Abstract
We have used in vitro evolution to probe the relationship between stability and activity in a mesophilic esterase. Previous studies of these properties in homologous enzymes evolved for function at different temperatures have suggested that stability at high temperatures is incompatible with high catalytic activity at low temperatures through mutually exclusive demands on enzyme flexibility. Six generations of random mutagenesis, recombination, and screening stabilized Bacillus subtilis p-nitrobenzyl esterase significantly (>14°C increase in Tm) without compromising its catalytic activity at lower temperatures. Furthermore, analysis of the stabilities and activities of large numbers of random mutants indicates that these properties are not inversely correlated. Although enhanced thermostability does not necessarily come at the cost of activity, the process by which the molecule adapts is important. Mutations that increase thermostability while maintaining low-temperature activity are very rare. Unless both properties are constrained (by natural selection or screening) the evolution of one by the accumulation of single amino acid substitutions typically comes at the cost of the other, regardless of whether the two properties are inversely correlated or not correlated at all.
Natural enzymes are poised on the brink of conformational instability, with native structures that walk a tightrope between large stabilizing and destabilizing forces. The molecular origins of enzyme stability are critical to understanding how proteins fold into their unique three-dimensional structures as well as to understanding the limits of life. The challenge, however, is daunting. Life on earth exists over a temperature range of nearly 200°C, yet proteins isolated from organisms inhabiting the very coldest and hottest environments show only subtle structural differences (1).
It has been long hoped that studies of naturally thermostable proteins would yield insights into their adaptive mechanisms as well as general rules that could be applied to stabilizing other, less stable proteins. Sequences of homologous enzymes from mesophilic organisms (optimal growth temperature ≈30–50°C) have been compared with those from thermophiles (≈50–80°C) and extreme thermophiles (≥80°C) in an effort to identify the interactions responsible for conferring enhanced thermostability (2–4). Numerous and intensive site-directed mutagenesis studies also have probed this issue (5–7). Despite these efforts, considerable disagreement remains over which forces dominate thermostabilization mechanisms, and no generally applicable rules have been established. Some of the confusion arises from the large evolutionary distances that separate thermophilic enzymes from their mesophilic homologs. The substantial sequence differences greatly complicate attempts to identify specific mutations responsible for changes in thermostability. Moreover, large changes in thermostability often reflect the cumulative small contributions of many mutations. In addition, the effects of temperature on the forces contributing to protein stability are many and highly complex (8). Rules for engineering protein stability by rational design are likely to be protein specific, and any such design effort would need to be guided by detailed structural information.
The design problem becomes even more challenging if improvements in thermostability are not to come at the cost of enzyme activity, particularly at reduced temperatures. The activities of thermophilic enzymes at lower temperature often are compromised with respect to their mesophilic or psychrophilic cousins (9–11). This has led to the suggestion that enzymes cannot be both highly active at low temperature and highly thermostable. This would be expected, for example, if thermostability and catalysis make mutually exclusive demands on enzyme flexibility. An alternate explanation for the often-observed trade-off between stability and activity in enzymes evolved for different temperatures, however, is not that these properties are incompatible or even inversely correlated, but simply that natural selection has exerted pressure on one but not both (12). Here we show that the thermal stability of an enzyme can be increased significantly without cost to its activity at lower temperature by directed evolution and screening for both properties.
MATERIALS AND METHODS
Random Mutagenesis.
Plasmid pNB106R (13) contains the 1.5-kb wild-type p-nitrobenzyl esterase (pNBE) gene from Bacillus subtilis under control of a temperature-sensitive promoter. Random mutations were introduced during mutagenic PCR (14). For the 2.0-kb fragment amplification no manganese was required to obtain the desired low level of mutagenesis (1–2 amino acid substitutions). Primers RM1A (5′-CAATTATCTAGACTACACGAG) and RM2A (5′-GGTGGCTGACACTCGGTGAGG) flank the gene beyond the XbaI and BamHI restriction sites present in the plasmid. Forty picomoles of each primer was mixed with 10 ng of the expression plasmid containing the parent esterase gene in a 100-μl PCR. The reaction conditions were: 50 mM KCl/10 mM Tris, pH 8.5/0.1% Triton X-100/7 mM MgCl2/1 mM dCTP/1 mM TTP/0.2 mM dATP/0.2 mM dGTP/5 units Taq polymerase (Promega). The reaction was thermocycled for 25 cycles of 94°C, 1 min; 45°C, 1 min; 72°C, 1 min. Amplification of the 2-kb product was checked by running a small aliquot of the reaction on an agarose gel.
