Abstract
Sirtuins are genes implicated in cellular and organismal ageing. Consequently, they are speculated to be involved in diseases of ageing including cancer. Various cancers with widely differing prognosis have been shown to have differing and characteristic expression of these genes; however, the relationship between sirtuin expression and cancer progression is unclear. In order to correlate cancer progression and sirtuin expression, we have assessed sirtuin expression as a function of primary cell ageing and compared sirtuin expression in normal, ‘nonmalignant’ breast biopsies to breast cancer biopsies using real-time polymerase chain reaction (PCR). Levels of SIRT7 expression were significantly increased in breast cancer (P<0.0001). Increased levels of SIRT3 and SIRT7 transcription were also associated with node-positive breast cancer (P<0.05 and P<0.0001, respectively). This study has demonstrated differential sirtuin expression between nonmalignant and malignant breast tissue, with consequent diagnostic and therapeutic implications.
Keywords: breast cancer, sirtuins, gene expression
Breast cancer is the most common cancer to affect women, with over 40 000 new cases being detected and around 13 000 women dying from the disease in Britain each year (CancerStats, 2005). The prognosis of breast cancer has traditionally been based on prognostic factors, such as tumour size, nodal status and histological grade, that have been derived from population-based studies. To improve the accuracy of predicting prognosis, scoring systems that combine these factors have been developed, such as the Nottingham Prognostic Index (NPI) (Galea et al, 1992). However, these prognostic factors are less accurate predictors of prognosis in an individual patient, owing to widely differing disease behaviour that ranges from slow-growing, nonmetastasising disease with an excellent prognosis, to fast-growing disease with a propensity for early metastasis and rapid death.
In contrast to the well-defined behaviour of breast cancer in a population, the molecular aetiology of cancer initiation and progression is not fully understood. Genes involved in biological ageing may be informative in this context, as the incidence of cancer increases as a function of chronological age. Specifically, genes involved in NAD+-dependent protein deacetylation may have a role to play in cancer pathogenesis (Marks et al, 2001; Johnstone, 2002; Neumeister et al, 2002). These genes comprise the sirtuins, orthologues of the yeast silent information regulator 2 (SIR2) family of genes. Sirtuins are a highly conserved set of genes found in organisms ranging from bacteria to man (Brachmann et al, 1995; Frye, 2000), involved in a variety of essential cell processes, including ageing, preventing differentiation, apoptosis and resistance to metabolic stress (Vaziri et al, 2001; Dryden et al, 2003). We have previously hypothesised that sirtuins, which link the functions of the mitochondrion, telomere nucleo-protein complex and ribosome production (the MTR), may be important contributors to a wide range of ageing related diseases including cancer (Shiels and Davies, 2003). Consequently, we have undertaken a study designed to investigate sirtuin expression in breast cancer. Seven sirtuins (SIRT1–7) have so far been reported in humans. Of these, SIRT2 and 7 have been associated with particular cancers with widely varying natural histories, SIRT2 with gliomas and SIRT7 with thyroid cancer (de Nigris et al, 2002; Frye, 2002; Hiratsuka et al, 2003). Given the association of sirtuin expression with clinical behaviour, we investigated sirtuin expression in breast cancer to determine if changes in the transcriptional expression of individual sirtuins correlated with malignancy and disease stage.
MATERIALS AND METHODS
Cell culture
Sirtuin expression was initially studied in replicatively ageing mammary epithelial cell lines. These included a primary mammary epithelial cell line (HMEC) obtained from Clonetics, UK, an immortalized nontumorigenic MCF-12A breast cell line from the Department of Pathology, University of Glasgow and a tumorigenic MCF-7 breast cancer cell line from the Department of Radiation Oncology, University of Glasgow. The HMECs were grown in mammary epithelial cell growth medium (MGEM) supplemented with bovine pituitary extract (BPE), epidermal growth factor, insulin, GA-1000 and hydrocortisone. The MCF-12A cells were grown in a 1 : 1 mixture of Dulbecco's modified Eagle's medium and Ham's F12 medium, supplemented with 5% foetal calf serum, 20 ng ml−1 epidermal growth factor, 100 ng ml−1 cholera toxin, 0.01 mg ml−1 bovine insulin and 500 ng ml−1 hydrocortisone. The MCF-7 cells were grown in RPMI medium with 10% foetal calf serum.
