Abstract
Type I interferons (IFNs) are helical cytokines that have diverse biological activities despite the fact that they appear to interact with the same receptor system. To achieve a better understanding of the structural basis for the different activities of α and β IFNs, we have determined the crystal structure of glycosylated human IFN-β at 2.2-Å resolution by molecular replacement. The molecule adopts a fold similar to that of the previously determined structures of murine IFN-β and human IFN-α2b but displays several distinct structural features. Like human IFN-α2b, human IFN-β contains a zinc-binding site at the interface of the two molecules in the asymmetric unit, raising the question of functional relevance for IFN-β dimers. However, unlike the human IFN-α2b dimer, in which homologous surfaces form the interface, human IFN-β dimerizes with contact surfaces from opposite sides of the molecule. The relevance of the structure to the effects of point mutations in IFN-β at specific exposed residues is discussed. A potential role of ligand–ligand interactions in the conformational assembly of IFN receptor components is discussed.
Interferons (IFNs) are important cytokines characterized by antiviral, antiproliferative, and immunomodulatory activities (1). These activities form a basis for the clinical benefits that have been observed in a number of diseases, including hepatitis, various cancers and, more recently, in multiple sclerosis (2). The IFNs are divided into the type I and type II classes. IFN-β belongs to the class of type I IFNs, which also includes IFN-α, -τ, and -ω, whereas IFN-γ is the only known member of the distinct type II class. It is thought that type I IFNs began diverging 250 million years ago when IFN-α (with its family of more than a dozen closely related intronless genes) became distinct from IFN-β (3).
Structurally, IFNs are members of the helical cytokine family, known also as the hematopoietic growth factor family. These proteins are characterized by a similar four-helical bundle topology (4). Human IFN-β (huIFN-β) is a 166-amino acid protein with 35% sequence identity to the consensus sequence of IFN-α and 50% sequence identity to murine IFN-β (muIFN-β). Unlike IFN-α, huIFN-β is glycosylated at a single site. Crystal structures have been determined for nonglycosylated muIFN-β at 2.15 Å (5) and more recently for huIFN-α2b at 2.9 Å (6). Other than IFNs, the family includes in part, growth hormone (GH), several interleukins (ILs), granulocyte colony-stimulating factor (G-CSF), granulocyte–macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (MCSF), ciliary neurotrophic factor (CNTF), leukemia inhibitory factor (LIF), erythropoietin, and leptin. The crystal structures determined for many of these helical cytokines, as well as biochemical studies, show that some of them form dimers (IFN-γ, IL-10, MCSF, IL-5) with topologies suggesting that rearrangements at the genetic level have occurred that preserve the dimerization potential (4).
The crystal structures of human growth hormone (HGH) complexed with its receptor (HGHR) (7) and of IFN-γ complexed with its high-affinity receptor chain (8) illustrate modes of interaction of members of the helical cytokine family with their receptors. The helical cytokine receptors have extracellular domains that typically contain tandem fibronectin-III-related repeats. Two fibronectin repeats form a module with a binding site for the cytokine. For convenience of discussion we term this module a cytokine-binding module (CBM). The HGH molecule is observed to be “cradled” between two identical receptor molecules, each one of which contains one CBM that interacts with different sites on the HGH molecule. In the case of the IFN-γ-receptor complex, two receptor molecules (each containing one CBM) bind the identical, twofold symmetry-related surface sites of the IFN-γ dimer. In both cases, it is believed that signal transduction is mediated by ligand-induced receptor dimerization.
Mechanistically, type I IFNs are thought to exert their biological effects through interaction with the common IFN α/β receptor (IFNAR) which then activates Janus kinases (jak1 and tyk2) and STAT transcription factors. The multimeric receptor is composed of at least two chains, IFNAR1 and IFNAR2 (9). Whereas IFNAR2 contains a single CBM, IFNAR1 carries two tandem CBMs, suggesting that it can possibly bind two ligand molecules simultaneously.
Several lines of evidence indicate that different type I IFNs may engage receptor components in a different way, possibly accounting for the marked differences in biological activities observed for different IFN subtypes (10). Mutant human cells lacking functional tyk2 retain substantial partial responsiveness and binding to IFN-β but are insensitive to IFN-α (11). In addition, certain human glioblastoma and melanoma cell lines show a much more dramatic response to IFN-β than to IFN-α (12, 13). More recently, it has been reported that when Daudi cells are stimulated with IFN-β, phosphotyrosine-modified IFNAR2 is detected after immunoprecipitation of IFNAR1 with anti-IFNAR1 antibodies. Stimulation with IFN-α did not result in this coimmunoprecipitation, suggesting that IFN-β causes an association of different strength or nature with IFNAR1 and IFNAR2 than does IFN-α (14–16).
