +/+ | +/+ | Small |
| 38 | +/+ | +/− | +/− | +/+ | +/+ | Microdeletion |
| 39 | +/+ | +/− | +/− | +/+ | +/+ | Microdeletion |
| 41 | +/+ | +/− | +/− | +/+ | +/+ | Microdeletion |
| 46 | +/+ | +/− | +/− | +/+ | +/+ | Microdeletion |
| 49 | +/+ | +/− | +/− | +/+ | +/+ | Microdeletion |
| 91 | +/+ | +/− | +/− | +/− | +/+ | Large |
| 184 | +/+ | +/− | +/− | +/+ | +/+ | Microdeletion |
| 28 | +/− | −/− | −/− | +/− | +/− | Microdeletionf largeg |
| 72 | +/+ | −/− | −/− | +/+ | +/+ | Microdeletion |
| 88 | +/− | −/− | −/− | +/− | +/− | Microdeletionf Largeg |
Abbreviations: BAC, bacterial artificial chromosome; FISH, fluorescence in situ hybridisation.
The summarised FISH analysis described the findings from the largest clonal cell population.
+/+ no copy change; +/− hemizygous deletion; −/− homozygous deletion.
Estimated microdeletion: ∼200 kb.
Estimated small deletion: ∼7–8 Mb.
Estimated large deletion: ∼19–21 Mb.
10q23.2 (88.2–88.6 Mb).
10q23.2 (89.4–89.5 Mb).
PTEN locus (89.6–89.7 Mb).
10q23.33 (96.2–96.6 Mb).
10q25.1 (108.9–109.3 Mb).
First event.
Second event.
DISCUSSION
The current challenge faced by prostate cancer researchers is to discover the critical genes and cognate molecular pathways responsible for the onset of neoplasia and disease progression, as well as the development of novel therapeutic strategies based on these discoveries. Initial progress in understanding the genetics of this disease started with cytogenetic and genomic analyses of primary prostate cancer tumours. Chromosomal losses of 10q suggested that PTEN at cytoband 10q23.3 might be a tumour suppressor gene involved in the development of prostate cancer (Whang et al, 1998). More recently, a single-nucleotide polymorphism mapping array in prostate cancer (Liu et al, 2006) has implicated the PTEN region to be the most frequently deleted in prostate cancer. The reported frequency of PTEN deletion in prostate cancer varies widely, most likely as a result of differences in tissue preparation, stage of disease, and the methodology used to detect molecular aberrations (Yoshimoto et al, 2006a). The heterogeneous nature of these studies has potentially obscured the clinical impact of PTEN loss in human prostate cancer. Our findings using interphase FISH analysis of prostatic adenocarcinoma TMAs have shown that PTEN deletion is an important event in tumour progression of prostate cancer. The value of analysing PTEN genomic losses by FISH methodologies is illustrated by its ability to distinguish both deletion events associated with homozygous PTEN losses in tumours. Moreover, our FISH analysis is able to predict that 70% of hemizygous PTEN deletion will involve an interstitial microdeletion within band 10q23.2–23.31 since flanking BAC probes were usually not deleted.
PTEN is a phosphoinositide 3-phosphatase which negatively regulates the PI3K/AKT signalling pathway (Ohigashi et al, 2005). Investigations to understand the role of PTEN loss have utilised a well-characterised animal model of human prostate cancer (Kwabi-Addo et al, 2001). Analysis of tumour progression in Pten (+/–) heterozygous mice, coupled with analysis of the PTEN gene and protein in the resulting tumours, has shown that haploinsufficiency of the PTEN gene promotes instability and the progression of prostate cancer (reviewed in Baker, 2007). Decreased PTEN activity has also been identified in several human cancers including prostate cancer (McMenamin et al, 1999; Koksal et al, 2004). A recent FISH and immunohistochemical PTEN analysis reported by our group showed deletions of PTEN at a very high frequency prostate cancer (Yoshimoto et al, 2006a). Other studies have demonstrated an association between decreased PTEN protein expression and a higher Gleason grade and advanced tumour stage (McMenamin et al, 1999; Koksal et al, 2004; Schmitz et al, 2007). Recently, complete loss of PTEN expression was observed in 26 of 112 (23%) of prostate cancer patients at the time of first diagnosis (Schmitz et al, 2007). Moreover, it was reported that 25 of 42 (59%) of both the neoplastic prostate glands and the invasive prostatic adenocarcinomas cells in the lymph node showed complete lack of PTEN expression, and of these 52% exhibited already loss of PTEN expression at first diagnosis. To determine whether PTEN genomic losses were also more prevalent in metastatic disease, as predicted by the above reports, we selected a small cohort of paired primary prostate adenocarcinomas and metastatic adenocarcinoma in the regional lymph nodes derived from the same patient. A very high frequency (90%) of PTEN deletion was evident in this study group, consistent with notion that loss of PTEN function is generally associated with more aggressive disease. Interestingly, two patients with hemizygous deletions in their primary tumours had homozygous deletions in their matched nodal metastatic sample. These data are in keeping with the idea that complete loss of PTEN function is associated with transition to more progressive disease (Verhagen et al, 2006; Schmitz et al, 2007).