Biopsies
Archival breast biopsies stored in liquid nitrogen and held by the Department of Surgery, Western Infirmary, Glasgow, were used for this study. These biopsies came from patients with potentially curable, invasive ductal adenocarcinomas who had given permission for the storage and future analysis of samples of tissue. All studies were undertaken with the approval of the local NHS ethics committee.
RNA extraction, DNase treatment and cDNA synthesis
RNA extraction was performed using TRIzol® (Life Technologies, UK) on both the mammary epithelial cells and biopsies. Breast glandular tissue was carefully dissected free from surrounding fatty tissue. TRIzol® (1 ml) was added per 50 mg of tissue. Tissue was then homogenised using a mechanical tissue homogenizer at 0°C. RNA was DNase treated with a commercially available kit (Ambion, UK) according to the manufacturer's recommendations. Then, 2 μg of RNA was reverse transcribed into cDNA using the Superscript™ First Strand Synthesis System (Invitrogen).
Taqman® polymerase chain reaction (PCR)
Taqman® PCR was carried out on duplicate cDNA samples using an ABI Prism 7700 sequence detector in a 25 μl reaction. The mixture consisted of 20 ng of cDNA, 300 nM of each forward and reverse primer, 200 nM Taqman® probe, 12.5 μl of Taqman® Universal PCR Master Mix (Eurogentech, UK) and H2O. Cycle conditions were 50°C for 2 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Sequences of forward and reverse primers and probes used are shown below (Table 1).
Table 1. The primer and probe sequences used in Taqman® PCR.
| Forward primer (5′–3′) | Reverse primer (5′–3′) | Probe (5′ FAM–TAMRA 3′) | |
|---|---|---|---|
| SIRT1 | TAGAGCCTCACATGCAAGCTCTA | GCCAATCATAAGATGTTGCTGAAC | ACTCCAAGGCCACGGATAGGTCCATATACTT |
| SIRT2 | CCTCGCCTGCTCATCAACA | TCCTCCGAGGCCCATAATC | TGGCCAGTCGGACCCTTT |
| SIRT3 | CATTCGGGCTGACGTGATG | AACCACATGCAGCAAGAACCT | TGCACCGGCGTTGTGAAGCC |
| SIRT7 | CGTCCGGAACGCCAAATAC | GACGCTGCCGTGCTGATT | TGGTCGTCTACACAGGC |
| p21 | GCAGACCAGCATGACAGATTTCTA | GCGGATTAGGGCTTCCTCTT | CACTCCAAACGCCGGCTGATCTTC |
| p16 | CATAGATGCCGCGGAAGT | CCCGAGGTTTCTCAGAGCCT | CCTCAGACATCCCCGATTGAAAGAACC |
| 18S | ACCTGGTTGATCCTGCCAGTAG | AGCCATTCGCAGTTTCACTGTAC | TCAAAGATTAAGCCATGCATGTCTAAGTACGCAC |
Senescence-associated β-galactosidase assay
The senescence-associated-β-galactosidase (SA-β-Gal) assay detects the presence of SA-β-Gal, specifically at pH 6, as an established histochemical marker of cellular senescence (Dimri et al, 1995). An assessment of the level of SA-β-Gal expression in all cultures was therefore determined at pH 6 following the method of Dimri et al (1995), with a pH 4 positive control included in all instances.
Statistical analysis
The analysis of variance (ANOVA) was used to determine whether changes in sirtuin expression with replicative ageing were significant. The comparative ΔΔCT method was employed to quantify relative gene expression for the Taqman® studies. Analysis of individual gene expression between each of biopsy groups (i.e., normal, and node-positive and negative-breast biopsies) was performed using ANOVA. Post hoc comparisons were performed using the independent Student's t-test. Differences in gene expression between the biopsy groups were calculated along with the 95% confidence interval (CI). Results are presented in figures displaying mean relative gene expression and the standard error of the mean.