Underlying structural differences between IFN-α and IFN-β must determine these differences in receptor engagement. Comparing the three-dimensional structures and mutational analyses of the IFNs should yield insights into the molecular features determining their distinct biological activities. Despite extensive attempts, no crystals of human IFN-β had been previously obtained because of the tendency of protein produced in Escherichia coli to aggregate (17). Here, we report the crystallization and structure determination of glycosylated recombinant huIFN-β, which was determined at 2.2-Å resolution by molecular replacement methods. A description of the structure as well as a comparison with other cytokines is presented. Existing mutagenesis data are analyzed in the context of the structure, and possible mechanisms of receptor signaling are discussed.
MATERIALS AND METHODS
Protein Preparation.
Recombinant huIFN-β was secreted from Chinese hamster ovary (CHO) cells and purified from clarified conditioned media by absorption to blue Sepharose followed by ion-exchange chromatography on SP-Sepharose. To reduce the complexity due to glycosylation, the material was further fractionated by Zn-chelating Sepharose chromatography. The resulting preparation contained mainly biantennary complex glycan structures. All agaroses were obtained from Pharmacia. For crystallization, the preparation was dialyzed into 25 mM sodium acetate, pH 5.0, and concentrated to 8 mg/ml by ultrafiltration in a Centricon-10 filter set-up (Amicon).
Crystallization.
Crystallization conditions were found by incomplete factorial screening (18) using the CrystalScreen kit from Hampton Research (Riverside, CA). Crystals were grown by the hanging drop vapor diffusion method (19) at room temperature (22°C). Typically for crystallization the well contained 14% PEG 4000, 0.1 M sodium acetate at pH 4.6, and 0.2 M ammonium acetate, and the drop contained 5 μl of protein solution mixed with an equal volume of well solution. Crystals grew in 2–3 days but were small. The size of the crystals increased considerably when they were grown in drops containing 5 μl of silica hydrogel (Hampton Research). The crystals are rod-shaped of dimensions 0.8 × 0.3 × 0.3 mm and reach their final size in about 5 days. Crystals were dissolved in water and analyzed by electrophoresis in the presence of SDS. The gel showed one major band of ≈23,000 Da representing the predominant biantennary glycoform and several weaker bands of higher molecular mass due to higher-order glycosylation.
Data Collection and Crystal Characterization.
Crystals were equilibrated in a cryoprotectant solution containing 20% (vol/vol) glycerol, 14% (wt/vol) PEG 4000, 0.1 M sodium acetate at pH 4.6, and 0.2 M ammonium acetate and were flash-frozen in a −165°C nitrogen stream. A full x-ray data set up to 2.2-Å resolution was collected from frozen crystals by using an R-AXIS II image plate system (Molecular Structure, The Woodlands, TX) mounted on a Rigaku RU-200 x-ray generator operated at 50 kV and 100 mA. An additional data set was collected at 2.6-Å resolution with a ×1000 Siemens multiwire area detector, mounted on an Elliott GX-13 generator, operated at 35 kV and 40 mA. The R-AXIS data were processed by the program biotex from Molecular Structure and the Siemens data set, with xds software (20). Data statistics are shown in Table 1. The unit cell is orthorhombic of dimensions a = 55.3 Å, b = 65.9 Å, and c = 121.5 Å. Observation of systematic absences suggested that the space group is P212121. The Matthews volume (21) is 4.83 Å3⋅Da−1, assuming a molecular mass of 22,400 Da, which suggested that there are two molecules (labeled A and B) in the asymmetric unit.
Table 1.
Crystallographic data statistics
| Measurement | Data set value
|
|
|---|---|---|
| R-AXIS | Siemens | |
| Symmetry | P212121 | P212121 |
| Unit cell, Å | a = 55.3, b = 65.9, c = 121.5 | a = 55.6, b = 66.3, c = 121.9 |
| Crystals, n | 1 | 1 |
| Resolution, Å | 2.2 | 2.6 |
| Reflections (measured) | 55,166 | 42,558 |
| Reflections (unique) | 18,405 | 13,956 |
| Rmerge, * % | 7.66 | 8.1 |
| Completeness, % | 83.1 | 84.2 |
| Completeness (2.2–2.3 Å), % | 59.5 | — |
Rmerge = ΣhΣi|Ihi − Ih|/ΣhiIhi.
Molecular Replacement.
For molecular replacement calculations the program xplor (22) was used unless otherwise noted. The data set collected with the Siemens detector was used. The search probe was a polyalanine model based on the muIFN-β Cα coordinates (Protein Data Bank entry code 1rmi) and generated by the program maxsprout (23). The correct rotation solution for molecule A was identified by Patterson correlation (PC) refinement to be the 7th highest peak of the cross-rotation function. Subsequent translation search, using the rotated probe, produced a peak of translation function, TF, = 0.177, which was 2σ higher than the next unrelated peak. The PC refinement showed no significant peak for molecule B. Molecule B was located by using the program AMoRe (24). The xplor translation function (data 10–3 Å) for the two correctly positioned models had a value of TF = 0.25, which was 6σ higher than the next unrelated peak. The correctly positioned models were subjected to rigid-body refinement. The R-factor was 50.3% and the Rfree (calculated with 10% of the data set) was 51.8% for 10–3 Å after the rigid-body refinenent (25).