In this study, we also demonstrate that both hemi- and homozygous PTEN loss is a highly significant prognostic marker for poor clinical outcome in prostate cancer. Prediction of 5-year biochemical risk failure analysis showed that PTEN status (not deleted, hemi- or homozygous deletion) correlated specifically with biochemical recurrence based on PSA levels. Our findings show a strong association between PTEN deletion and a shorter time interval to PSA recurrence. Significantly, homozygous PTEN deletion in five tumours was associated with a much earlier onset of biochemical recurrence based on PSA values. These tumours also had a median tumour volume typically >20%. Similar to the findings of (Schmitz et al, 2007) PTEN loss was not associated with Gleason score (Pearson's χ2 test, P=0.142). However, the absence of a statistical relationship in our study may have arisen because of the relatively small distribution of different Gleason scores in our cohort.
These novel findings implicate further an early role for PTEN haploinsufficiency and poor clinical outcome in prostate cancer. Our studies have demonstrated that the presence of PTEN genomic losses are frequent at diagnosis and are a significant prognostic marker for the subsequent development of clinically advanced disease. Previously, PTEN inactivation has been primarily been detected in locally advanced (Whang et al, 1998) or metastatic prostate cancer (Schmitz et al, 2007). Our new findings suggest that the acquisition of the deletion and concomitant loss of PTEN functional activity at an earlier phase in prostatic oncogenesis is an important determinant of the molecular pathways that govern a more aggressive tumour phenotype. Knowledge of the pathways downstream to PTEN and the genomic status of the PTEN gene in tumours must be considered in the design of future therapeutic trials of prostate cancer.
Acknowledgments
This research was supported by the National Cancer Institute of Canada (NCIC), Canadian Prostate Cancer Research Initiative (CPCRI) with funds from the Canadian Cancer Society. We would like to acknowledge the technical help of immunohistochemistry group in the Department of Pathology, Centro de Tratamento e Pesquisa, Hospital do Câncer, A.C. Camargo, São Paulo, Brazil; and Fundação de Amparo e Pesquisa de São Paulo (FAPESP) for infrastructure support for the sabbatical program of JS. Our thanks to Dr Georges Maire for his critical reading and helpful suggestions to improve this paper.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Ahlers CM, Figg WD (2006) ETS-TMPRSS2 fusion gene products in prostate cancer. Cancer Biol Ther 5: 254–255 [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Ludwig RL, Woods DB, Copeland TD, Weber HO, MacRae EJ, Vousden KH (2002) Phosphorylation of HDM2 by Akt. Oncogene 21: 1955–1962 [DOI] [PubMed] [Google Scholar]
- Baker SJ (2007) PTEN enters the nuclear age. Cell 128: 25–28 [DOI] [PubMed] [Google Scholar]
- Bertram J, Peacock JW, Fazli L, Mui AL, Chung SW, Cox ME, Monia B, Gleave ME, Ong CJ (2006) Loss of PTEN is associated with progression to androgen independence. Prostate 66: 895–902 [DOI] [PubMed] [Google Scholar]
- Besson A, Robbins SM, Yong VW (1999) PTEN/MMAC1/TEP1 in signal transduction and tumorigenesis. Eur J Biochem 263: 605–611 [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC (1998) Regulation of cell death protease caspase-9 by phosphorylation. Science 282: 1318–1321 [DOI] [PubMed] [Google Scholar]
- Cerveira N, Ribeiro FR, Peixoto A, Costa V, Henrique R, Jeronimo C, Teixeira MR (2006) TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia 8: 826–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241 [DOI] [PubMed] [Google Scholar]
- DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI (2003) Pathological and molecular aspects of prostate cancer. Lancet 361: 955–964 [DOI] [PubMed] [Google Scholar]
