Abstract
The three genes, gatC, gatA, and gatB, which constitute the transcriptional unit of the Bacillus subtilis glutamyl-tRNAGln amidotransferase have been cloned. Expression of this transcriptional unit results in the production of a heterotrimeric protein that has been purified to homogeneity. The enzyme furnishes a means for formation of correctly charged Gln-tRNAGln through the transamidation of misacylated Glu-tRNAGln, functionally replacing the lack of glutaminyl-tRNA synthetase activity in Gram-positive eubacteria, cyanobacteria, Archaea, and organelles. Disruption of this operon is lethal. This demonstrates that transamidation is the only pathway to Gln-tRNAGln in B. subtilis and that glutamyl-tRNAGln amidotransferase is a novel and essential component of the translational apparatus.
Since the early proposal in Crick’s Adaptor Hypothesis (1) that each amino acid is attached to tRNA by its own (cognate) aminoacyl-tRNA synthetase it has generally been assumed that every cell contains 20 aminoacyl-tRNA synthetases to produce the 20 different sets of aminoacyl-tRNAs that are required for faithful translation of the genetic message. The existence of an alternate pathway to aminoacylation was first suggested by the characterization of glutaminyl-tRNAGln (Gln-tRNAGln) formation in Bacilli (2). This pathway involves mischarging of a tRNA with a noncognate amino acid, and subsequent modification of the tRNA-bound amino acid. tRNA-dependent amino acid transformation has been reported for Gln-tRNA and asparaginyl-tRNA (Asn-tRNA), as well as for selenocysteinyl-tRNA and formylmethionyl-tRNA in diverse organisms (reviewed in ref. 3). The formation of mischarged tRNA and subsequent tRNA-dependent amino acid transformation are essential, normal routes to the synthesis of some aminoacyl-tRNAs; their evolutionary basis and connection to intermediary metabolism, where counterparts to the modification reactions are also present, is currently unexplained.
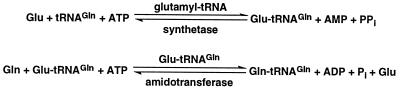
The biological significance of tRNA-dependent amino acid transformation became evident when it was shown that the transamidation pathway to Gln-tRNAGln synthesis is operative in a large part of the living kingdom: in the Archaea, Gram-positive eubacteria, mitochondria, and chloroplasts (4–6). In contrast, the standard aminoacylation pathway, using glutaminyl-tRNA synthetase (GlnRS) to attach glutamine directly to the cognate tRNA, is used to provide Gln-tRNAGln in Gram-negative eubacteria and in the cytoplasm of eukaryotic cells. The transamidation pathway (Fig. 1) is initiated by misacylation of tRNAGln by glutamyl-tRNA synthetase (GluRS) forming glutamyl-tRNAGln (Glu-tRNAGln) (reviewed in ref. 3). The mischarged tRNA may be excluded from protein synthesis because of poor recognition by elongation factor (EF)-Tu, as is the case in chloroplasts (7). In the presence of glutamine as amine donor and ATP, Glu-tRNAGln is then transamidated by Glu-tRNAGln amidotransferase (Glu-AdT), presumably through an activated γ-phospho-Glu-tRNAGln intermediate (8). Previous attempts to identify the enzyme responsible for this activity have not been successful (9).
Figure 1.
The transamidation pathway of Gln-tRNAGln formation
While the two pathways for Gln-tRNAGln formation are evolutionarily conserved, the reason for the existence and evolutionary maintenance of the different pathways is not yet known. It appears that this indirect pathway is the primary source of Gln-tRNAGln within the cells in which it operates and may act as a regulatory mechanism for glutamine metabolism. Evolutionarily, it has been suggested that glutamine was the last amino acid recruited for translation. Therefore, it may be postulated that cells that employ the transamidation pathway used the gene encoding GluRS to generate the Glu-AdT gene. Similarly, in the cells in which the direct glutaminylation pathway operates, the enzyme GlnRS may have evolved from a GluRS gene duplication (10). This scenario is reasonable because both enzymes are required to specifically recognize and bind tRNAGln and free glutamine. However, database searches, in particular of the Mycoplasma genome, the only Gram-positive genome sequence published to date (11), have shown no significant similarities of any protein likely to be Glu-AdT to GluRS or GlnRS. Thus, Glu-AdT may not have significant similarity to any known aminoacyl-tRNA synthetase.
Here we report the purification of the heterotrimeric Bacillus subtilis Glu-AdT, its gene arrangement, and a characterization of this enzyme.
MATERIALS AND METHODS
General.
The B. subtilis λgt11 library was a gift of C. W. Price (University of California, San Diego) (12). B. subtilis 168 DNA was prepared from the wild-type Marburg strain. RNA was prepared from B. subtilis BR151MA. The polyclonal rabbit antibodies used in our recent work were made against the heterotrimeric enzyme and were a gift of D. Beaulieu (Bristol–Myers Squibb). Oligonucleotides were synthesized by the Keck Foundation Research Biotechnology Resource Laboratory at Yale University.
Plasmids.
The 3.5-kb XbaI–XhoI fragment from pKS-CAB (carrying the gatC, gatA, and gatB genes; gat for Glu-tRNAGln amidotransferase) was cloned into pET-15b (13) resulting in the overexpression plasmid pCAB. The ORFs of the individual genes were also subcloned into the same vector to generate pA, pB, and pAB, whereas pCA and pC were cloned into pET-11a (13). The B. subtilis tRNAGln overexpression plasmid (pBtRNAQ) was made by insertion of the tRNAGln gene (14) isolated by PCR into plasmid pGFIB (15). Plasmid pIC56 (16) is an Escherichia coli plasmid unable to replicate autonomously in B. subtilis, which contains a spectinomycin resistance gene cassette selectable in single copy in B. subtilis. Plasmid pSPC-CA′ is a derivative of pIC56 containing the 5′ end of the transcriptional unit (starting with +1 of Fig. 2; positions 371-1111 in GenBank accession no. AF008553), whereas plasmid pSPC-A′ contains an internal fragment of the A gene (positions 1427–1917 in GenBank accession no. AF008553). DNA fragments were isolated by PCR using synthetic oligonucleotide primers flanking the desired region, which introduced appropriate restriction sites.
