Summary
Upon starvation Bacillus subtilis undergoes a developmental process involving creation of two cell types, the mother cell and forespore. A signal in the form of a serine protease, SpoIVB, is secreted from the forespore and leads to regulated intramembrane proteolysis (RIP) of pro-σK, releasing active σK into the mother cell. RIP of pro-σK is carried out by a membrane-embedded metalloprotease, SpoIVFB, which is inactive when bound by BofA and SpoIVFA. We have investigated the mechanism by which this complex is activated. By expressing components of the signalling pathway in Escherichia coli, we reconstructed complete inhibition of pro-σK RIP by BofA and SpoIVFA, and found that SpoIVB serine protease activity could partially restore RIP, apparently by targeting SpoIVFA. Pulse-chase experiments demonstrated that SpoIVFA synthesized early during B. subtilis sporulation is lost in a SpoIVB-dependent fashion, coincident with the onset of pro-σK RIP, supporting the idea that SpoIVB targets SpoIVFA to trigger RIP of pro-σK. Loss of BofA depended not only on SpoIVB, but also on CtpB, a serine protease secreted from the mother cell. CtpB appeared to cleave BofA near its C-terminus upon coexpression in E. coli, and purified CtpB degraded BofA. We propose that RIP of pro-σK involves a three-step proteolytic cascade in which SpoIVB first cleaves SpoIVFA, CtpB then cleaves BofA and finally SpoIVFB cleaves pro-σK.
Introduction
Communication between cells is crucial during the development of multicellular organisms, for interactions between hosts and pathogens or symbionts, and when microbial biofilms form. The endosporulation process of Bacillus subtilis provides an attractive experimental system to study signalling between two cell types. This starvation-induced developmental process (reviewed in Errington, 2003) involves formation of an asymmetrically located cell division septum, creating a larger mother cell and a smaller forespore, each with a copy of the genome (Fig. 1A). The two cell types differentiate further by activating different σ subunits of RNA polymerase, which direct transcription of different sets of genes (reviewed in Kroos and Yu, 2000; Piggot and Losick, 2002). Activation of σF in the forespore sends a signal that leads to activation of σE in the mother cell. Differential gene expression in the two cell types brings about engulfment of the fore-spore as the mother cell membrane migrates around the forespore, pinching it off as a protoplast within the mother cell (Fig. 1B). Hence, the forespore cytosol is separated from that of the mother cell by two membranes. Completion of engulfment activates σG in the forespore, initiating a signal transduction pathway that governs activation of σK in the mother cell (Fig. 1C). This signalling pathway, which has been called the σK checkpoint, is critical for efficient completion of endospore formation (Cutting et al., 1990). Components of the σK checkpoint have been identified, but questions remain about the mechanism of signalling.
Fig. 1. Morphological changes and cell–cell signalling pathways during B. subtilis sporulation, and reconstruction of the σK checkpoint in Escherichia coli.
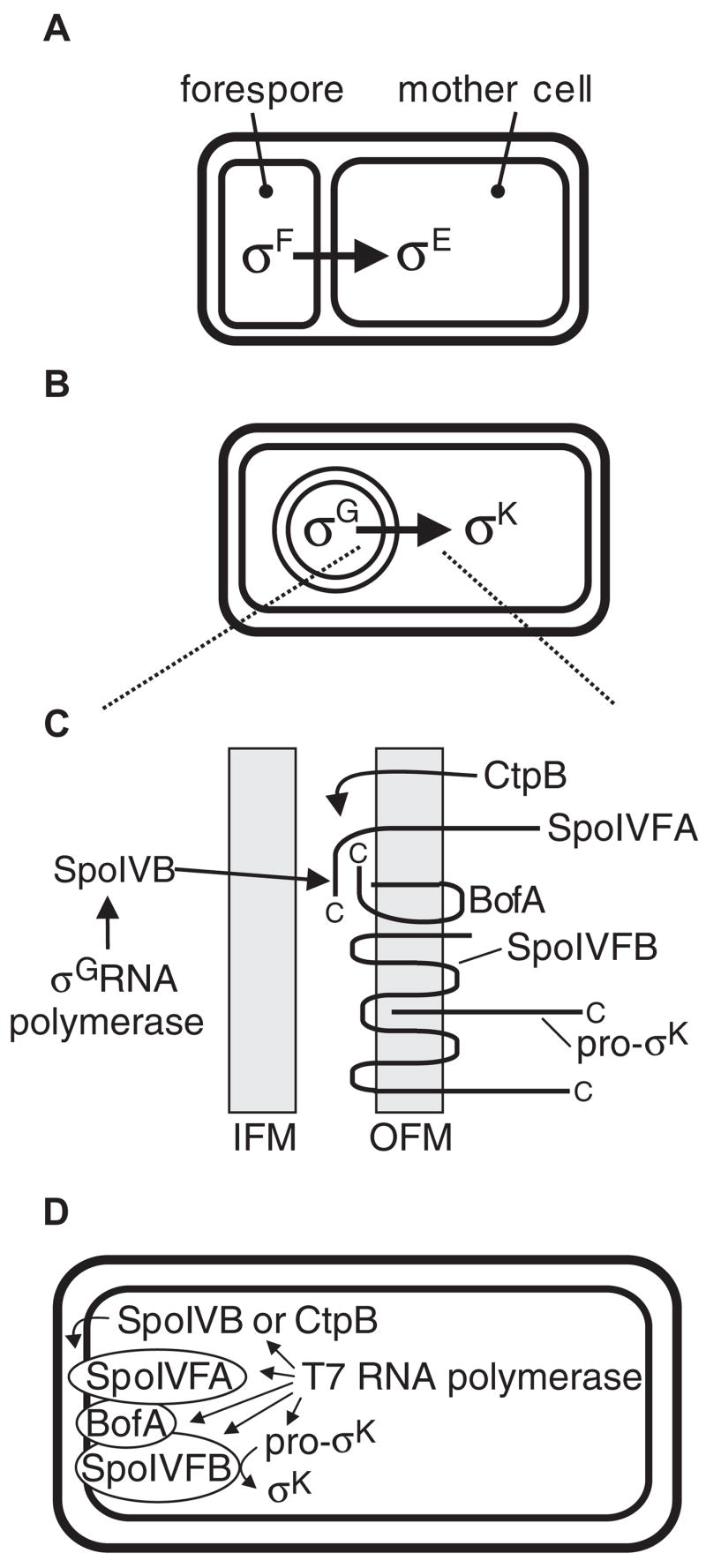
A. After the sporulation septum forms, σF RNA polymerase activity in the forespore generates a signal that leads to activation of σE in the mother cell.