RESULTS
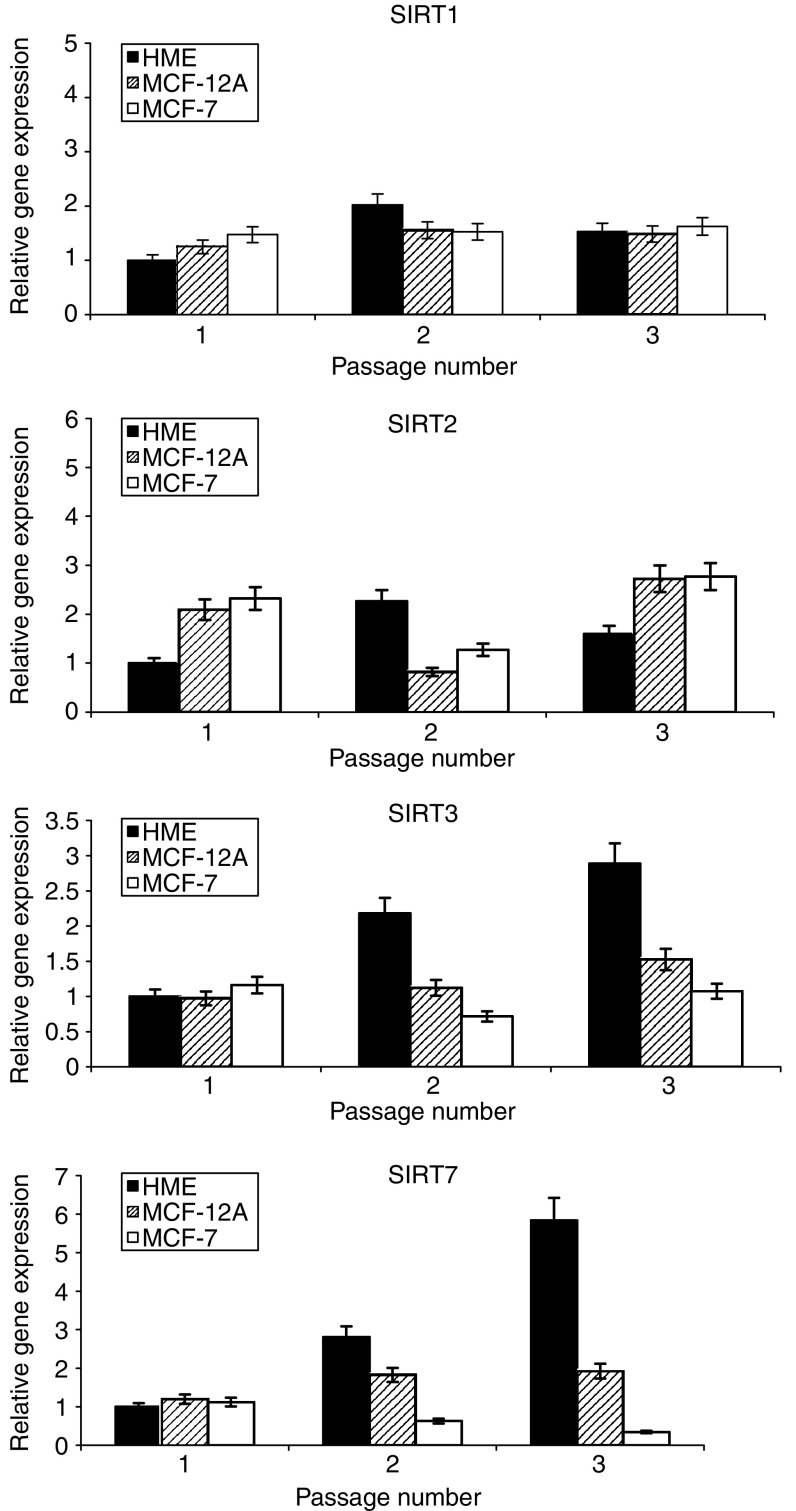
Sirtuin gene expression in primary and immortalised cells was observed for change over time in culture. Transcriptional expression for Sirtuins 1, 2, 3 and 7 was undertaken in serially passaged primary breast cells (HMECs) and in immortalised nontumorigenic (MCF-12A) and tumorigenic (MCF-7) mammary epithelial cell lines. This was performed with a view to determining the transcriptional profile of sirtuins as a function of time spent in culture and hence biological ageing. SIRT3 and SIRT7 demonstrated an increase in transcriptional expression with increasing passage in HMECs (Figure 1). This observation was not repeated for SIRT1 and SIRT2, nor did any sirtuin analysed showed a change in transcriptional expression in either of the immortalised cell lines (Figure 1).
Figure 1.
Sirtuin expression in serially passaged mammary epithelial cell cultures. The figure shows relative sirtuin gene expression in serially passaged cells relative to 18 s rRNA gene expression. Mean data are plotted ±s.d.