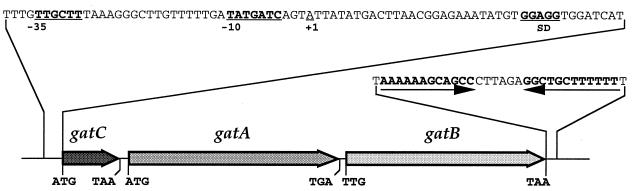
Figure 2.
Glu-AdT gene arrangement. The genomic organization of the Glu-AdT operon is shown. This includes −35 and −10 regions of the putative promoter, the Shine-Dalgarno (SD) sequence, and the putative transcription start +1.
Northern Blot Analysis.
Total RNA was isolated from exponential growth phase cells of strain BR151MA as described (17). RNA samples (10 μg) were electrophoresed, blotted, and hybridized with NorthernMax reagents (Ambion, Austin, TX) following the instructions of the manufacturer. Antisense biotinylated riboprobes were generated with the BrightStar BIOTINscript (Ambion) using T7 or T3 RNA polymerase. Plasmids pA and pB were linearized with BamHI and SacI, respectively, and transcribed with T7 RNA polymerase; plasmid pC was linearized with KpnI and transcribed with T3 RNA polymerase. Chemiluminescent signals were developed with BrightStar BioDetect (Ambion).
Purification of B. subtilis tRNAGln Expressed in Vivo in E. coli.
A 3-ml culture of E. coli DH5α/pBtRNAQ in Luria–Bertani (LB) medium (10 g bactotryptone/5 g yeast extract/10 g NaCl) with 50 μg/ml ampicillin and 10 μg/ml kanamycin was incubated at 37°C overnight. The culture was scaled up to 1 liter and overnight incubation was repeated. Cells (10 g) were harvested by centrifugation at 4,000 × g for 5 min at 4°C and resuspended in 10 ml lysis buffer (20 mM Tris⋅HCl, pH 7.4/20 mM magnesium chloride). Total nucleic acids were isolated by two sequential extractions with equal volumes of water-saturated phenol followed by isopropyl alcohol precipitation of the aqueous phase. The nucleic acids were collected by centrifugation at 10,000 × g for 15 min at 4°C. The pellet was resuspended in 5 ml of 200 mM Tris-acetate (pH 9.0) and incubated at 37°C for 1 hr to ensure complete deacylation of the tRNA. The nucleic acids were recovered by ethanol precipitation followed by centrifugation at 10,000 × g for 15 min at 4°C. The pellet was resuspended in 100 mM NaCl, incubated overnight at 4°C, and ethanol precipitated. The tRNAGln was purified by a two-step anion exchange chromatography protocol. The nucleic acids were resuspended in 5 ml of Buffer 1 (140 mM sodium acetate, pH 4.5), and 1 g of DE52 resin/100 A260 was added. The resin was washed with 200 ml buffer A and 150 ml Buffer 2 (140 mM sodium acetate, pH 4.5/300 mM NaCl) and the tRNA was eluted with 100 ml Buffer 3 (140 mM sodium acetate, pH 4.5/1 M NaCl). The nucleic acids were recovered by ethanol precipitation followed by centrifugation at 10,000 × g for 15 min at 4°C and resuspended in 20 mM triethanolamine⋅KOH (pH 7.5), 1 mM magnesium chloride, and 1 mM DTT and applied onto a Pharmacia Mono Q (HR 10/10) column. The tRNA was eluted with a gradient of 450–750 mM NaCl in 20 mM triethanolamine⋅KOH (pH 7.5), 1 mM magnesium chloride, and 1 mM DTT. Fractions containing the tRNAGln, based on ability to be aminoacylated with both Glu (by B. subtilis GluRS) and Gln (by E. coli GlnRS), were pooled and used in the amidotransferase assays. This purification protocol was also used to separate wild-type tRNAGlu and tRNAGln from B. subtilis DB431 cells.
Formation of Misacylated Glu-tRNAGln.
The tRNA (isolated as described above) was charged with radioactive glutamate by B. subtilis GluRS (partially purified by DEAE-Sepharose chromatography). tRNA charging was carried out at 37°C in a buffer consisting of 10 mM ATP, 50 mM Hepes⋅KOH (pH 7.0), 25 mM KCl, 15 mM magnesium chloride, and 5 mM DTT. The concentration of tRNAGln and of [14C]glutamate (300 mCi/mmol; 1 Ci = 37 GBq) was 10 μM. Complete charging usually took 30 min. Aliquots from this mixture were then added to the amidotransferase assay mixtures either directly or following extraction with water-saturated phenol, ethanol precipitation, and resuspension in the aminoacylation buffer.
Assay of Glu-AdT Activity.
This assay was adapted from ref. 18. Unless otherwise noted, these reactions were conducted at 37°C in a buffer consisting of 1 mM ATP, 5 mM Hepes⋅KOH (pH 7.0), 2.5 mM KCl, 1.5 mM magnesium chloride, and 0.5 mM DTT. The concentration of [14C]Glu-tRNAGln was 1 μM and that of unlabeled l-glutamine was 1 mM. Glu-AdT (200 ng of pure enzyme or correspondingly more from impure fractions) was added, and the mixture was incubated for various lengths of time depending upon the assay, followed by quenching with 10 μl of 3 M sodium acetate (pH 5.0). The mixture was extracted with an equal volume of water-saturated phenol, and the aqueous and organic phases were separated by centrifugation at 15,000 × g at room temperature for 1 min. The aqueous phase was removed, 3 vol of ethanol were added, and the tRNA was precipitated at −70°C for 15 min. The precipitated aminoacyl-tRNA was recovered by centrifugation at 15,000 × g at 4°C for 15 min. The pellet obtained was resuspended in 50 μl of 25 mM KOH and deacylated at 65°C for 10 min. The reaction was neutralized with 1.3 μl of 0.1 M HCl (to pH ≈ 6–7) and the solution was dried completely under vacuum. The dried pellet was resuspended in 3 μl of double-distilled water and spotted onto a cellulose TLC plate (Aldrich). Development was 3.5–5 hr in one of the following systems: (A) isopropyl alcohol:formic acid:water (20:1:5) and (B) ammonia:water:chloroform:methanol (2:1:6:6). The Rf values were: Glu (solvent A, 0.35; solvent B, 0.23) and Gln (solvent A, 0.23; solvent B, 0.42). The plate was dried at 85°C and exposed to an activated phosphorimaging plate for about 12 hr. The image was analyzed using macbas v2.0 software.
Purification of B. subtilis Glu-AdT.