DNA Shuffling.
In vitro recombination was based on the method described by Stemmer (15), with modifications in the fragmentation reaction as described by Lorimer and Pastan (16).
Mutant Library Construction.
Mutagenic PCR or shuffling reaction products were purified using a DNA purification column (Promega) and cloned (using the XbaI and BamHI restriction sites) back into the expression vector using standard molecular biology techniques (17). The resulting plasmids were transformed into freshly prepared electrocompetent TG1 cells, which were plated on Luria-Bertani agar plates containing 20 μg/ml tetracycline. After 24 hr of growth colonies were picked with sterile toothpicks into 96-well plates containing 200 μl of 2× YT medium with 20 μg/ml tetracycline. Plates were incubated at 30°C for 24 hr to allow cell growth. Plates were then duplicated by transferring 5 μl from each well into a new plate containing fresh media and antibiotic. The original plates were stored, and the duplicate plates were grown for an additional 14 hr, after which the temperature was raised to 42°C to induce esterase expression. After 8 hr, the plates were screened.
Thermostability Screening Assay.
For identification of thermostable pNBE variants, a 96-well plate thermostability screening assay was developed based on the esterase activity assay described (18). From each well of the induced 96-well plate, 20 μl of cell culture was transferred to the corresponding wells of two new plates. One of these plates was assayed directly for enzyme activity (Ai) by the addition of 100 μl of 0.25 mM p-nitrophenyl acetate (Sigma) in 0.1 M Tris⋅HCl, pH 7.5. Mutants displaying less than 20% of parent activity at 30°C were removed from further consideration. The duplicate plate was heat-treated for 10 min at a specific temperature (Tm of the parent), chilled on ice for 15 min, incubated for 30 min at room temperature, and assayed for residual activity (Ar). The esterase reaction was monitored in a 96-well plate reader at 405 nm. The ratio s = Ar/Ai was used to estimate thermostability. Enhanced thermostability was verified by measuring Ar over a range of temperatures.
Statistical Analysis.
Linear correlation coefficients (r) for the initial activities (A) and stabilities (s = Ar/Ai) of clones in each generation mutant library were calculated using (19)
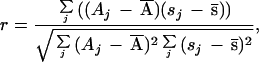 |
1 |
where Ā and s̄ are the mean values of the initial activities and stabilities, respectively, for the population. Positive values of r indicate that the two properties are correlated, while negative values indicate an inverse correlation. Clones displaying initial activities less than 20% of the parent value were not included in these calculations since low initial activities generate artificially high stability values. Only data collected on the same day were used to calculate r, as slightly different conditions can lead to significant variations in the absolute values. At least 200 mutants were used for each calculation.
Enzyme Purification.
Wild-type pNBE and variants were purified as described (18). A Q Sepharose column (Pharmacia) equilibrated with 20 mM triethanolamine, pH 7.5/50 mM NaCl/1 mM 2-mercaptoethanol/0.5 mM EDTA was used in place of the DEAE Sephadex column. The final protein pellet was resuspended in this same buffer before application to the column. Fractions exhibiting pNBE activity were analyzed by using SDS/PAGE, and samples showing 90% purity or higher were used in further studies. Enzyme activities measured before and after filtration indicated that at least 50% of wild-type esterase is inactivated during purification, while the thermostable variants remain stable during purification. Filtration does not affect the measurement of Tm by differential scanning calorimetry (DSC).
DSC and Enzyme Activity.
DSC was performed on a MicroCal MC-2 differential scanning microcalorimeter (MicroCal, Northampton, MA). A scanning rate of 1°C/min and protein concentrations of between 14.5 and 15.5 μM were used for all experiments. Before measurements, samples were dialyzed for at least 14 hr against 0.1 M Pipes (pH 7.0) with one change of buffer and degassed under stirring in vacuo. Experimental traces were corrected for the calorimeter baseline obtained by scanning the appropriate buffer solution in both cells of the calorimeter. Tm was determined from the maximum of the transition. Enzyme activity assays were performed using purified, filtered enzyme as described by Moore and Arnold (18).