B. Later during sporulation, the forespore is engulfed by the mother cell, σG becomes active in the forespore, and this signals activation of σK in the mother cell.
C. Expanded view of the σK checkpoint signalling pathway. See text for explanation. IFM, inner forespore membrane; OFM, outer fore-spore membrane. Proteins depicted in the OFM are labelled at their C-terminus and their topologies are based on analysis of lacZ and phoA fusions in E. coli (Varcamonti et al., 1997; Green and Cutting, 2000), with the exception of pro-σK, for which analysis of gfp fusions in B. subtilis suggests that its N-terminal prosequence somehow mediates membrane association (Zhang et al., 1998; Prince et al., 2005). A–C are adapted from the study by Prince et al. (2005).
D. Components of the σK checkpoint can be expressed from plasmids in E. coli by creating fusions to a T7 RNA polymerase promoter. SpoIVFA, BofA and SpoIVFB appear to insert into the inner membrane with the topologies depicted in C (Varcamonti et al., 1997; Green and Cutting, 2000) (i.e. with the E. coli cytosol equivalent to the B. subtilis mother cell cytosol and the E. coli periplasmic space equivalent to the space between the B. subtilis OFM and IFM). Pro-σK is predominantly membrane-associated (presumably with the inner membrane) when expressed in E. coli (Zhang et al., 1998; Zhou and Kroos, 2004). A SpoIVB-PhoA fusion protein appears to be translocated across the inner membrane to the periplasmic space (Wakeley et al., 2000) and the same localization is expected for CtpB, based on its predicted N-terminal secretion signal (Pan et al., 2003).
The σK checkpoint signal transduction pathway begins in the forespore when σG RNA polymerase transcribes the spoIVB gene, which encodes a serine protease with a PDZ domain (Cutting et al., 1991a; Wakeley et al., 2000). SpoIVB is believed to be secreted across the inner fore-spore membrane into the space between the two membranes surrounding the forespore (Wakeley et al., 2000). There, its PDZ domain has been proposed to target the protease to a complex of proteins located in the outer forespore membrane (Dong and Cutting, 2004). The complex includes SpoIVFA, SpoIVFB and BofA (Rudner and Losick, 2002). All three proteins are expressed in the mother cell under the control of σE RNA polymerase (Cutting et al., 1991b; Ricca et al., 1992). SpoIVFA is critical for assembly of the complex and its localization to the outer forespore membrane, which may involve initial insertion in the mother cell membrane, diffusion to the septum and capture in the outer forespore membrane during the process of engulfment (Rudner and Losick, 2002; Rudner et al., 2002; Doan et al., 2005). SpoIVFB appears to be a metalloprotease that catalyses regulated intramembrane proteolysis (RIP) of pro-σK (Rudner et al., 1999; Yu and Kroos, 2000), removing 20 amino acids from its N-terminal end (Kroos et al., 1989; Zhou and Kroos, 2004) and releasing active σK from the outer forespore membrane into the mother cell (Zhang et al., 1998). RIP is an important and widely conserved mechanism that controls signalling pathways in both prokaryotes and eukaryotes (reviewed in Brown et al., 2000; Wolfe and Kopan, 2004). It involves cleavage of a protein within a membrane or near the membrane surface. Hence, proteases that perform RIP are referred to as intramembrane-cleaving proteases (I-CLiPs). SpoIVFB is a founding member of the site-2 protease (S2P) family of I-CLiPs, named after human S2P, which cleaves transcription factors that regulate sterol and fatty acid biosynthesis, and the endoplasmic reticulum stress response (reviewed in Brown et al., 2000). BofA inhibits SpoIVFB protease activity, possibly by providing a metal ligand and blocking the active site (Zhou and Kroos, 2004) until the SpoIVB signal from the forespore relieves the inhibition.
One question that remains about the σK checkpoint is how SpoIVB relieves BofA-mediated inhibition of SpoIVFB. Recently, SpoIVB’s PDZ domain has been shown to interact with a C-terminal segment of BofA in vitro (Dong and Cutting, 2004). Also, SpoIVB’s serine protease activity can cleave SpoIVFA in vitro (Dong and Cutting, 2003). These findings suggest that SpoIVB might interact with the C-terminal segment of BofA in the space between the membranes surrounding the forespore (Fig. 1C), positioning SpoIVB to cleave SpoIVFA. After cleavage of SpoIVFA, the inhibitory effect of BofA on SpoIVFB would need to be relieved. We hypothesized that CtpB might be involved in this step. CtpB is a PDZ domain-containing serine protease expressed under σE control in the mother cell (Pan et al., 2003). Because CtpB has a predicted N-terminal secretion signal, it is likely translocated across the mother cell membrane and present in the space between the membranes surrounding the forespore after engulfment, positioning it to cleave the C-terminal end of BofA (Fig. 1C).
To further investigate the signalling pathway that governs pro-σK RIP, one of the approaches we have employed is to coexpress components of the pathway in different combinations in Escherichia coli (Fig. 1D). Previously, we showed that BofA markedly, but not completely, inhibits RIP of pro-σK by SpoIVFB in E. coli (Zhou and Kroos, 2004). Here, we show that adding SpoIVFA to the system completely inhibits RIP, and RIP is partially restored when SpoIVB is also added. Additional results provide evidence that SpoIVB targets SpoIVFA to relieve inhibition of SpoIVFB in E. coli, supporting a model proposed by Dong and Cutting (2003) based on the ability of SpoIVB to cleave SpoIVFA in vitro. Furthermore, we use pulse-chase analysis of sporulating B. subtilis to demonstrate that SpoIVFA is lost in a SpoIVB-dependent fashion coincident with the onset of pro-σK RIP. We also use pulse-chase analysis of sporulating B. subtilis, and coexpression of proteins in E. coli, to obtain evidence that CtpB plays a role in BofA degradation in vivo, and we demonstrate that purified CtpB degrades BofA in vitro. Our results support a new model for the σK checkpoint involving a cascade of three proteolytic events in which SpoIVB from the fore-spore first cleaves SpoIVFA in the space between the membranes surrounding the forespore (Fig. 1C), this exposes the C-terminal end of BofA to cleavage by CtpB from the mother cell, and finally SpoIVFB becomes active to perform RIP of pro-σK.