E. coli strain BL21(DE3)/pCAB was incubated overnight at 37°C in 3 ml LB medium containing 50 μg/ml ampicillin. The culture was scaled up to 1 liter and grown at 37°C overnight. The cells (16 g) were harvested by centrifugation (4,000 × g for 5 min at 4°C) and resuspended in 20 ml Buffer A (25 mM Hepes⋅KOH, pH 7.5/25 mM KCl/10 mM magnesium chloride/1 mM DTT). This step and all subsequent steps were performed at 4°C unless otherwise specified. The cells were lysed by sonication (4 × 15 sec) and centrifuged at 100,000 × g for 1 hr (fraction I). The supernatant was applied to a Q Sepharose Fast Flow (HR 16/10) column and Glu-AdT activity eluted with a linear gradient of 0–500 mM NaCl in Buffer A (fraction II). The active fractions were applied to a Superdex-200 (HR 26/100) column and the activity was eluted isocratically in Buffer A (fraction III). This fraction was applied to a MonoQ (HR 10/10) column and Glu-AdT activity eluted with a linear gradient of 150–300 mM NaCl in Buffer A. Active fractions (fraction IV) were pooled and dialyzed against a solution of Buffer A, 200 mM NaCl, and 50% glycerol for 12 hr and stored at −70°C.
Insertional Inactivation of the gat Genes.
DNA of plasmids pSPC-CA′ and pSPC-A′ was introduced into competent cells of B. subtilis strain BR151MA, as described (19). Chromosomal DNA from strain BR151T+spcA was used as a positive control for competence and selection for spectinomycin resistance. Spectinomycin-resistant transformants were selected on tryptose blood agar base medium (Difco) containing 200 μg/ml spectinomycin.
RESULTS
Cloning of the Genes Encoding Glu-AdT.
In an initial attempt to purify Glu-AdT from B. subtilis we isolated an active fraction consisting of a number of proteins against which polyclonal antibodies were made. Screening of a B. subtilis λgt11 expression library with these antibodies led to the isolation of a DNA fragment, which encoded an immunoreactive protein. Sequence analysis of a larger (4.3 kb) HindIII DNA fragment encoding this ORF revealed a B. subtilis operon comprised of three genes designated gatC, gatA, and gatB (Fig. 2; GenBank accession no. AF008553). The three ORFs are preceded by a normal vegetative (sigma A) promoter and a predicted factor-independent transcriptional terminator is located at the end of the gatB coding region. Northern hybridization analysis of B. subtilis total RNA with separate probes for the gatC, gatA and gatB genes revealed that all three ORFs are transcribed into one polycistronic mRNA approximately 3.5 kb in length (data not shown); the transcription start (indicated by +1 in Fig. 2) was determined by primer extension.
The predicted molecular masses for the three ORFs: GatA, 53,040 kDa; GatB, 53,535 kDa; GatC, 10,859 kDa. To characterize the role of these three genes in Glu-AdT function the individual ORFs were cloned into the pET expression vector system (13) and the following constructs were made: pCAB (encoding GatC, GatA, and GatB), pAB (encoding GatA and GatB), pCA (encoding GatC and GatA), pA (encoding GatA), pB (encoding GatB), and pC (encoding GatC).
Improved Assay for Glu-AdT Activity.
Glu-AdT converts misacylated Glu-tRNAGln to correctly charged Gln-tRNAGln. To purify and characterize the enzyme an assay system was required which would provide sufficient native tRNA substrate and could detect the subtle difference between the acidic amino acid glutamate and the amide amino acid glutamine. Since the cognate tRNAGln is present in low abundance in unfractionated tRNA, we generated this tRNA by in vivo transcription in E. coli of a B. subtilis tRNAGln gene. In vivo conditions were necessary to ensure the presence in tRNA (at position 34) of 5-methylaminomethyl-2-thiouridine, a modified nucleotide required as a recognition element for charging by B. subtilis GluRS (S.-i.K., unpublished results). This modified nucleoside is also essential for the glutamylation by E. coli GluRS (20). The B. subtilis tRNAGln gene was cloned into the pGFIB vector, which has been used successfully for E. coli tRNAGlu expression (20). Charging of this tRNA was carried out with partially purified B. subtilis GluRS. Glu-AdT activity was measured by the [14C]Glu-tRNAGln → [14C]Gln-tRNAGln conversion in the presence of Gln and ATP. The aminoacyl-tRNA was isolated and deacylated, the released amino acids were separated by thin-layer chromatography, and the radioactivity was detected and quantified by phosphorimaging. The assay is very sensitive and quantitative; 20 fmol can be detected if the specific activity of [14C]Glu is 300 cpm/pmol.
Purification of Glu-AdT.
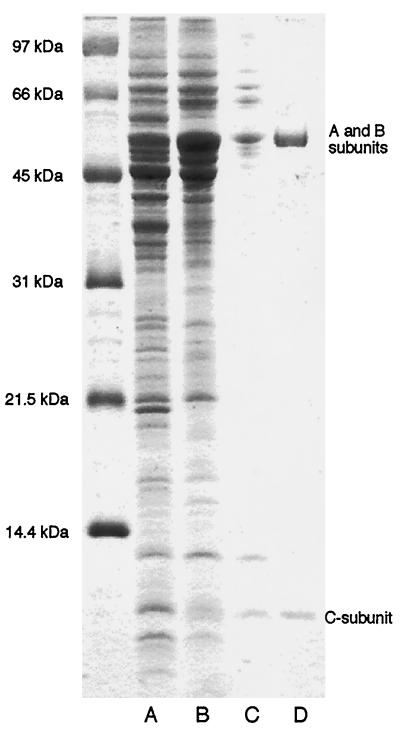
Glu-AdT was purified following overexpression of the pCAB clone in E. coli strain BL21(DE3) in four steps as summarized in Fig. 3 and Table 1. Anion exchange chromatography (Fig. 3, lane B) yielded a 3-fold increase in specific activity over the crude cell extract (Fig. 3, lane A). Gel filtration increased Glu-AdT-specific activity 13-fold (Fig. 3, lane C). Another anion exchange chromatographic step resulted in an 3.5-fold increase in specific activity. At this point the holoenzyme (containing all three subunits) appeared homogeneous as judged by SDS/PAGE (Fig. 3, lane D). The broad band at about 55 kDa represents the A (53.0 kDa) and B (53.5 kDa) subunits, whereas the single band of approximately 10 kDa is the C subunit (10.9 kDa).