RESULTS AND DISCUSSION
Directed Evolution of Thermostable Esterases.
B. subtilis pNBE, a monomer of 490 aa, hydrolyzes p-nitrobenzyl ester intermediates in the synthesis of cephalosporin-type antibiotics (13, 20). Wild-type pNBE has a Tm of 52.5°C (Fig. 1A). Thermostable variants were identified in a rapid screen of enzyme libraries expressed in Escherichia coli and picked into 96-well microtiter plates. Positives exhibiting high activity before and after incubation at high temperature were selected for purification and analysis by DSC. The gene encoding the mutant with the highest Tm was used to parent the next generation. Gene libraries were created by mutagenic PCR performed under conditions that average 1–2 amino acid substitutions per gene, conditions deemed optimal for the improvement of specific properties by mutagenesis and screening (21).
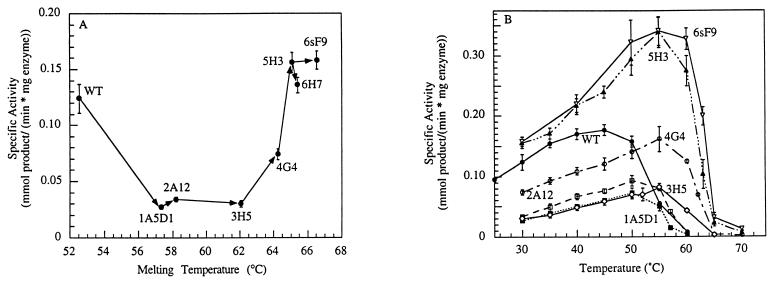
Figure 1.
Activities of purified wild-type (WT) and evolved esterases. (A) Evolutionary progress of the melting temperatures (Tm) and specific activities of evolved esterases. DSC was performed at least twice on each purified enzyme. (B) Activities of WT (•), and evolved esterases 1A5D1 (■), 2A12 (□), 3H5 (⋄), 4G4 (○), 5H3 (▴), and 6sF9 (▿), as a function of temperature. The temperature of optimal activity increases with increasing thermostability.
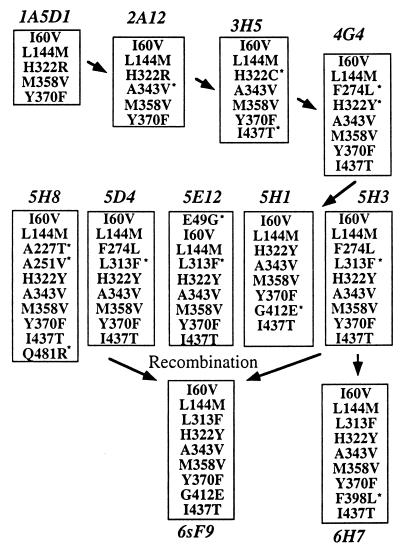
Two genes from the first random mutant library were recombined (using a unique XhoI site) to create gene 1A5D1, which encodes an esterase with a Tm of 57.3°C (Fig. 1A). Mutagenic PCR of gene 1A5D1 created a second generation library, from which mutant 2A12 with a Tm of 58.2°C was identified. This process of random mutagenesis and screening (500–2,000 clones per generation) was repeated a total of six times. Screening 1,500 clones from the random library prepared from fifth-generation gene 5H3 resulted in a single clone (6H7) with only marginally increased stability. Therefore, genes from the five best clones from the fifth-generation library were recombined by DNA shuffling to generate a second sixth-generation library. These genes contain new point mutations as well as novel combinations of the mutations from the five parental genes. Screening 1,500 clones yielded a mutant (6sF9) more stable than any of its parents. At 66.5°C, the Tm of 6sF9 is a full 14°C above wild type. This increase in thermostability is comparable to the difference between homologous mesophilic and thermophilic enzymes and is large compared with results typically obtained by site-directed mutagenesis. Site-directed mutagenesis has been similarly successful only where extensive structural and sequence data were available (22–25).
Relationship Between Stability and Activity in Evolved Esterases.