Results
Complete inhibition of pro-σK RIP by BofA and SpoIVFA in E. coli
Both BofA and SpoIVFA are necessary for complete inhibition of pro-σK RIP in sporulating B. subtilis, prior to fore-spore signalling that relieves the inhibition (Cutting et al., 1990; Cutting et al., 1991a,b; Ricca et al., 1992). To determine whether coexpression of BofA and SpoIVFA could bring about complete inhibition of pro-σK RIP in E. coli, we built on a system established previously (Zhou and Kroos, 2004). In this system, B. subtilis proteins are produced in E. coli under the control of T7 RNA polymerase (Fig. 1D). Coexpression of the tagged I-CLiP, H10SpoIVFB-GFP, with a C-terminally truncated substrate, pro-σK(1–126)H6, which makes it easier to separate the unprocessed and processed forms by SDS-PAGE, resulted in abundant RIP (Fig. 2, lane 1), as observed previously (Zhou and Kroos, 2004). Also, addition of SpoIVFA to the system had no effect (Fig. 2, lane 2), while GFPΔ27BofA, a functional fusion of green fluorescent protein (GFP) to amino acid 28 of BofA (Rudner and Losick, 2002), markedly, but not completely, inhibited RIP of pro-σK(1-126)H6 (Fig. 2, lane 3) (Zhou and Kroos, 2004). When both SpoIVFA and GFPΔ27BofA were simultaneously added to the system, we observed complete inhibition of RIP (Fig. 2, lane 4). Under these conditions, both SpoIVFA and GFPΔ27BofA were detected in complexes with H10SpoIVFB-GFP (data not shown). Complexes were isolated by cobalt affinity chromatography (to bind the H10-tag on H10SpoIVFB-GFP) after detergent (1% digitonin) solubilization of membranes. Using this technique, we showed previously that GFPΔ27BofA forms a complex with H10SpoIVFB-GFP in the absence of SpoIVFA, but SpoIVFA fails to form a stable complex with H10SpoIVFB-GFP in the absence of GFPΔ27BofA, when proteins are coexpressed in E. coli (Zhou and Kroos, 2004). Our new results suggest that SpoIVFA, GFPΔ27BofA and H10SpoIVFB-GFP can form a complex in E. coli, consistent with evidence that the three proteins form a complex in sporulating B. subtilis (Rudner and Losick, 2002). The enhanced inhibition of pro-σK(1–126)H6 RIP observed when SpoIVFA and GFPΔ27BofA were coexpressed in E. coli (Fig. 2, lane 4), relative to inhibition by GFPΔ27BofA alone (Fig. 2, lane 3), may reflect a larger number of inhibitory complexes being formed, greater stability of trimeric complexes than binary complexes of GFPΔ27BofA and H10SpoIVFB-GFP, and/or a direct effect of SpoIVFA on SpoIVFB I-CLiP activity when it is in a trimeric complex.
Fig. 2.
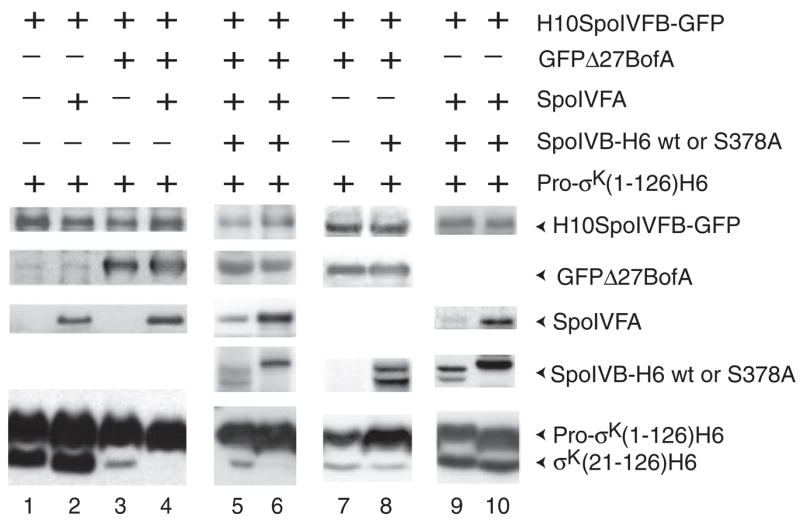
The combination of BofA and SpoIVFA completely inhibits RIP of pro-σK in E. coli, and SpoIVB partially restores RIP and can destabilize SpoIVFA. Samples were collected 2 h after IPTG induction of the indicated proteins in E. coli bearing pZR12 and pZR13 (lane 1), pZR12 and pZR33 (lane 2), pZR12 and pZR67 (lane 3), pZR12 and pZR69 (lane 4), pZR57 and pZR69 (lane 5), pZR71 and pZR69 (lane 6), pZR2 and pZR63 (lane 7), pZR57 and pZR67 (lane 8), pZR33 and pZR57 (lane 9), or pZR33 and pZR71 (lane 10). Whole-cell extracts were subjected to Western blot analysis using antibodies against GFP to detect H10SpoIVFB-GFP and GFPΔ27BofA, antibodies against SpoIVFA to detect SpoIVFA, and antibodies against penta-His to detect SpoIVB-H6, SpoIVB-H6 S378A, pro-σK(1-126)H6 and σK(21-126)H6.