Figure 3.
Analysis of Glu-AdT fractions during purification. Proteins were separated on SDS/16.5% PAGE in tricine (pH 8.4) of samples from the purification steps. Lanes: A, Fraction I [S100 extract of E. coli BL21(DE3)/pCAB]; B, Fraction II (Q Sepharose); C, Fraction III (gel filtration); D, Fraction IV (Mono Q).
Table 1.
Purification of B. subtilis Glu-tRNAGln amidotransferase
| Purification step | Total volume, ml | Total protein, mg | Total activity,* units × 103 | Specific activity, units/mg | Yield, % | Purification, -fold |
|---|---|---|---|---|---|---|
| S100 | 15 | 190 | 13.5 | 70 | 100 | 1† |
| Q Sepharose FF | 12 | 40 | 8.1 | 200 | 60 | 3 |
| Superdex-200 | 10 | 4.6 | 13.4 | 3,000 | 99 | 40 |
| Mono Q | 3.5 | 1.0 | 8.3 | 9,000 | 61 | 130 |
One unit is defined as 1 pmol glutamine produced per minute at 37°C under the assay conditions described in Materials and Methods. Protein concentrations were determined via the Bradford method.
Due to high background levels of amino and nucleic acids in the whole cell extracts, the activity of the Glu-AdT could not be determined. A 1-fold purification was assumed.
Biochemical Characterization of Glu-AdT.
The initial characterization (Table 2) of the enzyme was performed with the holoenzyme. The reaction requires Mg2+ and ATP; the other nucleoside triphosphates do not support the Glu → Gln conversion. As amino donor glutamine is better than asparagine or ammonium chloride. The enzyme also recognizes tRNAGln specifically, since [14C]Glu-tRNAGlu was not a substrate (Table 2). ATP is cleaved to ADP (data not shown), presumably in the process of phosphorylating the γ-carboxyl group of the glutamate attached to tRNAGln (8). As with some other amidases this activation may be required for the subsequent amidation (reviewed in ref. 21).
Table 2.
Requirements of the Glu-AdT reaction
| Enzyme/substrate combination* | pmol of glutamine recovered |
|---|---|
| Glu-AdT + Glu-tRNAGln + ATP + Gln + MgCl2 | 10.0 |
| Glu-AdT + Glu-tRNAGln + ATP + Asn + MgCl2 | 3.8 |
| Glu-AdT + Glu-tRNAGln + ATP + NH4Cl + MgCl2 | 2.0 |
| Glu-AdT + Glu-tRNAGlu + ATP + Gln + MgCl2 | <0.02† |
| Glu-AdT + Glu-tRNAGln + ATP + Gln | <0.02 |
| Glu-AdT + Glu-tRNAGln + CTP + Gln + MgCl2 | <0.02 |
| Glu-AdT + Glu-tRNAGln + GTP + Gln + MgCl2 | <0.02 |
| Glu-AdT + Glu-tRNAGln + UTP + Gln + MgCl2 | <0.02 |
| Glu-AdT + Glu-tRNAGln + ATP + Gln + MgCl2 + Ab‡ | <0.02 |
The assay conditions are described in Materials and Methods.
The amount of [14C]glutamine recovered was below the 0.02 pmol limit of detection of the assay protocol.
Ab, polyclonal antibodies raised against the heterotrimeric complex.
Polyclonal antibodies against the heterotrimeric Glu-AdT holoenzyme were raised. The antibodies were able to neutralize the activity of the pure heterotrimeric Glu-AdT and also destroy all enzyme activity in crude B. subtilis S100 extracts (data not shown). Because B. subtilis lacks GlnRS, the transamidation pathway catalyzed by Glu-AdT is the only route to Gln-tRNA in B. subtilis.
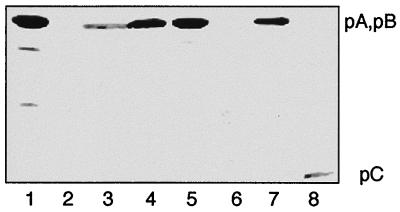
We also wanted to see whether the antibodies were able to recognize the individual Glu-AdT subunits and whether individual subunits are present in normal cell extracts. The immunoblot analysis of pure Glu-AdT protein is shown in Fig. 4 (lane 1), the minor bands are probably degradation products of A or B. All other lanes contain proteins of S100 cell extracts from wild-type B. subtilis (lane 3), E. coli BL21(DE3) containing the empty expression vector (lane 2), and E. coli cells containing pCAB (lane 4), pB (lane 7), or pC (lane 8). The results of expressing gatA are interesting. When gatA was expressed from pA alone, the A protein could not be detected by examination of a Coomassie blue-stained protein gel (data not shown), nor was it detected by immunoblot (lane 6). However, when gatA and gatC are simultaneously expressed from pCA, both the protein gel (not shown) and the immunoblot clearly showed the A protein (lane 5). These data suggest that the C subunit must be present to correctly fold or stabilize the A subunit to prevent its degradation in the cell extract.
Figure 4.
Immunoblot analysis of Glu-AdT expressed from various clones. Lane 1, purified Glu-AdT complex. S100 fractions from lane 2, E. coli BL21(DE3); lane 3, B. subtilis; lane 4, E. coli BL21(DE3)/pCAB; lane 5, E. coli BL21(DE3)/pCA; lane 6, E. coli BL21(DE3)/pA; lane 7, E. coli BL21(DE3)/pB; lane 8, E. coli BL21(DE3)/pC.
Are the Individual Subunits Active?
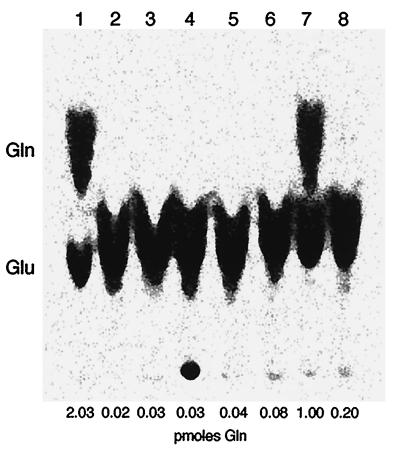
Activity of individual subunits was determined by measuring Glu-AdT activity in E. coli S100 extracts in which the subunits were overexpressed either individually or in various combinations (Fig. 5). When the subunits were expressed alone (A, B, or C) or in tandem (A+B or A+C) essentially no conversion was detectable (Fig. 5, lanes 2–6) and at most 4% of the total activity was seen (Table 1). However, good activity (50% compared with the holoenzyme) is observed when the A and C subunits were coexpressed and mixed in vitro with the individually expressed B subunit (Fig. 5, lane 7), whereas very little activity was detected when the three individually expressed subunits were mixed in vitro (Fig. 5, lane 8). These results show that the three subunits acting in concert are required for optimal enzyme activity and that the C subunit must be coexpressed with the A subunit to generate Glu-AdT activity.