Model Building and Refinement.
Initial stages of model building and refinement were done with the Siemens data set with a 2σ cutoff. Iterative cycles of model building and refinement were done using quanta (Molecular Simulations, San Diego, CA) and o (26) programs for molecular graphics manipulations and the xplor package for refinement. Initially, to minimize model bias, a partial model containing only residues from the α-helices was used for 3Fo − 2Fc map calculations. More atoms were added to the partial model as density was becoming interpretable. Occasionally, simulated annealing and restrained B-factor refinement were performed (22). No noncrystallographic symmetry restraints or constraints were included. Only manual structure modifications that resulted in lower Rfree after refinement were accepted. When Rfree reached 32%, the process was continued using the R-AXIS data set, which extended to 2.2-Å resolution. Selection of the same reflections up to 2.6 Å for calculation of Rfree was done for the new data set to ensure avoidance of model bias. For the final corrections of the model, simulated annealing omit maps were calculated consecutively by omiting 10 residues for each map (27). Water molecules were added by using the X-Solvate subroutine of quanta.
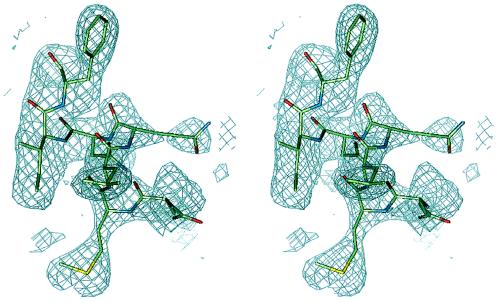
The current model of the crystal structure contains 3,521 protein atoms, 97 water molecules, and a zinc ion and has a crystallographic R factor of 22.3% and an Rfree of 28.3% for data between 100- and 2.2-Å resolution. Almost all residues are well defined in the final 2Fo − Fc electron density map. Fig. 1 shows a final 2Fo − Fc simulated annealing omit map for a representative region of the molecule. Residues 27–29 of molecule B and side chains of residues 71, 113, and 115–117 of molecule A and of residues 30 and 33 of molecule B are not included in the model because of weak electron density. In addition, some weak electron density in the region of probable carbohydrate has not been accounted for in the model because of uncertainty of the placement of these carbohydrate residues. The stereochemistry of the structure was analyzed with the program procheck (28). The rms deviations for bond lengths and angles are 0.013 Å and 1.58°, respectively, and the deviation of temperature factors in bonded atoms is 3.86 Å2. The Ramachandran diagram shows that 92.9% of all amino acid residues are in the most favored regions. The only residue in the disallowed region is Phe-111(A). Most of the residues in partially allowed regions are on the CD loop, which is associated with weak electron density. Atomic coordinates of huIFN-β have been deposited in the Brookhaven Protein Data Bank.
Figure 1.
Representative region of a simulated annealing omit map displayed in stereo for residues 61–67 of molecule A and contoured at 1σ. The figure was made with the program quanta.
RESULTS
Description of the Structure.
The crystal structure of huIFN-β shows it to be a protein molecule roughly cylindrical in shape with dimensions of approximately 20 × 30 × 40 Å. It adopts the standard fold of type I IFNs, which belong to the family of long-chain helical cytokines (4) (Fig. 2). It consists of five α-helices, labeled A (residues 2–22), B (residues 51–71), C (residues 80–107), D (residues 118–136), and E (residues 139–162) for molecule A. Helix D is six residues longer in molecule B (residues 112–136). Helices A, B, C, and E form a left-handed, type 2 (29) four-helix bundle (Fig. 2). There is a long overhand loop, the AB loop, that connects helices A and B and three shorter loops (named BC, CD, and DE) that connect the rest of the helices. All helices and a strand that constitutes most of the AB loop are roughly parallel to the long axis of the cylinder. There are several hydrophobic residues, such as Phe-70, Phe-154, Trp-79, and Trp-143, that are involved in interactions with each other that stabilize the core of the molecule. In addition, residues of the core form several hydrogen bonds such as between Gln-10 Oɛ1 and Gln-94 Nɛ2 and between Ser-118 Oγ and Thr-58 Oγ1.