SpoIVB partially restores RIP of pro-σK in E. coli and destabilizes SpoIVFA
SpoIVB has an N-terminal signal sequence capable of promoting translocation of alkaline phosphatase across the E. coli inner membrane (Wakeley et al., 2000). Therefore, we expected SpoIVB to be secreted into the periplasm upon expression in E. coli, placing it in a position to activate SpoIVFA/BofA/SpoIVFB complexes (Fig. 1D). We tagged SpoIVB at its C-terminus with hexahistidine to permit detection with anti-His antibodies. When SpoIVB-H6 was coexpressed with SpoIVFA, GFPΔ27BofA, H10SpoIVFB-GFP and pro-σK(1–126)H6 in E. coli, RIP was partially restored and the level of SpoIVFA was decreased (Fig. 2, lane 5). Coexpression of SpoIVB-H6 with just pro-σK(1–126)H6 did not result in RIP (data not shown). SpoIVB-H6 migrated as two or three distinct species in different experiments, consistent with the observation of multiple SpoIVB species in sporulating B. subtilis and evidence that SpoIVB synthesized in vitro self-cleaves at several distinct sites near its N-terminus (Wakeley et al., 2000). SpoIVB-H6 S378A, with a substitution of alanine for the putative catalytic serine residue of the protease (Wakeley et al., 2000), migrated as a single species of higher molecular weight than the multiple species observed for wild-type SpoIVB-H6, suggesting that the mutant enzyme fails to self-cleave (Fig. 2, lane 6). The mutant enzyme failed to restore RIP of pro-σK(1–126)H6, and SpoIVFA accumulated normally. These results suggest that SpoIVB serine protease activity can target SpoIVFA and partially relieve the complete inhibition of SpoIVFB I-CLiP activity brought about by the combined action of SpoIVFA and BofA in E. coli.
To further investigate the target of SpoIVB, we tested whether it could relieve the partial inhibition of RIP mediated by GFPΔ27BofA alone (Fig. 2, lane 3). In the absence of SpoIVFA, SpoIVB-H6 had no effect on RIP and no effect on the level of GFPΔ27BofA (Fig. 2, lanes 7 and 8). In contrast, SpoIVB-H6 had a dramatic effect on SpoIVFA accumulation in E. coli when GFPΔ27BofA was not coexpressed (Fig. 2, lane 9). This effect was not observed with the SpoIVB-H6 S378A mutant enzyme (Fig. 2, lane 10). Hence, SpoIVB-H6 serine protease activity appears to target SpoIVFA, not GFPΔ27BofA, in E. coli. This is consistent with in vitro transcription-translation experiments that suggest SpoIVB can cleave SpoIVFA but not BofA (Dong and Cutting, 2003). Based on their in vitro data, Dong and Cutting proposed that SpoIVB cleavage of SpoIVFA in complexes would relieve BofA-mediated inhibition of SpoIVFB. In support of this model, SpoIVB-H6 appears to target SpoIVFA in E. coli and reverse the extra inhibition of pro-σK RIP brought about by coexpressing SpoIVFA with GFPΔ27BofA, restoring RIP to a level similar to that observed when GFPΔ27BofA alone is expressed (Fig. 2, lanes 3–5). However, RIP is not restored to the level seen in the absence of GFPΔ27BofA (Fig. 2, lane 2), perhaps because some SpoIVFA persists (Fig. 2, lane 5) and/or another mechanism is involved in relieving BofA-mediated inhibition of SpoIVFB (see below).
Loss of SpoIVFA synthesized early during sporulation coincides with onset of pro-σK RIP
If SpoIVB cleavage of SpoIVFA leads to activation of SpoIVFB and RIP of pro-σK, one might expect that the steady-state level of SpoIVFA measured by Western blot analysis would decrease when pro-σK RIP begins during B. subtilis sporulation. This expectation appeared to be met in experiments reported previously (Kroos et al., 2002). Also as expected, the level of SpoIVFA did not decrease at the normal time and RIP of pro-σK did not commence in a spoIVB mutant (Kroos et al., 2002; Dong and Cutting, 2003). However, we found that in some experiments RIP of pro-σK appeared to precede loss of SpoIVFA (see below). Because the synchrony of sporulation can vary depending on experimental conditions, we reasoned that the lack of correlation between loss of SpoIVFA and the onset of pro-σK RIP in some experiments might be attributed to SpoIVFA synthesis in cells that are delayed in the sporulation process relative to other cells in the population.
To focus on SpoIVFA synthesized early during the period of σE RNA polymerase activity in the population, we pulse-labelled proteins at 1.75 h post starvation by adding [35S]methionine for 5 min. This was followed by incubation in the presence of excess unlabelled methionine and collection of samples for immunoprecipitation of SpoIVFA at different times during the chase period. Samples were also collected to monitor pro-σK RIP and the steady-state level of SpoIVFA by Western blot analysis. During the chase with excess unlabelled methionine, the amount of labelled SpoIVFA rose initially, perhaps because of increasing SpoIVFA synthesis in the population and incomplete dilution of labelled methionine (Fig. 3). Even so, in wild-type cells, SpoIVFA decreased markedly between 75 and 105 min after labelling, coincident with the onset of pro-σK RIP, and while the steady-state level of SpoIVFA increased. By 135 min post labelling, SpoIVFA decreased further, RIP of pro-σK was substantial, and the steady-state level of SpoIVFA was unchanged. In a spoIVB mutant, SpoIVFA decreased little even 135 min after labelling and RIP of pro-σK was not observed. We conclude that SpoIVFA synthesized early during the period of σE RNA polymerase activity is cleaved in a SpoIVB-dependent fashion at a time close to the onset of pro-σK RIP, consistent with the model that SpoIVB cleavage of SpoIVFA triggers RIP of pro-σK.
Fig. 3.

Loss of SpoIVFA coincides with onset of pro-σK RIP during B. subtilis sporulation. Wild-type B. subtilis PY79 and spoIVB mutant LK1 were induced to sporulate by resuspending growing cells in SM medium, pulse-labelled for 5 min with [35S]methionine at 1.75 h after sporulation induction, and chased with a 2000-fold molar excess of unlabelled methionine for the indicated time. Samples were immunoprecipitated with antibodies against SpoIVFA, subjected to SDS-PAGE, visualized by fluorography, and labelled SpoIVFA was quantified (numbers below fluorograms indicate the per cent relative to the amount at the beginning of the chase for each strain). Samples were also collected at the indicated times for preparation of whole-cell extracts that were subjected to Western blot analysis with antibodies against pro-σK and SpoIVFA.