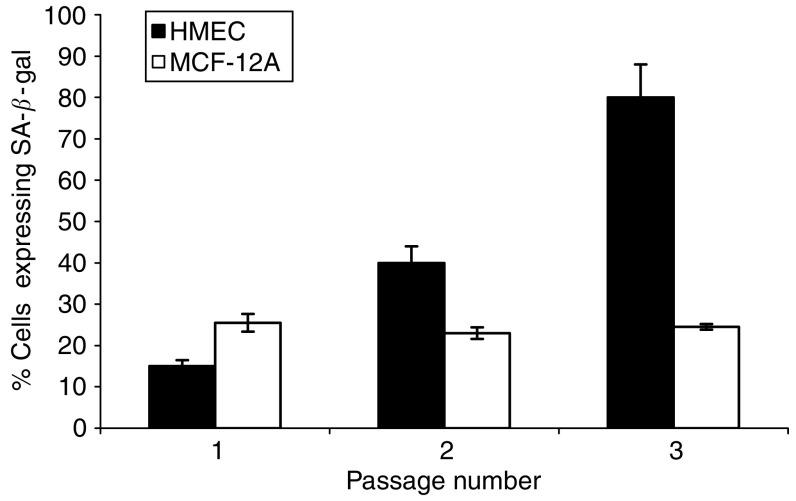
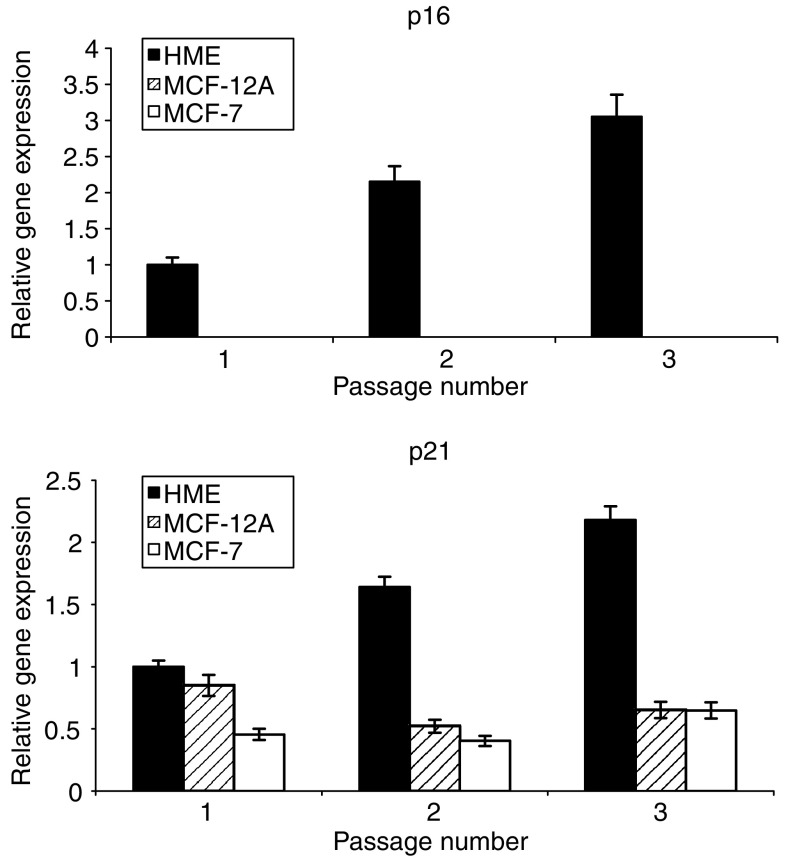
The increase in SIRT3 and SIRT7 expression was concurrent with an increase in SA-β-Gal expression in the culture with increasing passage, indicating the presence of increasing amounts of senescent cells in the population (Figure 2). Neither of the immortalised cell lines displayed a similar increase in SA-β-Gal expression. The acquisition of the senescent phenotype by the HMECs in association with serial passage was confirmed by a transcriptional analysis of the cellular stress and senescence genes p16INK4a and p21CIP1 (Figure 3). The expression of p16INK4a increased with serial passage only in the primary cell line, as did expression for p21CIP1, indicating that the transcriptional expression data for the sirtuins was associated with the generation of a senescent state in these cells.
Figure 2.