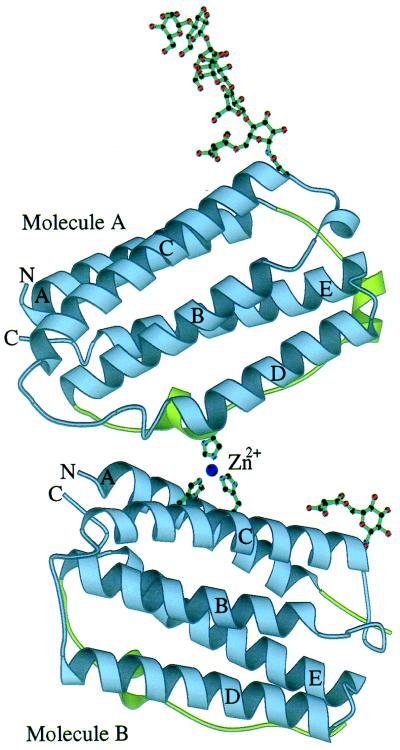
Figure 2.
Schematic representation of the crystallographic dimer of huIFN-β. The modeled portion of the carbohydrates and part of the zinc-binding site are also shown. The sphere corresponds to the zinc ion. Helices and N and C termini are labeled. The AB loop is colored green. The figure was made with the program molscript (44).
The AB loop can be conceptually subdivided into three segments: the AB1 (residues 23–35), the AB2 (residues 36–40), and the AB3 (residues 41–50) loops. The AB1 and AB3 loops connect the relatively straight AB2 loop with helices A and B, respectively. Residues 29–35 of the AB1 loop form two turns of a 310 helix, and residues 42–46 of the AB3 loop form one and a half turn of an α-helix. Cys-31 of the 310 helix forms a disulfide bridge with Cys-141 of the DE loop and plays an important role in the stabilization of the AB1 loop (30). Additional interactions that appear to stabilize the AB1 loop are hydrogen bonds between Tyr-132 OH and Asp-34 O and between Arg-147 Nη1 and Leu-24 O. There is also a free cysteine residue (Cys-17) on helix A that is proximal to the surface but buried. Its proximity to the surface is consistent with the intermolecular disulfide bond formation under certain partially denaturing conditions or after prolonged storage of the molecule (unpublished data).
A single glycosylation site exists at residue Asn-80 that involves a biantennary complex-type carbohydrate chain. Electron density is relatively interpretable for the carbohydrate on molecule A, and seven hexose rings have been included in the model (Fig. 2). On molecule B only the first two hexose rings are well defined. The carbohydrate chains from both molecules A and B are accommodated within a large solvent channel, about 45 Å wide, formed by the crystal structure. Despite the apparent absence of noncovalent interactions with neighboring molecules in the crystal, the carbohydrate of molecule A has an extended conformation. The only interactions that may be stabilizing the carbohydrate conformation are those between the core α1–6 fucose residue with the side-chain atoms of residues Asn-86 and Gln-23 in both molecules A and B and also at least two internal hydrogen bonds. Glycosylation of huIFN-β is likely to play an important role in protein solubility and stability (31). It has been observed that nonglycosylated huIFN-β is much more susceptible to aggregation (17). Inspection of the crystal structure shows that there are a relatively high number of surface-exposed hydrophobic residues in the vicinity of the glycosylation site. A possible role of the carbohydrate might be to “shield” those residues from the solvent.
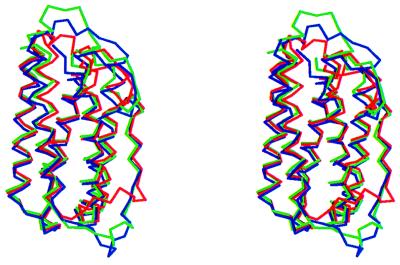
Superposition of molecules A and B (Fig. 3) shows that the conformation of certain loops is highly dependent on the immediate environment in the crystal. Thus, helix D is longer by six residues in molecule B than in molecule A, a fact that is presumably related to the extensive participation of helix D and loop CD in a crystal contact formed between molecules A and B. The biggest differences in loop conformation are observed in loops AB1 and the CD loop (the rms deviation of Cα atoms for AB1 loop is 3.04 Å and for the CD loop it is 4.08 Å). The overall rms deviation for molecules A and B is 2.42 Å and for the Cαs it is 1.83 Å. The high B-factors and the weak electron density for residues 108–117 suggest that the CD loop is very mobile. Examination of crystal contacts shows that the AB and CD loops in both molecule A and molecule B are involved in many interactions with symmetry-related molecules and therefore the conformations observed may be very much dependent on this crystal form.
Figure 3.
Superposition of Cα traces of molecules A (in green), B (blue), and muIFN-β (red) in stereo. The figure was made with the program molscript (44).
Comparison with Other IFNs.