Loss of GFPΔ27BofA also correlates with onset of pro-σK RIP
How does cleavage of SpoIVFA relieve BofA-mediated inhibition of SpoIVFB? One possibility is that loss of SpoIVFA from complexes with BofA and SpoIVFB, makes BofA susceptible to proteolysis. To test whether GFPΔ27BofA is lost in a SpoIVB-dependent fashion, B. subtilis expressing GFPΔ27BofA in a wild-type or spoIVB mutant background were treated with [35S]methionine to pulse-label proteins at hour 2 of sporulation and analysed as described above, except using antibodies against GFP to immunoprecipitate GFPΔ27BofA or detect it by Western blot analysis. For comparison, labelled SpoIVFA was also immunoprecipitated. The level of labelled SpoIVFA decreased markedly between 1 and 2 h after labelling (Fig. 4), in agreement with experiments like that shown in Fig. 3. Labelled GFPΔ27BofA showed a similar pattern of decrease as SpoIVFA in wild type, but neither GFPΔ27BofA nor SpoIVFA decreased much even 3 h after labelling in the spoIVB mutant (Fig. 4). RIP of pro-σK commenced as the levels of labelled SpoIVFA and GFPΔ27BofA decreased in wild type. As observed for the steady-state level of SpoIVFA (Fig. 3), the steady-state level of GFPΔ27BofA (determined by Western blot analysis) did not decrease when σK first appeared (Fig. 4), perhaps because of asynchronous sporulation. We conclude that like SpoIVFA, GFPΔ27BofA made early during the period of σE RNA polymerase activity is degraded in a SpoIVB-dependent fashion at a time close to the onset of pro-σK RIP. This is consistent with the idea that BofA in complexes with SpoIVFA and SpoIVFB becomes susceptible to proteolysis after SpoIVB cleaves SpoIVFA. Loss of BofA would relieve inhibition of the SpoIVFB I-CLiP, allowing RIP of pro-σK.
Fig. 4.
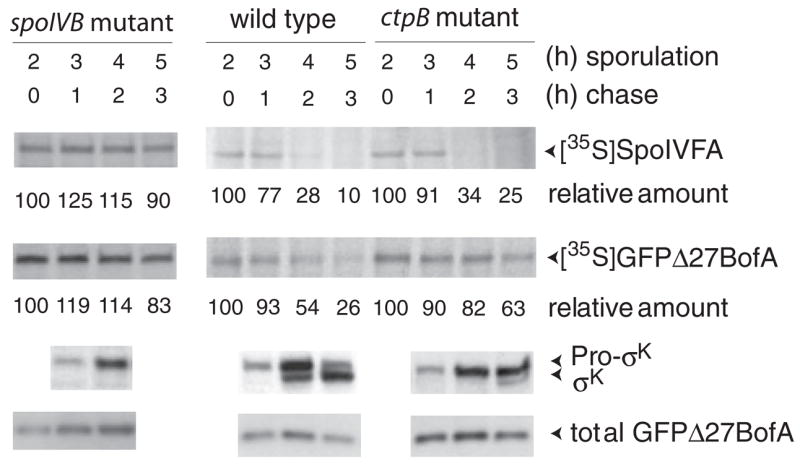
Loss of GFPΔ27BofA correlates with onset of pro-σK RIP in wild-type B. subtilis and both are delayed in a ctpB mutant. B. subtilis BDR954, ZR266 and ZR368 express GFPΔ27BofA but not native BofA in otherwise wild-type, spoIVB mutant, and ctpB mutant backgrounds respectively. These strains were induced to sporulate by resuspending growing cells in SM medium, pulse-labelled for 5 min with [35S]methionine at 2 h after sporulation induction, and chased with a 2000-fold molar excess of unlabelled methionine for the indicated time. Samples were immunoprecipitated with antibodies against SpoIVFA or GFP (to immunoprecipitate GFPΔ27BofA), subjected to SDS-PAGE, visualized by fluorography, and labelled proteins were quantified (numbers below fluorograms indicate the per cent relative to the amount at the beginning of the chase for each strain). Samples were also collected at the indicated times for preparation of whole-cell extracts that were subjected to Western blot analysis with antibodies against pro-σK and GFP (to detect total GFPΔ27BofA).
Loss of GFPΔ27BofA, but not loss of SpoIVFA, is delayed in a ctpB mutant that is delayed for RIP of pro-σK
The SpoIVB serine protease did not appear to target GFPΔ27BofA in E. coli (Fig. 2, lanes 7 and 8) or in vitro (Dong and Cutting, 2003). How, then, might SpoIVB cleavage of SpoIVFA render BofA susceptible to proteolysis? CtpB, like SpoIVB, is a PDZ-domain-containing serine protease that has been shown to influence RIP of pro-σK (Pan et al., 2003). A ctpB mutant exhibits delayed pro-σK RIP. The target of CtpB’s protease activity is unknown, but it is likely that CtpB acts in the space between the membranes surrounding the forespore as it has a predicted N-terminal signal sequence for secretion (Pan et al., 2003). In Fig. 1C, CtpB is depicted being translocated across the outer forespore membrane after engulfment. Alternatively or in addition, prior to engulfment, CtpB is likely translocated across the mother-cell membrane, may diffuse into the asymmetric septum, and be captured between the membranes surrounding the forespore during engulfment. We hypothesized that CtpB in the space between the membranes surrounding the forespore targets the C-terminal region of BofA.
To test whether loss of GFPΔ27BofA and/or SpoIVFA is delayed in a ctpB mutant, we performed pulse-chase, immunoprecipitation experiments as described above. In the CtpB mutant, labelled SpoIVFA decreased normally, but loss of labelled GFPΔ27BofA was delayed; its level changed little between 1 and 2 h after labelling (Fig. 4). The onset of pro-σK RIP was also delayed in the ctpB mutant by about 1 h, being first observed at hour 5 of sporulation, rather than at hour 4 as in wild type. The correlation between delayed loss of labelled GFPΔ27BofA and delayed onset of pro-σK RIP in the ctpB mutant is consistent with the idea that CtpB targets BofA, relieving its inhibitory effect on SpoIVFB and allowing RIP of pro-σK. Loss of SpoIVFA at the normal time was insufficient to trigger pro-σK RIP at the normal time in the ctpB mutant. This finding, taken together with the observation that neither SpoIVFA nor GFPΔ27BofA decreased normally in the spoIVB mutant, strongly suggests that SpoIVB must cleave SpoIVFA before CtpB can cleave BofA, which in turn allows SpoIVFB to cleave pro-σK.