Senescence associated-β-galactosidase expression in serially passaged primary and immortalised mammary epithelial cells. Mean SA-β-Gal expression is shown ±s.d.
Figure 3.
p16 and p21 expression in successively passaged breast cells. Histograms show the relative gene expression for p16 and p21 relative to 18 s rRNA expression in primary cell cultures and immortalized breast cell lines. Data is plotted as means±s.em.
Biopsies
The pathological characteristics of the breast biopsies used in this study were selected to include equal distribution by nodal status (12 lymph node positive and 12 lymph node negative), and tumour grade (Table 2). A total of 24 breast cancer samples were chosen and matched with 21 ‘normal’, nonmalignant breast tissue biopsies from the breasts of cancer patients. These were then assessed for sirtuin expression levels.
Table 2. Summary of pathological prognostic factors of breast biopsies.
|
ER status
|
Lvi |
Grade
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No | Age years (s.e.m.) | Nodal status | Size mm (s.e.m.) | +ve | −ve | II | III | NPI (s.e.m.) | ||
| Normal | 21 | 61.4 (3.1) | N/A | N/A | N/A | N/A | N/A | N/A | ||
| Cancer | 24 | 62.3 (3.1) | +ve | 30.7 (3.1) | 9 | 3 | 6 | 4 | 8 | 4.1 (0.2) |
| −ve | 35.7 (3.1) | 8 | 4 | 5 | 5 | 6 | 6.0 (0.3) | |||
s.e.m.=standard error of the mean; N/A=not applicable; ER=oestrogen receptor; LVi=lymphovascular invasion; NPI=Nottingham Prognostic Index.
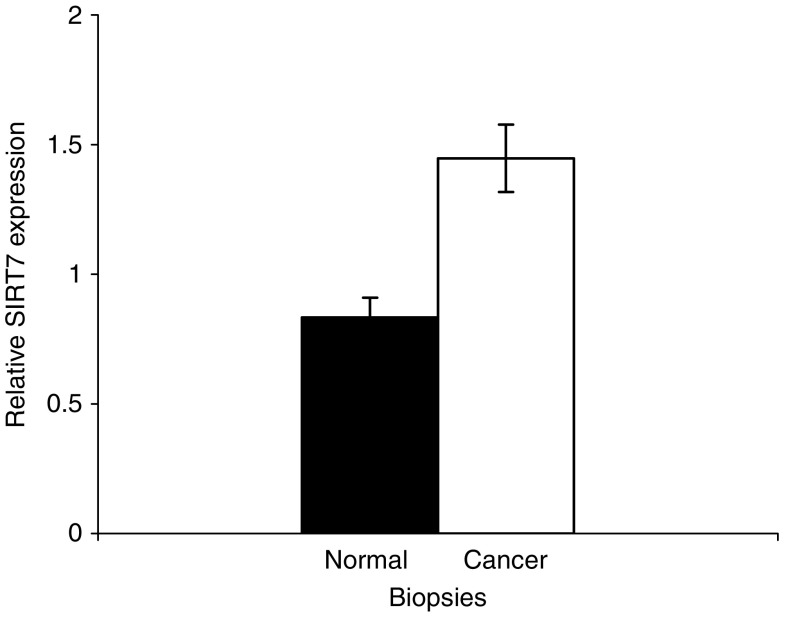
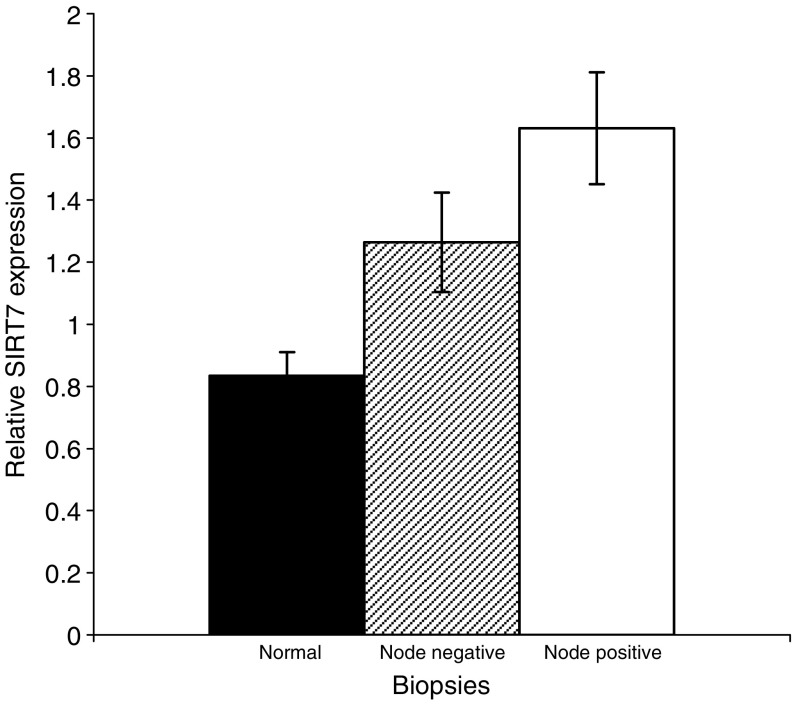
Relative SIRT7 gene expression was significantly greater in biopsies from breast cancers compared to ‘normal’, nonmalignant breast tissue (1.74 (95% CI 1.37–2.10), P<0.001, Figure 4). The relative expression of SIRT7 was also significantly higher in node-positive compared to node-negative cancers (1.96, (95% CI 1.54–2.38) vs 1.52 (95% CI 1.13–1.90), P<0.001, Figure 5).