Superposition of huIFN-β (molecule A) with muIFN-β Cα coordinates shows that the molecules have very similar structures (Fig. 3). The rms deviation for the Cα atoms of 136 structurally homologous residues is 0.71 Å. The overall rms deviation is 2.49 Å. The biggest differences are in the AB1 and CD loops (rms deviations are 6.28 and 5.68 Å, respectively). There are three more residues on the AB1 loop in huIFN-β and a 310 helix (residues 29–35) that is not observed in muIFN-β. Formation of this helix may be related to the existence of the disulfide bond in this region, which is absent from muIFN-β. A similar arrangement exists in huIFN-α2b (6), where two turns of 310 helix (helices 310A and 310B), a disulfide bridge, and one additional residue (Ser-28) on the AB1 loop are observed. The interaction between Arg-147 Nɛ and Leu-24 O in huIFN-β is conserved in muIFN-β and appears to play a role in the stabilization of the AB1 loop.
As predicted (5), the buried hydrogen bond network involving Tyr-125, Tyr-126, Asn-153, and Glu-149 is also conserved. Specifically, there are three hydrogen bonds between Tyr-125 OH⋅⋅⋅Asn-1 Nδ2, Tyr-126 OH⋅⋅⋅Asn-153 Oδ1, and Tyr-125 OH⋅⋅⋅Glu-149 Oɛ2. This network facilitates interactions between helices D and E. Another hydrogen bond network described in muIFN-β (5) and conserved in huIFN-β involves Arg-128, which interacts with main-chain carbonyl oxygens from residues Phe-38 and Asp-39. Nearby, His-121 forms a hydrogen bond with Glu-43, which itself forms a hydrogen bond with its main-chain amide nitrogen. Several water molecules form hydrogen bonds with the aforementioned residues. This network appears to stabilize the extended conformation of the AB2 loop.
As would be expected from the very high degree of sequence conservation in its region, the B helix has a structure very similar to that of muIFN-β. This is in contrast to huIFN-α2b, for which a break in the B helix was observed. As predicted (6), helix E is very similar to that of muIFN-β. The rotation of helix E around the helix axis observed in huIFN-α2b is not observed in huIFN-β.
In huIFN-β, the CD loop is the region with the highest mobility. This is similar to what has been seen in muIFN-β and even more in huIFN-α2b, where no electron density has been observed for this loop. In the case of muIFN-β the CD loop is packed tightly against the rest of the molecule but in huIFN-β the CD loop appears to be positioned away from the rest of the molecule, resulting in a lack of contacts between residues 106–115 of the CD loop and the rest of the molecule. Unlike what has been observed for muIFN-β, in huIFN-β there is no observed helical structure for the CD loop. In the muIFN-β crystal structure the CD loop appears to be involved minimally in crystal contacts and may therefore have been allowed to adopt the helical conformation. If that is the case, then there may also be a helical conformation possible for this region of the huIFN-β molecule in solution. Such a difference in the secondary structure of the CD loop has also been observed in the case of HGH when bound to its receptor (7).
Dimerization.
A zinc ion is observed to exist at the interface between molecules A and B (Fig. 2). It is coordinated in a tetrahedral manner by His-121 of molecule A and His-93 and His-97 of molecule B. A water molecule occupies the fourth coordination site. A network of hydrogen bonds formed between His-121 and Glu-43 (molecule A) and between His-97 and Gln-94 (molecule B) appears to assist in the stabilization of the zinc-binding site. The interface between molecules A and B covers an area of ≈300 Å2 and involves hydrophobic and polar residue contacts from helices A (residues 1–12) and C (residues 93–104) of molecule B and helix D (residues 113–124) and the AB3 loop (residues 42–53) of molecule A. The carbohydrate chain of molecule B lies close to the dimerization interface and could possibly interact with molecule A. It appears that formation of the interface causes the unfolding and shortening of the D helix of molecule A by six residues. The D helix, at its full length, would have been in steric conflict with molecule B. These observations taken together with the recent crystallographic observation of the existence of IFN-α2b as a zinc-mediated dimer (6) raise the possibility that a physiological huIFN-β dimer may exist. The huIFN-β dimer is different from the huIFN-α2b dimer in that the two molecules are not related by a twofold axis but by a rotation of about 44° along an axis approximately perpendicular to the plane defined by helices C, B, and D. Thus whereas huIFN-α2b dimerizes with the same contact surface (helix D and AB loop), huIFN-β dimerizes with contact surfaces from opposite sides of the molecule. It is interesting to note that none of the solutions used for the crystallization of huIFN-β contained zinc, a fact which suggests that traces of zinc remained from an earlier purification step involving the zinc-chelate chromatography column (more details are in Materials and Methods) and were in amounts sufficient for incorporation into the zinc-binding site. It should also be pointed out that huIFN-α2b crystals were grown out of solutions containing 40 mM Zn(OAc)2. Gel filtration experiments show no evidence of dimerization of huIFN-β in solution by the addition of up to 5× molar excess of zinc chloride (unpublished data).