- Elo JP, Visakorpi T (2001) Molecular genetics of prostate cancer. Ann Med 33: 130–141 [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL (2004) Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest 113: 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goberdhan DC, Wilson C (2003) PTEN: tumour suppressor, multifunctional growth regulator and more. Hum Mol Genet 12(Spec No 2): R239–R248 [DOI] [PubMed] [Google Scholar]
- Graff JR, Konicek BW, McNulty AM, Wang Z, Houck K, Allen S, Paul JD, Hbaiu A, Goode RG, Sandusky GE, Vessella RL, Neubauer BL (2000) Increased AKT activity contributes to prostate cancer progression by dramatically accelerating prostate tumor growth and diminishing p27Kip1 expression. J Biol Chem 275: 24500–24505 [DOI] [PubMed] [Google Scholar]
- Hughes S, Yoshimoto M, Beheshti B, Houlston RS, Squire JA, Evans A (2006) The use of whole genome amplification to study chromosomal changes in prostate cancer: insights into genome-wide signature of preneoplasia associated with cancer progression. BMC Genomics 7: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ (2006) Cancer statistics, 2006. CA Cancer J Clin 56: 106–130 [DOI] [PubMed] [Google Scholar]
- Kasahara K, Taguchi T, Yamasaki I, Kamada M, Yuri K, Shuin T (2002) Detection of genetic alterations in advanced prostate cancer by comparative genomic hybridization. Cancer Genet Cytogenet 137: 59–63 [DOI] [PubMed] [Google Scholar]
- Kawai T, Hiroi S, Nakanishi K, Sakurai Y, Torikata C (2004) Abnormalities in chromosome 17 and p53 in lung carcinoma cells detected by fluorescence in situ hybridization. Pathol Int 54: 413–419 [DOI] [PubMed] [Google Scholar]
- Koksal IT, Dirice E, Yasar D, Sanlioglu AD, Ciftcioglu A, Gulkesen KH, Ozes NO, Baykara M, Luleci G, Sanlioglu S (2004) The assessment of PTEN tumor suppressor gene in combination with Gleason scoring and serum PSA to evaluate progression of prostate carcinoma. Urol Oncol 22: 307–312 [DOI] [PubMed] [Google Scholar]
- Korshunov A, Sycheva R, Gorelyshev S, Golanov A (2005) Clinical utility of fluorescence in situ hybridization (FISH) in nonbrainstem glioblastomas of childhood. Mod Pathol 18: 1258–1263 [DOI] [PubMed] [Google Scholar]
- Kwabi-Addo B, Giri D, Schmidt K, Podsypanina K, Parsons R, Greenberg N, Ittmann M (2001) Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc Natl Acad Sci USA 98: 11563–11568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, Li T, Yan G, Sun J, Sun J, Adams TS, Turner AR, Kim JW, Meyers DA, Zheng SL, Isaacs WB, Xu J (2006) Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer 45: 1018–1032 [DOI] [PubMed] [Google Scholar]
- Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, Loda M, Lane HA, Sellers WR (2004) mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med 10: 594–601 [DOI] [PubMed] [Google Scholar]
- McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR (1999) Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res 59: 4291–4296 [PubMed] [Google Scholar]
- Merscher S, Marondel I, Pedeutour F, Gaudray P, Kucherlapati R, Turc-Carel C (1997) Identification of new translocation breakpoints at 12q13 in lipomas. Genomics 46: 70–77 [DOI] [PubMed] [Google Scholar]
- Mezzelani A, Alasio L, Bartoli C, Bonora MG, Pierotti MA, Rilke F, Pilotti S (1999) c-erbB2/neu gene and chromosome 17 analysis in breast cancer by FISH on archival cytological fine-needle aspirates. Br J Cancer 80: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GJ, Brawer MK, Sakr WA, Thrasher JB, Townsend R (2001) Prostate cancer: serum and tissue markers. Rev Urol 3(Suppl 2): S11–S19 [PMC free article] [PubMed] [Google Scholar]
- Ohigashi T, Mizuno R, Nakashima J, Marumo K, Murai M (2005) Inhibition of Wnt signaling downregulates Akt activity and induces chemosensitivity in PTEN-mutated prostate cancer cells. Prostate 62: 61–68 [DOI] [PubMed] [Google Scholar]
- Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA (2006) TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res 66: 8337–8341 [DOI] [PubMed] [Google Scholar]
- Qian J, Jenkins RB, Bostwick DG (1998) Determination of gene and chromosome dosage in prostatic intraepithelial neoplasia and carcinoma. Anal Quant Cytol Histol 20: 373–380 [PubMed] [Google Scholar]