Figure 4.
SIRT7 expression in normal and breast cancer biopsies. HIstograms show relative sirtuin gene expression in nonmalignant and malignant breast biopsies, relative to an 18 s rRNA control. Data show mean sirtuin expression ±s.e.m.
Figure 5.
SIRT7 expression in breast biopsies by nodal status. The histogram shows relative sirtuin gene expression in nonmalignant and lymph node-positive and -negative breast cancer biopsies. Data are plotted as mean sirtuin expression ±s.e.m.
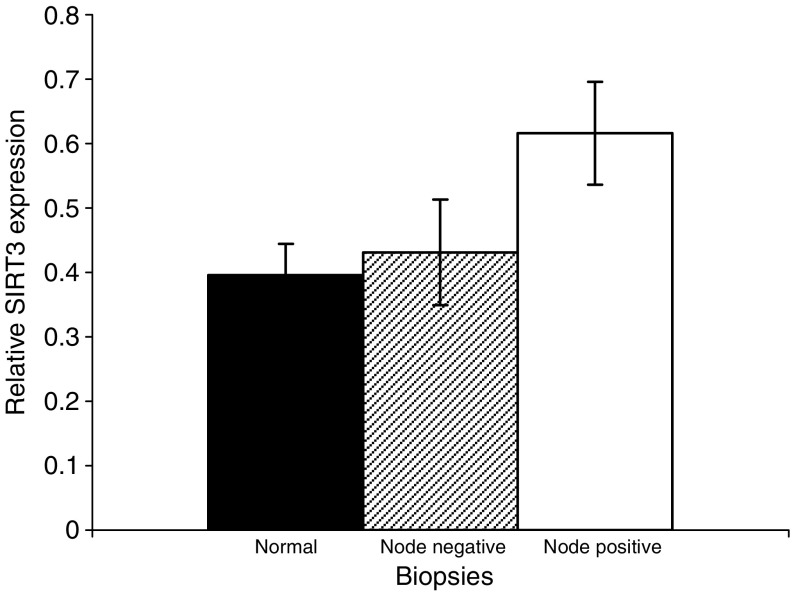
No significant differences in SIRT1, SIRT2 and SIRT3 gene expression were observed in a simple comparison of biopsies from breast cancers with ‘normal’ breast tissue (data not shown). However, analogous to SIRT7 expression, SIRT3 expression was also significantly higher in lymph node-positive breast cancer biopsies when compared to ‘normal’ breast biopsies (1.56, 95% C.I. 1.10–2.01, P<0.05) (Figure 6).
Figure 6.
SIRT3 expression in breast biopsies by nodal status. The histogram shows relative sirtuin gene expression in nonmalignant and lymph node-positive and -negative breast cancer biopsies. Data are plotted as mean sirtuin expression ±s.e.m.
Sirtuin expression was similarly compared by grade, oestrogen receptor status and the presence of lymphovascular invasion in both node-positive and node-negative cancers. No significant differences in sirtuin expression were seen using these factors (data not shown).