Structure–Function Studies.
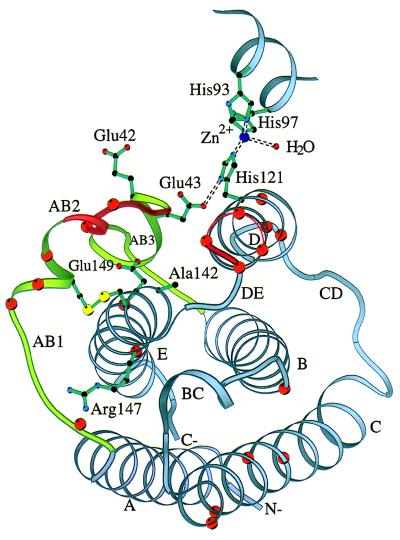
Although extensive structure-function studies of α-IFNs employing mutagenesis and construction of hybrid molecules have been reported (reviewed in ref. 17), similar studies on IFN-β up to now have been limited to chemical mutagenesis (32) and alanine-scanning mutagenesis of loops AB (Arg-33 to Glu-41) and DE (Lys-134 to Ser-137) (unpublished data cited in ref. 5) (Fig. 4). Many of the substitutions involve charged residues. Mutations Glu-42 → Lys or Glu-43 → Lys result in loss or partial-loss of function, respectively (32). Glu-42 and Glu-43 are exposed residues of the AB3 loop and lie in a region of the sequence that has several acidic residues in most type I IFNs. Glu-43 is involved in stabilizing the conformation of His-121 which participates in the formation of the zinc-binding site (Fig. 4). Glu-43 has also been identified as part of an antigenic epitope critical for function. Studies of monoclonal antibodies that neutralize huIFN-β activity showed that residues 41–43 and 46 are important for the binding of these antibodies (33). On helix E, mutations Ala-142 → Thr, Glu-149 → Lys (32), and Arg-147 → Ala (unpublished data, cited in ref. 5) also result in loss of function. Ala-142 is a buried residue at the end of helix E whose side chain makes van der Waals contacts with the side chain of Ser-139. Residues Arg-147(144) and Glu-149(146) are conserved in IFN-α and are involved in IFN-α activity. As discussed earlier, Arg-147 appears to play a role in the stabilization of the AB1 loop; however, it is also partially exposed to the solvent and could possibly be accessible to a long side chain from a receptor residue. Glu-149 is an almost completely buried residue whose side chain forms a hydrogen bond with the OH group of Tyr-125.
Figure 4.
Ribbon diagram of huIFN-β Cα backbone with side chains of residues known to be important for activity. The yellow spheres represent the sulfur atoms of the disulfide bridge. The ribbon is colored red at positions of the alanine-scanning mutagenesis cited in ref. 5. Orange spheres correspond to Cα atoms of residues homologous to those of IFN-α that are important for activity. Part of the zinc-binding site is also shown. The figure was made with the program molscript (44).
DISCUSSION
The three-dimensional structure of human IFN-β presented here reveals many similarities with structural determinants of murine IFN-β and human IFN-α2b but also displays several strikingly different features. Such a feature is the formation of an asymmetric, zinc-mediated dimer that presents contact surfaces opposite to those found in the IFN-α2b crystal structure, raising questions regarding their implications for receptor binding. The biological significance of these dimers is unclear. Evidence for the existence of an active huIFN-β dimer, gathered by using the method of radiation target analysis, has been reported (34). There are many additional cases of noncovalent or covalent dimerization of helical cytokines such as the well documented cases of HGH (35), ciliary neurotrophic factor (CNTF) (36), IFN-γ (37), and IL-10 (38). Typically, in many cases of helical cytokine dimerization the association constant is low and the residues responsible for the dimerization are not conserved between different sequences. Moreover, the relative orientations of monomers that constitute the dimers are also not conserved among the different members of the family, which suggests that there is no conservation of the dimerization interface. For instance, His-121 is conserved in muIFN-β and bovine IFN-β but not in horse IFN-β. There is no obvious conservation of His-93 and His-97 between murine, bovine, and equine IFN-βs; however, there seems to be a frequent occurrence of a histidine at the adjacent position (His-88 in muIFN-β) and an acidic residue (Asp or Glu) in the His-97 position.
IL-6 is known to form a multimeric ligand–receptor complex involving two molecules each of IL-6, α and β receptor chains (39). Recently, the crystal structure of IL-6 was reported (40). The authors proposed a model, based on mutational analyses and the IL-6 crystal structure, that predicts dimerization of IL-6 upon receptor binding. Although there is no adequate evidence for the existence of type I IFN dimers in solution, the possibility that such dimers form upon receptor binding on the cell surface cannot be excluded. Indeed, there are cases in which the environment of the crystal is known to “trap” proteins in assemblies that are thermodynamically favored only when these proteins are complexed with other molecules (41).