Figure 5.
Activity of Glu-AdT holoenzyme and subunits. Phosphoroimage of thin-layer chromatography separation of 14C-labeled Gln and Glu derived from deacylation of aminoacyl-tRNA recovered from the assay (see text). Glu-AdT was from S100 extracts of E. coli BL21(DE3) harboring: Lane 1, pCAB; lane 2, pAB; lane 3, pCA; lane 4, pA; lane 5, pB; lane 6, pC; lane 7, S100 extracts from pCA and pB mixed in vitro; lane 8, S100 extracts from pA, pB, and pC mixed in vitro.
Glu-AdT has glutaminase activity in its route of generating the amide donor ammonia. Thus, we tested the holoenzyme and its subunits for the ability to cleave glutamine. The holoenzyme and the A subunit (coexpressed A+C) carry out the Gln → Glu conversion; ATP is not required for this reaction. The B subunit is not active in this process (data not shown).
Insertional Inactivation of gat Genes.
To determine whether the gatCAB genes in the above arrangement encode all the Glu-AdT activity of the organism, a gene disruption experiment was conducted. Introduction of a plasmid lacking a B. subtilis origin of replication results in stable transformants only if the plasmid can integrate into the chromosome by homologous recombination. If a fragment of B. subtilis chromosomal DNA is introduced into the plasmid, integration into the chromosomal locus can occur. A fragment containing the 5′ end of the transcriptional unit will permit integration without disruption of the gene, whereas an internal fragment will result in insertional inactivation of the target gene in the chromosome. Spectinomycin-resistant transformants were recovered using plasmid pSPC-CA′, containing the 5′ portion of the gatCAB operon; however, no transformants were recovered with plasmid pSPC-A′, containing an internal portion of gatA. This provides a strong indication that disruption of the gatCAB transcriptional unit is a lethal event, and that the gatCAB genes are essential for cell viability.
Arrangement of the Glu-AdT Operon.
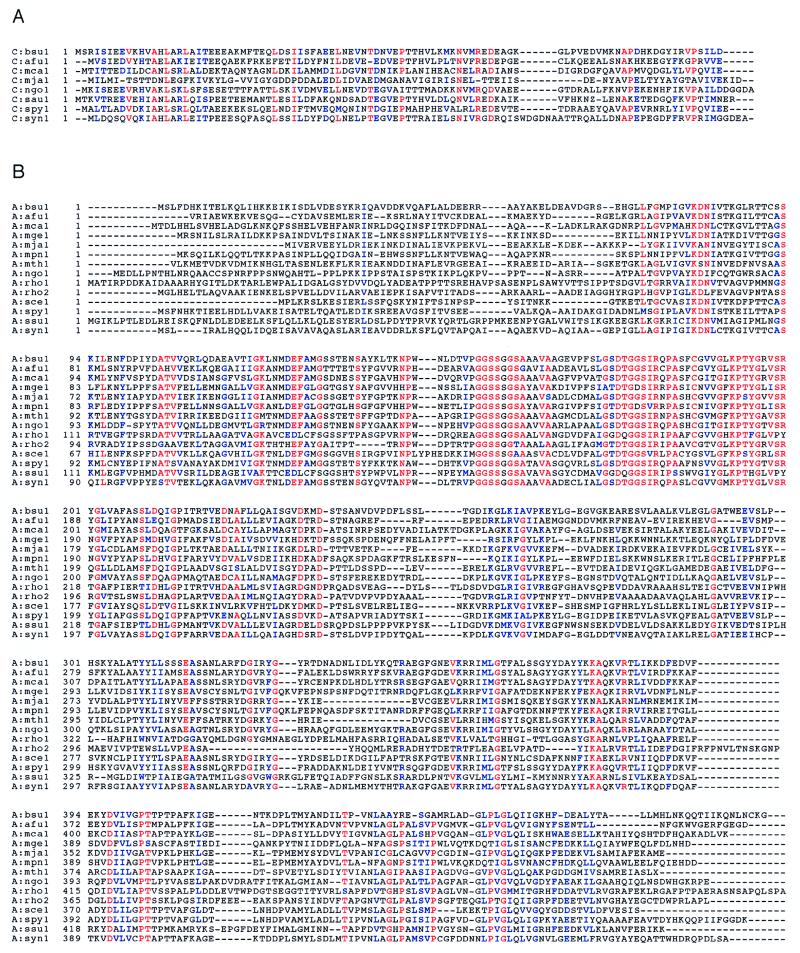
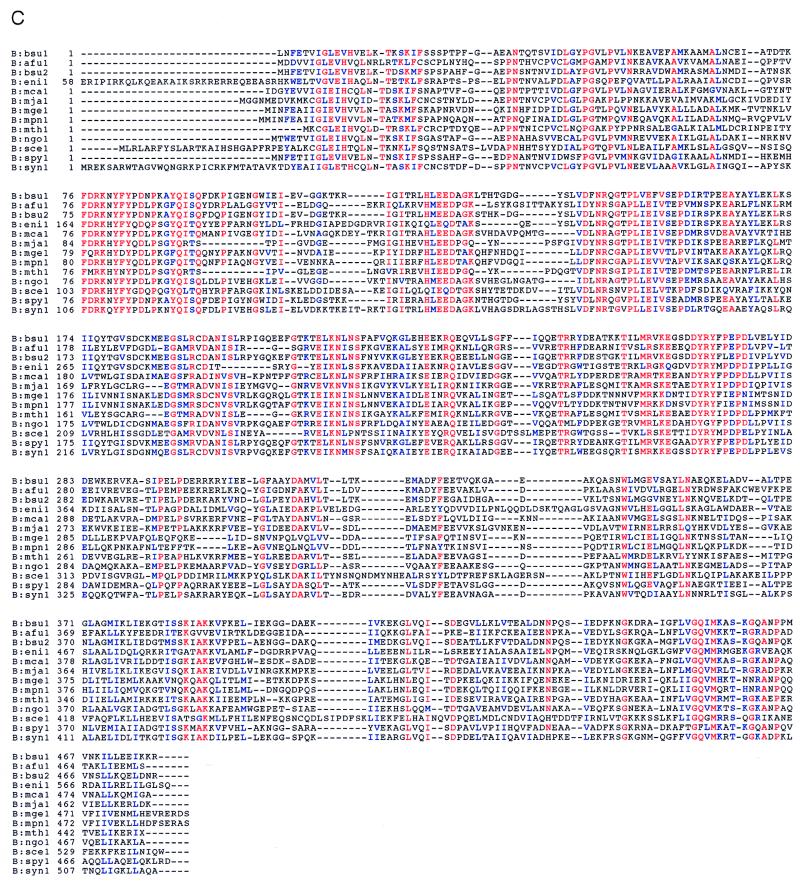
As shown above (Fig. 1), the B. subtilis gatCAB operon encodes the three Glu-AdT subunits. The predicted amino acid sequences of these genes were used in tblastn database searches to identify similar sequences. The complete ORFs thus identified have been aligned and are displayed in Fig. 6. While paralogs of each of the individual subunits can be identified in several different species, true orthologous three-subunit operons are found only in two species of Gram-positive eubacteria, B. subtilis and S. pyogenes, and in the cyanobacterium Synechocystis. The same arrangement may also be in other organisms (e.g., S. aureus), where partial sequences are in the database. While genes encoding the three subunits are found in the Gram-negative proteobacteria M. catarrhalis and N. gonorrhoeae, the putative operon is interrupted by a β-lactamase gene in the former and a gene of unknown function in the latter. This may lead to a nonfunctional gene arrangement, which will not produce active Glu-AdT. Biochemical analysis (J. Lapointe, personal communication) showed that this M. catarrhalis strain possesses GlnRS activity, the normal route to Gln-tRNA in Gram-negative eubacteria, but not Glu-AdT. A clone (GenBank accession no. U49790) from another B. subtilis strain (168M) contains a gatB and part of a gatA gene; however, the DNA fragment sequenced is too small to indicate if this is also part of a trimeric operon.
Figure 6.