DISCUSSION
We undertook this study to assess the possible association of changes in the transcriptional expression of individual sirtuin genes with breast cancer. Firstly, we investigated sirtuin gene expression in both primary and immortalised cells with a view to determining how cellular ageing and immortalisation influenced their transcription profile. Consequently, we observed SIRT3 and SIRT7 transcription to increase in primary cells as they approach senescence (Figure 1), concurrent with an increase in p16INK4a and p21CIP1 expression (Figure 3) and the accumulation of SA-β-Gal-containing cells (Figure 2). Secondly, we demonstrated a significant in vivo association between altered SIRT3 and SIRT7 transcription and malignant breast disease (Figures 4, 5 and 6).
A link between the transcriptional expression of genes involved in biological ageing and cancer is pertinent. Individual sirtuins are respondents to intracellular redox changes and cell stress and hence biological ageing. In yeast, SIR2 provides a functional link between the telomere and ribosomal DNA cluster integrity, thus impacting on ribosome production and enabling the cell to respond to stress induced redox state changes in the mitochondrion (Guarente, 2000). Consequently, damage responses in the cell can be modulated to match the degree of insult. In man, individual sirtuins may provide separate functional components of this process (Shiels and Davies, 2003). This is intriguing, in the light of the observations that SIRT7 localises to the nucleolus and SIRT3 localises to the mitochondrion (Onyango et al, 2002; Michishita et al, 2005). These subcellular locations, along with the telomere nucleoprotein complex, provide the components of a functional trinity, we have termed the MTR, that senses, assesses and signals damage (Shiels and Davies, 2003; Shiels and Jardine, 2003). Our observations that SIRT3 and SIRT7 expression is increased as primary mammary epithelial cells approach senescence and are also increased in node-positive breast cancer is in keeping with the MTR hypothesis.
As SIRT7 expression is invariably elevated in thyroid cancer cell lines and biopsies, increased expression of SIRT7 may represent an important step in malignant transformation (de Nigris et al, 2002; Frye, 2002). Indeed, SIRT7 has recently been shown to activate Pol I encouraging growth and proliferation (Ford et al, 2006). In this study, we have shown that increased SIRT7 expression is also observed in breast cancer, and may therefore be postulated, in a similar manner, to be required for malignant transformation (Figure 4). Furthermore, SIRT7 expression is also associated with nodal invasion and therefore locally aggressive disease (Figure 5). Whether this increased expression contributes to the increased proliferative potential of mammary cancer cells or reflects a molecular change necessary for breast cancer tumorigenesis is undetermined. Alternatively, it may reflect a compensatory mechanism as a consequence of the increased proliferative state. This is supported by observations on the activity of the yeast sirtuin orthologue, SIR2. Nucleolar SIR2 activity is a limiting factor for determining when the cell undergoes senescence (Guarente, 1999). The high proliferative rate in many tumours often results in them having shortened telomeres as a consequence of rapid cell turnover outstripping telomerase-mediated telomere repair. This may, in analogy with the situation in yeast (Guarente, 1999), lead to an upregulation of sirtuin activity in an attempt to stabilise the nucleolus. The increased SIRT3 and SIRT7 transcription in primary mammary epithelial cells, as a consequence of increased oxidant load with in vitro growth, is supportive of such a scenario. The association of elevated SIRT7 expression in node-positive tumours, which have a greater recurrence and poorer survival, suggests that this gene may prove to be a good marker of disease progression and tumour behaviour.
There was no overall difference observed in SIRT3 expression between breast cancer and normal breast tissue. However, on subdividing the cancer biopsies by nodal status, SIRT3 expression, like that of SIRT7, was greater in node-positive breast cancer compared to normal breast tissue (Figure 6). SIRT3 has been shown to specifically targeted and converted into its active form within the mitochondria (Dryden et al, 2003; North et al, 2003). Cumulative mitochondrial damage contributes to a fall in relative nicotinamide adenine dinucleotide (NAD) levels, and concomitant fall in SIRT3 activity, and is associated with growth arrest, senescence and apoptosis (Shiels and Davies, 2003). Analogous to SIRT1 and SIRT2, SIRT3 may also function to provide a growth and survival advantage. Indeed, variability of the SIRT3 gene has been linked to survival in the elderly (Rose et al, 2003). The increased expression of SIRT3 seen in lymph node-positive tumours may, therefore, contribute to survival of these more aggressive tumours.