Structure–function studies of IFN-α have defined several distinct sequence regions that may define surfaces involved in either receptor–ligand or ligand–ligand interactions. For α-IFNs it has been suggested that IFNAR1 interactions occur through exposed C helix residues (9), and it has been further proposed that exposed residues of the A helix on the C helix side of the molecule also bind IFNAR1 (42). Mutational studies on the opposite molecular face in the AB loop, D helix, and DE loop region implicate this region as the IFNAR2 binding site. Supporting evidence for the importance of this region comes from alignment of 45 type I IFN sequences, which reveals that this surface region is highly conserved (5). Comparison of the huIFN-β structure with a stereo diagram of huIFN-α2b (6) shows that the residues of the conserved region also share similar position, conformation, and accessibility (Fig. 4).
Nonetheless, IFN-α and IFN-β do not share all activities. Differential engagement of IFN receptor subunits by α and β IFNs is implicated from observations on tyk2 mutant cell lines (43) and by the observation that IFN-β, but not IFN-α, can induce a coimmunoprecipitable complex of phosphotyrosine-modified IFNAR1 and IFNAR2 (14–16). Results from studies with other members of the helical cytokine family suggest that there is a large variability in the ways that these molecules interact with their receptors. Major differences between HGH–receptor and IFN-γ–receptor complexes are observed in their crystal structures. In addition, although helical cytokines share the same four-helix-bundle fold architecture, there are many differences in the tertiary structure (e.g., HGH and IFN-β) and quaternary structure (e.g., IFN-γ and IL-10). These differences are related to low sequence homology as well as differences in the intron–exon structure of these cytokines at the gene level. Such variability in ligand structure may also require large variability in the mode of ligand–receptor interactions. Therefore it appears that the IFN–receptor complex may not be adequately described by a model similar to the HGH–receptor complex, a conclusion also reached in ref. 17 after examining mutagenesis data for HGH and granulocyte–macrophage colony-stimulating factor.
The crystal structure of huIFN-β will allow the design of key mutations to test the functional relevance of dimerization, and it should provide additional evidence for differential receptor subunit assemblies proposed for different type I IFN subtypes. However, a complete understanding of the system may require the determination of the crystal structures of the complexes of IFNs with their receptors.
Acknowledgments
We gratefully acknowledge Stephen C. Harrison for helpful discussions, Joe Rosa for encouragement and discussions, and Laura Runkel and Eric Mogensen for discussion and editing of the manuscript. M.N and W.N.L were supported by National Institutes of Health Grant M06920 to W.N.L.
ABBREVIATIONS
- IFN
interferon
- huIFN
human IFN
- muIFN
murine IFN
- IL
interleukin
- HGH
human growth hormone
- IFNAR
IFN α/β receptor
- CBM
cytokine-binding module
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1au1).
References
- 1.Gutterman J U. Proc Natl Acad Sci USA. 1994;91:1198–1205. doi: 10.1073/pnas.91.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeMaeyer E, Galasso G, Schellekens H. The Biology of the Interferon System. Amsterdam: Elsevier/North–Holland Biomedical; 1981. [Google Scholar]
- 3.Weissmann C, Weber H. Prog Nucleic Acid Res Mol Biol. 1986;33:251–300. doi: 10.1016/s0079-6603(08)60026-4. [DOI] [PubMed] [Google Scholar]
- 4.Sprang S R, Bazan J F. Curr Opin Struct Biol. 1993;3:815–827. [Google Scholar]
- 5.Senda T, Saitoh S, Mitsui Y. J Mol Biol. 1995;253:187–207. doi: 10.1006/jmbi.1995.0544. [DOI] [PubMed] [Google Scholar]
- 6.Radhakrishnan R, Walter L J, Hruza A, Reichert P, Trotta R P, Nagabhushan T L, Walter M R. Structure. 1996;4:1453–1463. doi: 10.1016/s0969-2126(96)00152-9. [DOI] [PubMed] [Google Scholar]
- 7.De Vos A M, Ultsh M, Kossiakoff A A. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 8.Walter M R, Windsor W T, Nagabhushan T L, Lundell D J, Lunn C A, Zauodny P J, Narula S K. Nature (London) 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 9.Uzé G, Lutfalla G, Mogensen K E. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- 10.Zoon K C, Arnheiter H. Pharmacol Ther. 1984;24:259–278. doi: 10.1016/0163-7258(84)90037-8. [DOI] [PubMed] [Google Scholar]