Increases in thermostability were accepted in generations 1–3 at the cost of an increased Km and decreased kcat at 30°C (Fig. 1 and Table 1). In the fourth generation, Km recovers to wild-type levels. The wild-type activity level is reached again in mutant 5H3 because of a further decrease in Km, and this activity is preserved in 6sF9. Although we did not screen for activity at high temperature, an increase in Tm always resulted in an upward shift in the temperature optimum for activity, as shown in Fig. 1B. Furthermore, the activity increases with temperature until the enzyme denatures. Thus, it appears that simultaneously screening for maintenance of low-temperature activity and improved thermostability is sufficient to obtain enzyme variants that are highly active over a wide temperature range.
Table 1.
Kinetic parameters for pNB esterase wild type and mutants in 0.1 M Pipes, pH 7.0, at 30°C
| Generation | Km, mM | kcat, s−1 | kcat/Km, M−1⋅s−1 × 105 |
|---|---|---|---|
| WT | 1.9 (0.3) | 720 (70) | 3.9 |
| 1A5D1 | 4.4 (1.2) | 390 (70) | 0.89 |
| 2A12 | 3.5 (0.5) | 400 (60) | 1.1 |
| 3H5 | 3.1 (0.1) | 320 (30) | 1.1 |
| 4G4 | 2.1 (0.2) | 470 (20) | 2.2 |
| 5H3 | 0.71 (0.08) | 430 (30) | 6.1 |
| 6sF9 | 0.60 (0.07) | 470 (60) | 7.8 |
Reported are the mean (SD) from at least two experiments.
The existence of enzyme variants that are significantly more thermostable without losing low-temperature activity indicates that activity and thermostability are at least partially independent properties and can be optimized in the same enzyme. Improvements in catalytic activity are particularly easy to acquire when the enzyme is poorly optimized toward a particular substrate (26) or under nonnatural conditions (27). The natural substrate(s) of this enzyme is not known.
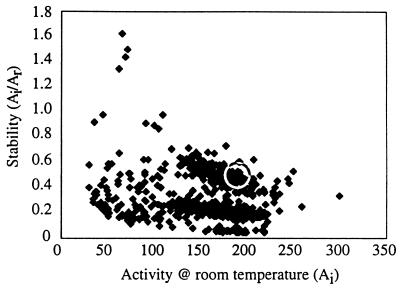
Fig. 2 shows the activities and stabilities (as measured by the ratio between retained activity after incubation and initial activity) of all the clones from a single generation. While there are variants that are more stable and variants that are more active than the parent, none of the 1,100 clones picked from this particular library is both more stable and more active. Whether these properties are inversely correlated for a given mutant library can be discerned from the linear correlation coefficient calculated from the measured activities and stabilities (see Materials and Methods). Although the values of the correlation coefficients vary somewhat from library to library, the mean value is −0.09 and the median is 0.02 (with an SD of 0.3), indicating that over the entire population of random pNBE mutants catalytic activity and stability are not (strongly) correlated. Because the mutations are distributed over the full length of the gene, inverse correlations that exist for specific residues, such as those in the active site (9), are not identifiable.
Figure 2.
Results of screening 1,100 clones picked from the second-generation library. Values from multiple screens of the parent enzyme (1A5D1) fall within the circle. Variants with less than 20% of the initial activity of 1A5D1 have been removed.
Mutations that improve both properties simultaneously are rare. Thus, the process by which a trait is acquired becomes particularly important in determining the product enzyme. Since most mutations are neutral or deleterious, an increase in any one property is likely to come at the cost of another if the experiment, natural or laboratory, does not constrain both (and the two properties are not positively correlated). Thus, two properties need not be physically coupled to show an apparent trade-off during evolution. Extensive screening during laboratory evolution allowed us to identify and incorporate rare mutations that allow thermostability to increase at no cost to activity. Less stringent natural conditions, however, would allow the fixation of the more frequent mutations that sacrifice stability for increased activity (and vice versa). For example, if life began at high temperature (28) and the ancestors of today’s mesophiles were thermophilic (29), thermostability likely drifted down while enhanced activity at low temperatures was acquired. If thermostability is drifting in mesophilic (or psychrophilic) enzymes, sequence comparisons of homologous enzymes are unlikely to provide useful rules for engineering protein stability.
By selecting only highly active mutants to parent subsequent generations, we significantly restricted the available evolutionary paths to thermostability. This may explain our difficulty (especially in later generations) in finding mutants that could meet all criteria. By removing the requirement for high activity at room temperature, we should be able to identify enzymes with activity profiles shifted to even higher temperatures. Because single mutations that improve two or more properties are rare, it should be more efficient to direct the evolution of multiple independent properties by recombining those properties that evolved separately (30).