Alignment of gatA, gatB, and gatC genes with related sequences. Alignments were created with the Genetics Computer Group, clustalw, and boxshade programs. The database accession numbers are shown in parentheses. Red represents residues that are completely conserved in at least 85% of the sequences, blue those residues that are functionally conserved within the sequences, and black the other residues. (A) gatC genes. afu1, Archaeoglobus fulgidus (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-tigrbl); bsu1, B. subtilis (AF008553); mca1, M. catarrhalis (U49269); mja1, Methanococcus jannaschii (U67558) and also MJ0243; ngo1, Neisseria gonorrhoeae (http://dna1.chem.uoknor.edu/gono.html); sau1, Staphylococcus aureus (U06451); spy1, Streptococcus pyogenes (http://dna1.chem.uoknor.edu/strep.html); syn1, Synechocystis sp. PCC6803 (D90913). (B) gatA genes. afu1, Archaeoglobus fulgidus (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-tigrbl); bsu1, B. subtilis (AF008553); mca1, M. catarrhalis (U49269); mge1, Mycoplasma genitalium (U39689); mja1, Methanococcus jannaschii (U67558); mth1, Methanococcus thermoautotrophicum (22); mpn1, Mycoplasma pneumoniae (AE000058); ngo1, N. gonorrhoeae (http://dna1.chem.uoknor.edu/gono.html); rho1, Rhodococcus sp. N-774 (X54074); rho2, Rhodococcus sp. (M74531); sce1, Saccharomyces cerevisiae (L22072); spy1, S. pyogenes (http://dna1.chem.uo-knor.edu/strep.html); ssu1, Sulfolobus solfataricus (Y08256); syn1, Synechocystis sp. PCC6803 (D90913). (C) gatB genes. afu1, A. fulgidus (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-tigrbl); bsu1, B. subtilis 168 (AF008553); bsu2, B. subtilis 168M (U49790); eni1, Emericella nidulans (U62332). To condense the alignment figure, the N-terminal 57 amino acids (part of a mitochondrial targeting sequence) were omitted; mca1, M. catarrhalis (U49269), this sequence was edited in two places to generate a protein of 483 aa instead of the 419 aa protein reported; mge1, M. genitalium (U39689); mja1, M. jannaschii (U67473); mth1, M. thermoautotrophicum (22); mpn1, M. pneumoniae (AE000058); ngo1, N. gonorrhoeae (http://dna1.chem.uoknor.edu/gono.html); sce1, S. cerevisiae (D90913); spy1, S. pyogenes (http://dna1.chem.uoknor.edu/strep.html); syn1, Synechocystis sp. PCC6803 (D90913). The B. subtilis 168M clone shows only 75% amino acid identity with our B. subtilis 168 gatB gene. Because 168M is a cross of 168 with Bacillus W23, it may well be that the U49790 gatB gene originates from W23. This is likely, because the two gatB genes differ in their G+C content (bsu1, 46.5%; bsu2, 33.3%).
Analysis of the deduced protein sequences by Beauty (23) was carried out to search for domains of known function. The C subunit (Fig. 6A) shares no similarity with any sequences in the Conserved Regions Data Base (23). Thus, the only function we currently can assign to this subunit is its essentiality for stable expression of the A subunit. As seen in Fig. 6B, the A subunit belongs to a family of amidase proteins that share a highly conserved glycine, serine, and alanine signature motif found between residues 152 and 183 of the B. subtilis sequence. The A subunit also may have a P-loop-type ATP binding motif. Finally, the B subunit (Fig. 6C) shares similarity with a class of proteins known as Pet112-like proteins (24), which are found in eukaryotic mitochondria, eubacteria, and the Archaea. As can be seen in Fig. 6C, the Pet112-like proteins are a related family likely to represent Glu-AdT B subunits. In accordance with this it is pertinent to note that all organisms (with known genomic sequence) that possess B subunits also have the A subunit ortholog and thus the potential for Glu-AdT activity.