CtpB causes BofA cleavage in E. coli and degrades H6GFPΔ27BofA in vitro
To test the idea that CtpB cleaves BofA, we expressed tagged versions of the two proteins in E. coli. CtpB was tagged with hexahistidine at its C-terminus (CtpB-H6). As a control, CtpB-H6 S309A, with a substitution that inactivates the serine protease (Pan et al., 2003), was also tested. The mutant form accumulated much better than wild-type CtpB-H6 (Fig. 5A). This does not appear to be attributed to poor expression of the wild-type enzyme, as an abundant 20 kDa polypeptide was detected with antibodies against the C-terminal H6 tag (data not shown). Rather, CtpB appears to undergo autoproteolysis, as observed for SpoIVB (Wakeley et al., 2000) (Fig. 2). Coexpression of GFPΔ27BofA with wild-type CtpB-H6, but not CtpB-H6 S309A, resulted in a decrease in the level of full-length GFPΔ27BofA and the appearance of at least one and possibly two lower molecular weight species (Fig. 5A, lane 2). To ask whether the lower molecular weight species is attributed to cleavage of BofA or GFP, we tagged GFPΔ27BofA at its N-terminus with hexahistidine and coexpressed H6GFPΔ27BofA with CtpB-H6 in E. coli. Western blot analysis with anti-His antibodies yielded a similar result as shown in Fig. 5A (lane 2) (data not shown), suggesting that H6GFPΔ27BofA is cleaved near its C-terminus. The lower band could be purified by affinity chromatography with cobalt resin (which binds H6) and then detected by anti-GFP antibodies (data not shown), demonstrating that it was not a breakdown fragment of CtpB-H6. These results suggest that BofA is cleaved near its C-terminus in a CtpB-dependent fashion when the two proteins are coexpressed in E. coli. Because at least the C-terminal 29 amino acids of BofA appear to be present in the periplasm when BofA-alkaline phosphatase fusion proteins are expressed in E. coli (Varcamonti et al., 1997), CtpB would presumably have to be secreted into the periplasm in order to directly cleave BofA near its C-terminus (Fig. 1D). As noted previously (Pan et al., 2003), CtpB has an N-terminal amino acid sequence that could serve as a signal sequence for secretion. In contrast, CtpB-H6 had no effect on SpoIVFA when the two proteins were coexpressed in E. coli (Fig. 5B, lanes 1 and 2). As SpoIVFA, like BofA, has a C-terminal periplasmic domain when expressed in E. coli (Green and Cutting, 2000), we conclude that CtpB specifically causes BofA to be cleaved.
Fig. 5. Serine protease CtpB causes BofA cleavage in E. coli and purified CtpB degrades H6GFPΔ27BofA in vitro.
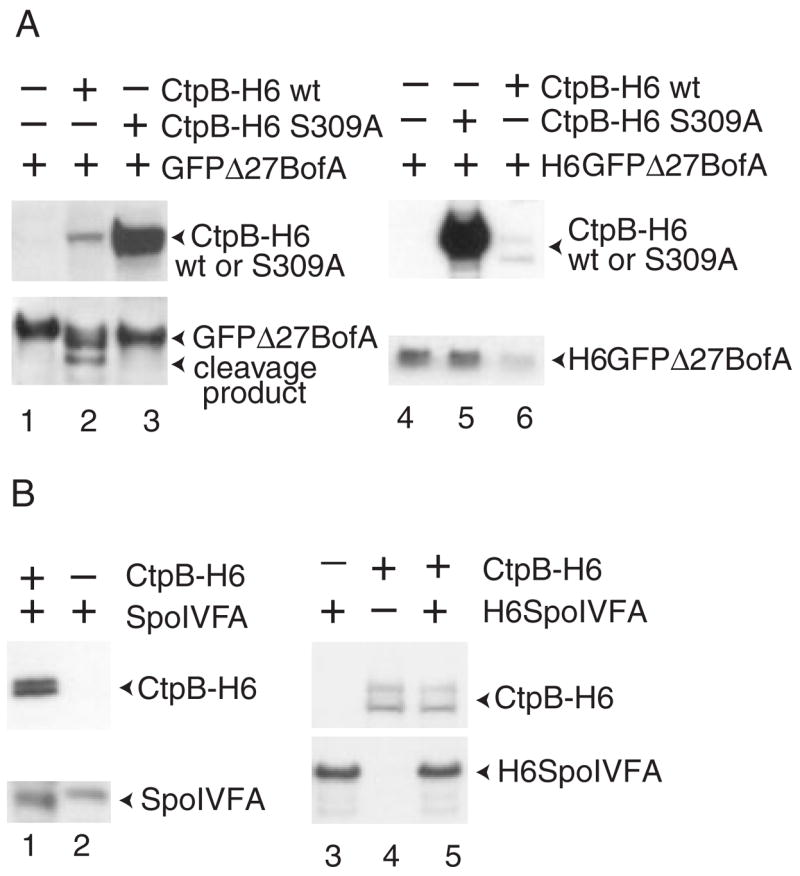
A. Effect of CtpB on GFPΔ27BofA in E. coli and in vitro. Samples were collected 2 h after IPTG induction of the indicated proteins in E. coli bearing pZR62 alone (lane 1), pZR62 and pZR114 (lane 2), or pZR62 and pZR119 (lane 3). Whole-cell extracts were subjected to Western blot analysis using antibodies against penta-His to detect CtpB-H6 and antibodies against GFP to detect GFPΔ27BofA. For in vitro analysis, purified H6GFPΔ27BofA was incubated at 37°C for 2 h alone (lane 4) or in combination with purified CtpB-H6 S309A mutant enzyme (lane 5) or wild-type CtpB-H6 (lane 6). After incubation, Western blot analysis was performed using antibodies against penta-His.