Amino Acid Substitutions in Thermostable Esterases.
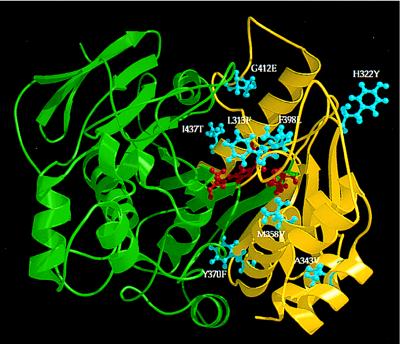
Thermostabilizing mutations were identified by sequencing the genes encoding thermostable esterases (Fig. 3). Where there were ambiguities because of two or more amino acid substitutions appearing in a single generation, mutations were incorporated individually into the wild-type enzyme by site-directed mutagenesis. The eight confirmed thermostabilizing mutations, Leu-313 → Phe, His-322 → Tyr, Ala-343 → Val, Met-358 → Val, Tyr-370 → Phe, Phe-398 → Leu, Gly-412 → Glu, and Ile-437 → Thr, are shown in Fig. 4 on a structural model of the pNB esterase (18). All the stabilizing mutations are clustered in what may be a flexible C-terminal domain. A long stretch of sequence (from residue 1 to 312) corresponding to the N-terminal domain and extending into the C-terminal domain (green in Fig. 4) yielded no thermostabilizing mutations, although numerous silent mutations and mutations that are neutral with respect to stability did appear in this region. This indicates that the C-terminal domain limits the thermostability of wild-type pNB esterase.
Figure 3.
Amino acid mutations in the most thermostable variants from each generation (∗ indicates a new mutation). With the exception of 6sF9, which was generated by DNA shuffling of five parents, all variants were generated by random mutagenesis of a single parent.
Figure 4.
Model of pNB esterase constructed based on homology to esterases of known structure (18), showing positions of thermostabilizing mutations (blue). The model is displayed using molscript (31). Active-site residues are shown in red. Residues 1–312 are shown in green. Thermostabilizing mutations are clustered in the C-terminal domain.
Preliminary crystallographic data for a pNB esterase mutant indicate that the model structure generally is reliable (B. Spiller and R. C. Stevens, personal communication). Based on the model (18), the solvent accessibility and secondary structure were assigned for the eight thermostabilizing mutations. Seven are at least partially solvent accessible: five (322, 343, 358, 370, and 437) are on the surface, and two (313 and 398) are in the active-site cleft. Single buried mutations that improve stability appear to be rare, presumably because buried mutations are not easily made without compensating changes. Two of the mutations are in regions of undefined or loop-like structure (313 and 398), one (412) is in a turn, and the remaining mutations are in helices (322, 343, 358, 370, and 437).
Conclusions.
Many enzymes are believed to have evolved via gene duplication and mutation. This laboratory evolution experiment mimics natural evolution in that it decouples the enzyme from its biological function and accumulates primarily single-amino-acid substitutions. The cumulative contributions of a relatively small number of substitutions (only 7 of 490 in 6sF9) dramatically increase esterase thermostability. Engineering enzyme properties such as thermostability and catalytic activity, which reflect the cumulative influences of many small contributions distributed over large portions of the protein, presents a significant challenge to rational protein design. The evolutionary design process, however, circumvents the need for detailed structural and mechanistic information. The database of thermostabilizing mutations that can be generated rapidly during laboratory evolution largely is free from the confounding effects of neutral evolution and conflicting evolutionary requirements that produced extant proteins. Furthermore, the ability to evolve features or combinations of features not required in nature allows us to probe important features of protein structure and function.
Acknowledgments
We thank Dr. Litian Fu for helpful discussions and Candace Chang for excellent technical assistance. This research is supported by the Department of Energy and by the Army Research Office.