- 11.Pellegrini S, John J, Shearer M, Kerr I M, Stark G R. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenblum M G, Yung W K A, Kelleher P J, Ruzicka F, Steck P A, Borden E C. J Interferon Res. 1990;10:141–151. doi: 10.1089/jir.1990.10.141. [DOI] [PubMed] [Google Scholar]
- 13.Garbe C, Krasagakis K. J Invest Dermatol. 1993;100:239S–244S. [PubMed] [Google Scholar]
- 14.Constantinescu S N, Croze E, Murti A, Wang C, Basu L, Hollander D, Russell-Harde D, Betts M, Garcia-Martinez V, Mullerman J E, Pfeffer L M. Proc Natl Acad Sci USA. 1995;92:10487–10491. doi: 10.1073/pnas.92.23.10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Platanias L C, Udin S, Domanski P, Colamonici O. J Biol Chem. 1996;271:23630–23633. doi: 10.1074/jbc.271.39.23630. [DOI] [PubMed] [Google Scholar]
- 16.Croze E, Russell-Harde D, Wagner T C, Pu H, Pfeffer L M, Perez H D. J Biol Chem. 1996;271:33165–33168. doi: 10.1074/jbc.271.52.33165. [DOI] [PubMed] [Google Scholar]
- 17.Mitsui Y, Senda T, Shimazu T, Matsuda S, Utsumi J. Pharmacol Ther. 1993;58:93–132. doi: 10.1016/0163-7258(93)90068-o. [DOI] [PubMed] [Google Scholar]
- 18.Jancarik J, Kim S H. J Appl Crystallogr. 1991;24:409–411. [Google Scholar]
- 19.McPherson A. In: Preparation and Analysis of Protein Crystals. Glick D, editor. New York: Wiley; 1982. pp. 82–159. [Google Scholar]
- 20.Kabsch W. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 21.Matthews B W. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A T, Kuriyan J, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. J Mol Biol. 1991;218:183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 24.Navaja J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 25.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 26.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Hodel A, Kim S-H, Brünger A T. Acta Crystallogr A. 1992;48:851–858. [Google Scholar]
- 28.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 29.Presnel S R, Cohen F E. Proc Natl Acad Sci USA. 1989;86:6592–6596. doi: 10.1073/pnas.86.17.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepard H M, Leung D, Stebbing N, Goeddel D V. Nature (London) 1981;294:563–565. doi: 10.1038/294563a0. [DOI] [PubMed] [Google Scholar]
- 31.Conradt H S, Egge H, Peter-Katalinic J, Reiser W, Siklosi T, Schaper K. J Biol Chem. 1987;262:14600–14605. [PubMed] [Google Scholar]
- 32.Stewart A G, Adair J R, Catlin G, Hynes C, Hall J, Davies J, Dawson K, Porter A G. DNA. 1987;6:119–128. doi: 10.1089/dna.1987.6.119. [DOI] [PubMed] [Google Scholar]
- 33.Redlich P N, Hoeprich P D, Jr, Colby C B, Grossberg S E. Proc Natl Acad Sci USA. 1991;88:4040–4044. doi: 10.1073/pnas.88.9.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestka S, Kelder B, Familletti P C, Noschera J A, Crowl R, Kempner E S. J Biol Chem. 1983;258:9706–9709. [PubMed] [Google Scholar]
- 35.Cunningham B C, Mulkerrin M G, Wells J A. Science. 1991;253:545–548. doi: 10.1126/science.1907025. [DOI] [PubMed] [Google Scholar]
- 36.McDonald N Q, Panayotatos N, Hendrickson W A. EMBO J. 1995;14:2689–2699. doi: 10.1002/j.1460-2075.1995.tb07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ealick S E, Cook W J, Vijay-Kumer S, Carson M, Nagabushan T L, Trotta P P, Bugg C E. Science. 1991;252:698–702. doi: 10.1126/science.1902591. [DOI] [PubMed] [Google Scholar]
- 38.Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A. Structure. 1995;3:591–601. doi: 10.1016/s0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- 39.Ward L D, Howlett G J, Discolo G, Yasukawa K, Hammacher A, Moritz R L, Simpson R J. J Biol Chem. 1994;269:23286–23289. [PubMed] [Google Scholar]
- 40.Somers W, Stahl M, Seehra J S. EMBO J. 1997;16:989–997. doi: 10.1093/emboj/16.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, Kwong P D, Hendrickson W A. Nature (London) 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 42.Uzé G, Di Marco S, Mouchel-Vielh E, Monneron D, Bandu M-T, Horisberger M A, Dorques A, Lutfalla G, Mogensen K E. J Mol Biol. 1994;243:245–257. doi: 10.1006/jmbi.1994.1651. [DOI] [PubMed] [Google Scholar]
- 43.Velazquez L, Mogensen K E, Barbieri G, Fellous M, Uzé G, Pellegrini S. J Biol Chem. 1995;270:3327–3334. doi: 10.1074/jbc.270.7.3327. [DOI] [PubMed] [Google Scholar]
- 44.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]