B. Effect of CtpB on SpoIVFA in E. coli and in vitro. Samples were collected 2 h after IPTG induction of the indicated proteins in E. coli bearing pZR73 and pZR114 (lane 1) or pZR73 alone (lane 2). Whole-cell extracts were subjected to Western blot analysis using antibodies against penta-His to detect CtpB-H6 and antibodies against SpoIVFA to detect SpoIVFA. For in vitro analysis, the indicated purified proteins were incubated at 37°C for 2 h (lanes 3–5). After incubation, Western blot analysis was performed using antibodies against penta-His to detect CtpB-H6 and antibodies against SpoIVFA to detect H6SpoIVFA.
To test whether CtpB could proteolyse BofA in vitro, we purified wild-type CtpB-H6 and the S309A mutant enzyme from E. coli. Although it was difficult to purify much of the full-length, wild-type enzyme, presumably because of autoproteolysis, it caused a marked decrease in the level of H6GFPΔ27BofA upon co-incubation, whereas a much larger amount of the S309A mutant enzyme had no effect (Fig. 5A, lanes 4–6). Wild-type CtpB-H6 had no effect on the level of H6SpoIVFA upon co-incubation (Fig. 5B, lanes 3–5). These results support the idea that CtpB serine protease activity can specifically target BofA.
Discussion
Our results support a new model for events governing RIP of pro-σK (Fig. 6). In agreement with a model proposed previously based on in vitro data (Dong and Cutting, 2003), our findings in E. coli suggest that SpoIVB serine protease activity targets SpoIVFA in complexes that otherwise inhibit pro-σK RIP. Careful examination revealed a good correlation between loss of SpoIVFA and onset of pro-σK RIP during B. subtilis sporulation (Fig. 3), consistent with the idea that SpoIVB from the forespore initiates events leading to RIP of pro-σK by cleaving SpoIVFA (Fig. 6A). We also found that loss of GFPΔ27BofA depends on SpoIVB (Fig. 4), yet SpoIVB failed to cleave BofA in E. coli (Fig. 2) or in vitro (Dong and Cutting, 2003). Our evidence suggests that CtpB cleaves BofA in the second step of a proteolytic cascade (Fig. 6B). Loss of BofA would free SpoIVFB from inhibition, allowing RIP of pro-σK (Fig. 6C). Below, we discuss further the evidence for this three-step cascade model and its implications.
Fig. 6. Model for events governing RIP of pro-σK.
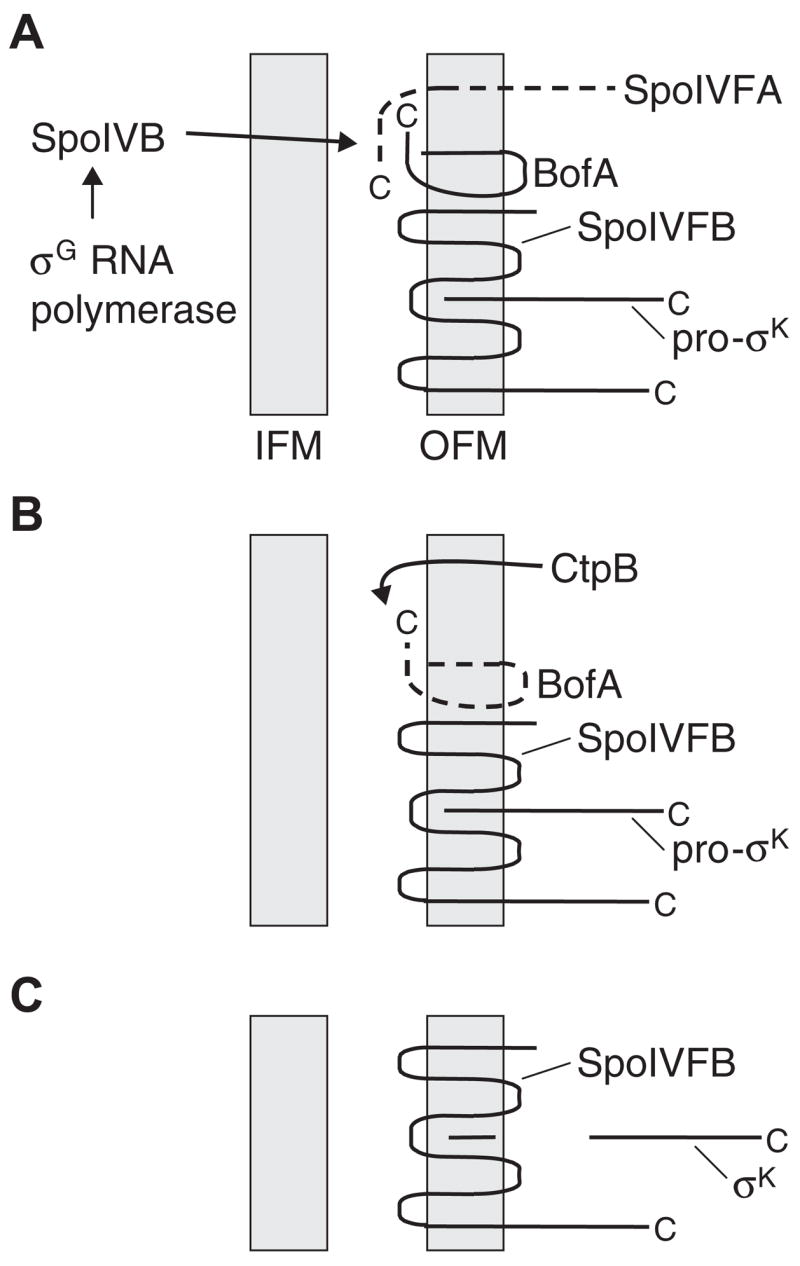
A. SpoIVB from the forespore initiates a proteolytic cascade by cleaving SpoIVFA, leading to its loss from complexes with BofA and SpoIVFB.
B. In the second step, CtpB from the mother cell normally cleaves BofA near its C-terminus, leading to its loss from complexes with SpoIVFB. In a ctpB null mutant, another protease(s) causes loss of BofA, albeit less efficiently.