Our data showed that the expression of SIRT1 did not differ between normal and malignant breast biopsies. This suggests that transcriptional changes for SIRT1 may not be implicated in breast cancer pathogenesis. This observation may be pertinent to the p53 status of breast cancers, as p53 is a substrate for SIRT1 (Luo et al, 2001). Unlike the upregulation of telomerase, abnormalities in p53 are seen in only 20–40% of breast cancers, suggesting that disruption of p53 pathways may be of lesser importance in breast cancer pathogenesis (Mokbel et al, 2000; Borresen-Dale, 2003). These data do not, however, negate a role for SIRT1 in breast cancer pathogenesis, as we looked at overall sirtuin transcriptional expression levels and not at any putative post-transcriptional regulation.
The lack of transcriptional change observed for SIRT2 in this study may be a reflection of a cell cycle dependency in its expression (Dryden et al, 2003; North et al, 2003). It may be that more dynamic evaluation of changes in SIRT2 expression at specific stages of the cell cycle may reveal abnormal patterns of SIRT2 expression in breast cancer.
The clinical significance of the differences we have observed in sirtuin expression needs further investigation. Our study has indicated the potential utility of sirtuins as prognostic markers in breast cancer. The observation that SIRT7 is increased in breast cancer tissue compared to normal breast tissue suggests that it may be related to breast cancer tumorigenesis. The discovery of a greater expression of SIRT3 and SIRT7, within the prognostically poorer lymph node-positive breast cancers, suggests that overexpression of these sirtuins may be related to dissemination.
In summary, the heterogeneity of breast cancer behaviour is paralleled and may be explained by sirtuin expression. Specifically, molecular changes in SIRT3 and SIRT7 expression may contribute to tumour development and disease progression. This study shows that the study of sirtuins has a potential application in breast cancer diagnosis and prognosis, as well as in understanding disease biology.
Acknowledgments
PS, WDG and NA were supported by a grant from the Marjorie Wilkinson Bequest. We thank Dr Joanne Edwards and Dr Donald Mac Millan for critical reading of the manuscript.
References
- Borresen-Dale AL (2003) TP53 and breast cancer. Hum Mutat 21: 292–300 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9: 2888–2902 [DOI] [PubMed] [Google Scholar]
- CancerStats (2005) (http://info.cancerresearchuk.org/cancerstats). In Cancer Research UK
- de Nigris F, Cerutti J, Morelli C, Califano D, Chiariotti L, Viglietto G, Santelli G, Fusco A (2002) Isolation of a SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma cell lines and tissues. Br J Cancer 86: 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith OPeacocke M, Campisi J (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92: 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA (2003) Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol 23: 3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R (2002) ‘SIRT8’ expressed in thyroid cancer is actually SIRT7. Br J Cancer 87: 1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273: 793–798 [DOI] [PubMed] [Google Scholar]
- Galea MH, Blamey RW, Elston CE, Ellis IO (1992) The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 22: 207–219 [DOI] [PubMed] [Google Scholar]
- Guarente L (1999) Diverse and dynamic functions of the Sir silencing complex. Nat Genet 23: 281–285 [DOI] [PubMed] [Google Scholar]
- Guarente L (2000) Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 14: 1021–1026 [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, Oshimura M (2003) Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun 309: 558–566 [DOI] [PubMed] [Google Scholar]
- Johnstone RW (2002) Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 1: 287–299 [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W (2001) Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107: 137–148 [DOI] [PubMed] [Google Scholar]
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK (2001) Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer 1: 194–202 [DOI] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16: 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokbel K, Ghilchik M, Williams G, Akbar N, Parris C, Newbold R (2000) The association between telomerase activity and hormone receptor status and p53 expression in breast cancer. Int J Surg Investig 1: 509–516 [PubMed] [Google Scholar]
- Neumeister P, Albanese C, Balent B, Greally J, Pestell RG (2002) Senescence and epigenetic dysregulation in cancer. Int J Biochem Cell Biol 34: 1475–1490 [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444 [DOI] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP (2002) SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA 99: 13653–13658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G (2003) Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol 38: 1065–1070 [DOI] [PubMed] [Google Scholar]
- Shiels PG, Davies RW (2003) Ageing and Death in Neurons. The Molecular Biology of the Neurone. 2nd edn., pp 435–464
- Shiels PG, Jardine AG (2003) Dolly, no longer the exception: telomeres and implications for transplantation. Cloning Stem Cells 5: 157–160 [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149–159 [DOI] [PubMed] [Google Scholar]