- Ribeiro FR, Diep CB, Jeronimo C, Henrique R, Lopes C, Eknaes M, Lingjaerde OC, Lothe RA, Teixeira MR (2006a) Statistical dissection of genetic pathways involved in prostate carcinogenesis. Genes Chromosomes Cancer 45: 154–163 [DOI] [PubMed] [Google Scholar]
- Ribeiro FR, Henrique R, Hektoen M, Berg M, Jeronimo C, Teixeira MR, Lothe RA (2006b) Comparison of chromosomal and array-based comparative genomic hybridization for the detection of genomic imbalances in primary prostate carcinomas. Mol Cancer 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M, Grignard G, Margue C, Dippel W, Capesius C, Mossong J, Nathan M, Giacchi S, Scheiden R, Kieffer N (2007) Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int J Cancer 120: 1284–1292 [DOI] [PubMed] [Google Scholar]
- Sirvent N, Forus A, Lescaut W, Burel F, Benzaken S, Chazal M, Bourgeon A, Vermeesch JR, Myklebost O, Turc-Carel C, Ayraud N, Coindre JM, Pedeutour F (2000) Characterization of centromere alterations in liposarcomas. Genes Chromosomes Cancer 29: 117–129 [DOI] [PubMed] [Google Scholar]
- Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I (2006) Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer 45: 717–719 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM (2006) TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res 66: 3396–3400 [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648 [DOI] [PubMed] [Google Scholar]
- van Dekken H, Paris PL, Albertson DG, Alers JC, Andaya A, Kowbel D, van der Kwast TH, Pinkel D, Schroder FH, Vissers KJ, Wildhagen MF, Collins C (2004) Evaluation of genetic patterns in different tumor areas of intermediate-grade prostatic adenocarcinomas by high-resolution genomic array analysis. Genes Chromosomes Cancer 39: 249–256 [DOI] [PubMed] [Google Scholar]
- Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, Siebert R (2006) FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn 8: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen PC, van Duijn PW, Hermans KG, Looijenga LH, van Gurp RJ, Stoop H, van der Kwast TH, Trapman J (2006) The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol 208: 699–707 [DOI] [PubMed] [Google Scholar]
- Verhagen PC, Zhu XL, Rohr LR, Cannon-Albright LA, Tavtigian SV, Skolnick MH, Brothman AR (2000) Microdissection, DOP-PCR, and comparative genomic hybridization of paraffin-embedded familial prostate cancers. Cancer Genet Cytogenet 122: 43–48 [DOI] [PubMed] [Google Scholar]
- Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP (1995) In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 9: 401–406 [DOI] [PubMed] [Google Scholar]
- Wang J, Cai Y, Ren C, Ittmann M (2006) Expression of VARIANT TMPRSS2/ERG fusion messenger rnas is associated with aggressive prostate cancer. Cancer Res 66: 8347–8351 [DOI] [PubMed] [Google Scholar]
- Whang YE, Wu X, Suzuki H, Reiter RE, Tran C, Vessella RL, Said JW, Isaacs WB, Sawyers CL (1998) Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci USA 95: 5246–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Mousses S, Hautaniemi S, Karhu R, Huusko P, Allinen M, Elkahloun A, Monni O, Chen Y, Kallioniemi A, Kallioniemi OP (2004) High-resolution analysis of gene copy number alterations in human prostate cancer using CGH on cDNA microarrays: impact of copy number on gene expression. Neoplasia 6: 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Cutz JC, Nuin PA, Joshua AM, Bayani J, Evans AJ, Zielenska M, Squire JA (2006a) Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet 169: 128–137 [DOI] [PubMed] [Google Scholar]
- Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, Zielenska M, Squire JA (2006b) Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia 8: 465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzelsberger H, Engert D, Walch A, Kulka U, Aubele M, Hofler H, Bauchinger M, Werner M (2001) Chromosomal changes during development and progression of prostate adenocarcinomas. Br J Cancer 84: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.