DISCUSSION
Several distinct enzymatic reactions constitute the mechanism of Glu-tRNAGln → Gln-tRNAGln conversion by Glu-AdT: (i) Gln is hydrolyzed to Glu; (ii) ATP is cleaved to ADP to form the carboxyl phosphate intermediate; (iii) Glu-tRNA is amidated to Gln-tRNAGln. In addition, Glu-AdT displays the remarkable RNA discrimination capacity of the aminoacyl-tRNA synthetases, because only Glu-tRNAGln and not Glu-tRNAGlu is recognized. Glu-AdT has been classified as a glutamine amidotransferase (21), enzymes that catalyze amide nitrogen transfer from glutamine to acceptor substrates (25). These enzymes are involved in many biosynthetic pathways (e.g., amino acid, nucleotide, and amino sugar formation) and are ubiquitous in the living kingdom. Amide transfer proceeds in a two-step mechanism: glutamine hydrolysis yielding glutamate and enzyme-sequestered ammonia, followed by ammonia-dependent synthesis of the product.
Possible Roles of Individual Subunits.
In converting Glu-tRNAGln → Gln-tRNAGln, Glu-AdT mechanistically resembles carbamoyl phosphate synthetase; ATP is used to form a carboxyl phosphate intermediate in the amidation process with the release of ADP (reviewed in ref. 21). However, despite these similarities, there is no obvious sequence relationship between either the Triad class or the Ntn class glutamine amidotransferases (25) and any of the subunits of Glu-AdT. On the other hand, there is significant amino acid sequence similarity (over the entire protein, see Fig. 6B) of the A subunit of B. subtilis Glu-AdT with an amidase from the Gram-positive organism Rhodococcus (26). The amidase signature sequence present in the A subunit (from position 152 to position 183) is GGSSGGSAAAVAAGEVPFSLGSDTGGSIRQPA (Fig. 6B). Amidases of this class have been studied from Rhodococcus, Brevibacterium, and Pseudomonas; these enzymes are important in the industrial conversion of certain amides into acids (26, 27). Interestingly, glutamine or asparagine are not substrates for the Rhodococcus rhodochrous enzyme (27). Thus, the amidotransferase function of Glu-AdT may have evolved from an amidase family distinct from the known glutamine amidotransferases and acquired new substrate specificity. Structural studies on Glu-AdT and comparison with the known glutamine amidotransferase structure will be illuminating on this last point.
The B subunit of Glu-AdT is related to a class of proteins defined on the basis of similarity to the yeast Pet112 gene product. Mutations in this gene in yeast result in a petite phenotype, and it has been suggested that Pet112 plays a role in mitochondrial translation (24). The B subunit ortholog from B. subtilis strain 168M, which is very similar to the gatB gene product described here (Fig. 6C, bsu1 vs. bsu2), complements the petite phenotype of a yeast Pet112 mutant (28). We conclude that as in B. subtilis the Pet112 encodes a part of the yeast mitochondrial Glu-AdT enzyme, and that this enzyme is essential for mitochondrial function. This result supports the idea that Glu-AdT enzymes are highly conserved and are critically important in the systems in which they are found. In addition to the B subunit ortholog, Archaea (M. jannaschii, M. thermoautotrophicum, A. fulgidus) also contain a paralog with reasonable amino acid identity. Because Archaea also form Asn-tRNAAsn by transamidation of Asp-tRNAAsn, and because it is known that the Glu-AdT and Asp-tRNAAsn amidotransferase share both glutamine and asparagine as donors (6, 18), it may well be that in Archaea Asp-tRNAAsn amidotransferase and Glu-AdT use the same amidase subunit (e.g., the A subunit ortholog), whereas Glu-AdT uses the B subunit ortholog to recognize Glu-tRNA, the B subunit paralog in Archaea may be the Asp-tRNA recognizing protein. It will be of great interest to further analyze the biochemistry of these aminotransferase systems.
As previously proposed, Glu-AdT occurs in Archaea, Gram-positive eubacteria, and organelles. A search of the database showed the presence of the genes encoding orthologs for the A and B subunits in all organisms belonging to these classes, but the genes encoding the C subunit were only found in a subset of these species. It is possible that only A and B subunits are required for Glu-AdT activity, and the role of subunit C in the formation of active subunit A can be supplanted by modifications in A itself or by other cellular proteins. Alternatively, the C subunit may be widely divergent among unrelated species. Thus, in mitochondria (e.g., yeast) only orthologs of the A and B subunit are found.
What Is the Danger of the Misacylated tRNA for the Cell?
Utilization of Glu-tRNAGln in protein synthesis would cause lethality. However, it was shown in the chloroplast system that EF-Tu rejects Glu-tRNAGln (7) so that the misacylated tRNA is not brought to the ribosome. It is interesting that EF-Tu discriminates against the only aminoacylated tRNA that is a substrate for Glu-AdT. Possibly the same unique determinant can be used by Glu-AdT for acceptance and by EF-Tu for discrimination. A similar mechanism may also be employed by other organisms using Glu-AdT, as well as those using Asp-AdT.
What Is the Phylogenetic Occurrence of the Transamidation Pathway of Gln-tRNA Formation Compared with That of Direct Acylation?
Based on the limited sampling of a number of organisms, we predicted (4) that the transamidation route occurs only in Gram-positive eubacteria, organelles, and Archaea. While this is generally true, there are a number of exceptions: mitochondria of Leishmania tarentolae possess the direct acylation pathway (29), whereas Rhizobium shows Glu-AdT activity (30). In addition, genomics suggest the occurrence of the transamidation genes in M. catarrhalis and N. gonorrhoeae. However, as discussed above, there is a disruption in the operon that may lead to loss of Glu-AdT activity. This underscores the need for a functional analysis of genes (ORFs) that are found in genomic sequences, because these genes may be inactive having been acquired coincidentally with the transfer of unrelated genes from another organism. There is also the very real likelihood that the distribution of the transamidation pathway may have to be reassessed once many more organisms have been investigated.
Is the Transamidation Pathway Apt to Represent the More Ancient Route to Gln-tRNAGln Formation?
This is plausible because it has been postulated that glutamine was among the last amino acids to be added to the repertoire of 20 amino acids presently employed. Thus, it may be reasonable to suggest that, early on, misacylated Glu-tRNAGln was converted to properly charged Gln-tRNAGln by enzymatic activities within the cell, which were recruited for this process. These activities could then have evolved into Glu-AdT. For instance, the progenitor to the A subunit may have included the ancestors of any of the paralogs outlined in Fig. 6B. Eventually, the proposed gene duplication of GluRS occurred that gave rise to the present day GlnRS, which is used in those systems that lack the transamidation pathway. There are several possible reasons for the retention of the Glu-AdT pathway. Glu-AdT activity may be required as a feedback control mechanism to regulate intracellular concentrations of glutamic acid and glutamine in intermediary metabolism in these species. Alternatively, the larger framework of an aminoacylated tRNA may be necessary for discrimination between Glu and Gln for proper tRNA charging (3). Either of these two scenarios are plausible because several organisms, the Gram-positive bacteria in particular, which use Glu-AdT, have unusually elevated glutamic acid concentrations (31).