ABBREVIATIONS
- pNBE
p-nitrobenzyl esterase
- DSC
differential scanning calorimetry
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Jaenicke R, Schurig H, Beaucamp N, Ostendorp R. Adv Protein Chem. 1996;48:181–269. doi: 10.1016/s0065-3233(08)60363-0. [DOI] [PubMed] [Google Scholar]
- 2.Argos P, Rossman M G, Grau U M, Zuber H, Frank G, Tratschin J D. Biochemistry. 1979;18:5698–5703. doi: 10.1021/bi00592a028. [DOI] [PubMed] [Google Scholar]
- 3.Imanaka T, Shibazaki M, Takagi M. Nature (London) 1986;324:695–697. doi: 10.1038/324695a0. [DOI] [PubMed] [Google Scholar]
- 4.Guez-Ivanier V, Hermann M, Baldwin D, Bedouell H. J Mol Biol. 1993;234:209–221. doi: 10.1006/jmbi.1993.1575. [DOI] [PubMed] [Google Scholar]
- 5.Matthews B W. Annu Rev Biochem. 1993;62:139–160. doi: 10.1146/annurev.bi.62.070193.001035. [DOI] [PubMed] [Google Scholar]
- 6.Matthews B W. Adv Protein Chem. 1995;46:249–278. doi: 10.1016/s0065-3233(08)60337-x. [DOI] [PubMed] [Google Scholar]
- 7.Jaenicke R. FASEB J. 1996;10:84–92. doi: 10.1096/fasebj.10.1.8566552. [DOI] [PubMed] [Google Scholar]
- 8.Jaenicke R. FEMS Microbiol Rev. 1996;18:215–224. doi: 10.1111/j.1574-6976.1996.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 9.Shoichet B K, Baase W A, Kuroki R, Matthews B W. Proc Natl Acad Sci USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somero G N. J Exp Zool. 1975;194:175–188. doi: 10.1002/jez.1401940111. [DOI] [PubMed] [Google Scholar]
- 11.Meiering E M, Serrano L, Fersht A R. J Mol Biol. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber G, Buckle A M, Fersht A R. Structure. 1994;2:945–951. doi: 10.1016/s0969-2126(94)00096-4. [DOI] [PubMed] [Google Scholar]
- 13.Zock J, Cantwell C, Swartling J, Hodges R, Pohl T, Sutton K, Rosteck P, Jr, McGilvray D, Queener S. Gene. 1994;151:37–43. doi: 10.1016/0378-1119(94)90630-0. [DOI] [PubMed] [Google Scholar]
- 14.Cadwell R C, Joyce G F. PCR Methods Appl. 1994;3:S136–S140. doi: 10.1101/gr.3.6.s136. [DOI] [PubMed] [Google Scholar]
- 15.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 16.Lorimer I A, Pastan I. Nucleic Acids Res. 1995;23:3067–3068. doi: 10.1093/nar/23.15.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 18.Moore J C, Arnold F H. Nat Biotech. 1996;14:458–467. doi: 10.1038/nbt0496-458. [DOI] [PubMed] [Google Scholar]
- 19.Snedecor G W, Cochran W G. Statistical Methods. 6th Ed. Ames: Iowa State Univ. Press; 1967. [Google Scholar]
- 20.Chen Y-R, Usui S, Queener S W, Yu C-A. J Ind Microbiol. 1995;15:10–18. [Google Scholar]
- 21.Arnold F H, Moore J C. Adv Biochem Eng Biotechnol. 1997;58:1–14. doi: 10.1007/BFb0103300. [DOI] [PubMed] [Google Scholar]
- 22.Kotik M, Zuber H. Eur J Biochem. 1993;211:267–280. doi: 10.1111/j.1432-1033.1993.tb19895.x. [DOI] [PubMed] [Google Scholar]
- 23.Stearman R S, Frankel A D, Freire E, Liu B, Pabo C O. Biochemistry. 1988;27:7571–7574. doi: 10.1021/bi00419a059. [DOI] [PubMed] [Google Scholar]
- 24.Van den Burg B, Vriend G, Veltman O R, Venema G, Eijsink V G H. Proc Natl Acad Sci USA. 1998;95:2056–2060. doi: 10.1073/pnas.95.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold F H. Proc Natl Acad Sci USA. 1998;95:2035–2036. doi: 10.1073/pnas.95.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J-H, Dawes G, Stemmer W P C. Proc Natl Acad Sci USA. 1997;94:4504–4509. doi: 10.1073/pnas.94.9.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K, Arnold F H. Proc Natl Acad Sci USA. 1993;90:5618–5622. doi: 10.1073/pnas.90.12.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woese C R. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes S M, Delwiche C F, Palmer J D, Pace N R. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchner O, Arnold F H. Trends Biotechnol. 1997;15:523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]