Genomic analysis suggests that organisms exist that may have kept both pathways of Gln-tRNAGln formation. This may indicate a particular position in the evolutionary stage, or it may provide the organism with increased flexibility/adaptability to survive in different metabolic environments. One may argue that the transamidation pathway is the earlier (unrefined) way of Gln-tRNA formation. Why then is it kept in so many branches of the living kingdom? Given the fact that it uses one additional molecule of ATP to make its final product (compared with direct glutaminylation of tRNA) one wonders what advantage it may provide for the organism. Clearly, it would be interesting to see what metabolic consequences would ensue if E. coli GlnRS would be replaced with the B. subtilis machinery.
The Glu-AdT CAB complex represents the first member of a novel family of proteins in the translational machinery with unique biochemical features. They provide a means of integrating functions of intermediary metabolism (transamidation) and translation (protein:RNA recognition). It will be interesting to see how structural studies and genomic analysis will show the evolution/origin of these enzymes.
Acknowledgments
We are indebted to J. Lapointe for sharing unpublished results, Danielle Beaulieu for providing antibodies and steady encouragement, and Debra Tumbula, Sunil Kochhar, and Michael Ibba for incisive discussions. This work was supported by grants from the National Institute of General Medical Sciences and from Bristol–Myers Squibb Corporation.
ABBREVIATIONS
- GlnRS
glutaminyl-tRNA synthetase
- GluRS
glutamyl-tRNA synthetase
- EF
elongation factor
- Glu-AdT
Glu-tRNAGln amidotransferase
- Gln-tRNAGln
glutaminyl-tRNAGln
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF008553).
A commentary on this article begins on page 11761.
References
- 1.Crick F H C. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 2.Wilcox M, Nirenberg M. Proc Natl Acad Sci USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibba M, Curnow A W, Söll D. Trends Biochem Sci. 1997;22:39–42. doi: 10.1016/s0968-0004(96)20033-7. [DOI] [PubMed] [Google Scholar]
- 4.Schön A, Kannangara S, Gough S, Söll D. Nature (London) 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 5.Schön A, Hottinger H, Söll D. Biochimie. 1988;70:391–394. doi: 10.1016/0300-9084(88)90212-x. [DOI] [PubMed] [Google Scholar]
- 6.Curnow A W, Ibba M, Söll D. Nature (London) 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 7.Stanzel M, Schön A, Sprinzl M. Eur J Biochem. 1994;219:435–439. doi: 10.1111/j.1432-1033.1994.tb19956.x. [DOI] [PubMed] [Google Scholar]
- 8.Wilcox M. Eur J Biochem. 1969;11:405–412. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 9.Strauch M A, Zalkin H, Aronson A I. J Bacteriol. 1988;170:916–920. doi: 10.1128/jb.170.2.916-920.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers K C, Söll D. J Mol Evol. 1995;40:476–481. doi: 10.1007/BF00166615. [DOI] [PubMed] [Google Scholar]
- 11.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 12.Suh J W, Boylan S A, Price C W. J Bacteriol. 1986;168:65–71. doi: 10.1128/jb.168.1.65-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 14.Wawrousek E F, Narasimhan N, Hansen J N. J Biol Chem. 1984;259:3694–3702. [PubMed] [Google Scholar]
- 15.Normanly J, Masson J M, Kleina L G, Abelson J, Miller J H. Proc Natl Acad Sci USA. 1986;83:6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmetz M, Richter R. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 17.Wu J J, Howard M G, Piggot P J. J Bacteriol. 1989;171:692–698. doi: 10.1128/jb.171.2.692-698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahn D, Kim Y C, Ishino Y, Chen M W, Söll D. J Biol Chem. 1990;265:8059–8064. [PubMed] [Google Scholar]
- 19.Henkin T M, Chambliss G H. Mol Gen Genet. 1984;193:364–369. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- 20.Sylvers L A, Rogers K C, Shimizu M, Ohtsuka E, Söll D. Biochemistry. 1993;32:3836–3841. doi: 10.1021/bi00066a002. [DOI] [PubMed] [Google Scholar]
- 21.Zalkin H. Adv Enzymol. 1993;66:203–309. doi: 10.1002/9780470123126.ch5. [DOI] [PubMed] [Google Scholar]
- 22.Smith, D. G., D. R., Doucette–Stamm, L. A., Deloughery, C., Lee, M., Dubois, J., et al. (1997) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 23.Worley K C, Wiese B A, Smith R F. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]
- 24.Mulero J J, Rosenthal J K, Fox T D. Curr Genet. 1994;25:299–304. doi: 10.1007/BF00351481. [DOI] [PubMed] [Google Scholar]
- 25.Zalkin, H. & Smith, J. L. (1997) Adv. Enzymol., in press.
- 26.Mayaux J F, Cerbelaud E, Soubrier F, Yeh P, Blanche F, Petre D. J Bacteriol. 1991;173:6694–6704. doi: 10.1128/jb.173.21.6694-6704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi M, Komeda H, Nagasawa T, Nishiyama M, Horinouchi S, Beppu T, Yamada H, Shimizu S. Eur J Biochem. 1993;217:327–336. doi: 10.1111/j.1432-1033.1993.tb18250.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim S I, Stange-Thomann N, Martins O, Hong K W, Söll D, Fox T D. J Bacteriol. 1997;179:5625–5627. doi: 10.1128/jb.179.17.5625-5627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nabholz C E, Hauser R, Schneider A. Proc Natl Acad Sci USA. 1997;94:7903–7908. doi: 10.1073/pnas.94.15.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagnon Y, Lacoste L, Champagne N, Lapointe J. J Biol Chem. 1996;271:14856–14863. doi: 10.1074/jbc.271.25.14856. [DOI] [PubMed] [Google Scholar]
- 31.Tempest D W, Meers J L, Brown C M. J Gen Microbiol. 1970;64:171–185. doi: 10.1099/00221287-64-2-171. [DOI] [PubMed] [Google Scholar]