C. Free SpoIVFB performs intramembrane cleavage of pro-σK, releasing σK into the mother cell to direct transcription. See the Fig. 1 legend for additional explanation.
Reconstruction of the σK checkpoint in E. coli
Expressing components of the σK checkpoint in different combinations in E. coli has proven to be very effective for elucidating the function of proteins in this signal transduction pathway (Zhou and Kroos, 2004), especially as reconstitution of pro-σK RIP in vitro has not yet been successful. Expressing SpoIVB-H6 in E. coli along with other components of the σK checkpoint provided evidence that SpoIVB-H6 targets SpoIVFA to partially relieve the complete inhibition of H10SpoIVFB-GFP brought about by the combined action of SpoIVFA and GFPΔ27BofA (Fig. 2). The model in Fig. 6 may explain why pro-σK RIP was only partially restored. As CtpB was not coexpressed, GFPΔ27BofA remains, and can substantially inhibit RIP on its own. In addition, GFPΔ27BofA appeared to partially protect SpoIVFA from SpoIVB-dependent proteolysis in E. coli (Fig. 2). Perhaps in sporulating B. subtilis, interactions between SpoIVFA and other proteins (Doan et al., 2005) render it more susceptible to cleavage by SpoIVB.
When we attempted to coexpress CtpB-H6 along with other components of the σK checkpoint in E. coli, we obtained a surprising result. σK was abundantly produced in a SpoIVFB-independent fashion (data not shown). This depended on CtpB serine protease activity, as it was not observed for CtpB-H6 S309A (data not shown). If CtpB-H6 is secreted across the E. coli inner membrane into the periplasm, it is unlikely that CtpB-H6 directly cleaves pro-σK. Perhaps periplasmic CtpB-H6 somehow leads to activation of RseP (formerly YaeL), an E. coli homologue of SpoIVFB (Alba et al., 2002; Kanehara et al., 2002; Akiyama et al., 2004). In any case, we were unable to investigate the role of CtpB in overcoming the complete inhibition of pro-σK RIP mediated by the combined action of GFPΔ27BofA and SpoIVFA in E. coli, or the partial inhibition of pro-σK RIP mediated by GFPΔ27BofA alone.
Evidence that CtpB cleaves BofA in the second step of a proteolytic cascade
Loss of labelled SpoIVFA occurred at the normal time in the ctpB mutant, yet RIP of pro-σK was delayed (Fig. 4). This implies that the first step of the cascade, SpoIVB cleavage of SpoIVFA leading to its loss, is intact in the ctpB mutant, but is insufficient to allow RIP of pro-σK. Loss of labelled GFPΔ27BofA was delayed in the ctpB mutant, and coincided with onset of pro-σK (Fig. 4). This suggests a second step in the cascade, CtpB cleavage of BofA leading to its loss. Support for this idea was obtained by coexpressing CtpB-H6 and GFPΔ27BofA in E. coli (Fig. 5A). Also, by coexpressing N-terminally tagged H6GFPΔ27BofA with CtpB-H6, we obtained evidence that BofA is cleaved near its C-terminus in a CtpB-dependent fashion (data not shown). Interestingly, CtpB shares sequence similarity with members of the carboxyl-terminal processing protease family (Pan et al., 2003). One member of the family, Tsp of E. coli, has been shown to use its PDZ domain to recognize non-polar C-terminal residues of its substrates (Keiler and Sauer, 1996; Beebe et al., 2000). As BofA has non-polar C-terminal residues (Ricca et al., 1992), it is tempting to speculate that CtpB uses its PDZ domain to recognize these amino acids. Deletion of as little as three amino acids from its C-terminal end inactivates BofA (Varcamonti et al., 1997), so these residues are also important for BofA-mediated inhibition of pro-σK RIP. Except for its C-terminal tail, the rest of GFPΔ27BofA is expected to be inaccessible to CtpB when the proteins are coexpressed in E. coli, based on membrane topological analysis of BofA fusion proteins in E. coli (Varcamonti et al., 1997) and assuming that CtpB’s predicted N-terminal signal sequence (Pan et al., 2003) mediates its translocation across the inner membrane to the periplasm (Fig. 1D). This might explain the limited digestion of GFPΔ27BofA in E. coli coexpressing CtpB-H6 (Fig. 5A). In contrast, purified CtpB-H6 caused a loss of full-length H6GFPΔ27BofA (Fig. 5A) and partial degradation products were not detected under the conditions we tested (data not shown), perhaps because H6GFPΔ27BofA had been purified after detergent solubilization from membranes. On the other hand, SpoIVFA was not recognized by CtpB in E. coli or in vitro (Fig. 5B). It is noteworthy that SpoIVFA has a charged residue at its C-terminus (Cutting et al., 1991b), which might prevent its recognition by CtpB. Taken together, our in vivo experiments in sporulating B. subtilis and in E. coli, and our in vitro experiments with purified CtpB-H6 strongly support the idea that CtpB cleaves BofA after SpoIVB has cleaved SpoIVFA.
Loss of BofA would allow the third step in the cascade, RIP of pro-σK by SpoIVFB. Loss of GFPΔ27BofA and RIP of pro-σK are not absolutely dependent on CtpB, but both are significantly delayed in a ctpB null mutant (Fig. 4), resulting in a twofold reduction in sporulation efficiency (Pan et al., 2003). This implies that there is a CtpB-independent mechanism(s) capable of ridding SpoIVFB of its inhibitor, BofA, albeit less efficiently. It is unlikely that CtpA, a paralogue of CtpB, is completely responsible for BofA proteolysis in the absence of CtpB, because a ctpA ctpB double mutant is only slightly more defective in sporulation than a ctpB mutant (Pan et al., 2003).
CtpB-H6 was extremely unstable when synthesized in E. coli (Fig. 5). It is attractive to speculate that CtpB is also unstable in sporulating B. subtilis, because it provides a rationale for the second step of the cascade; namely, that the second step monitors ongoing protein synthesis capability of the mother cell. CtpB, like SpoIVFA, BofA and SpoIVFB, is expressed in the mother cell under the control of σE RNA polymerase (Cutting et al., 1991b; Ricca et al., 1992; Pan et al., 2003). If CtpB is less stable than the other mother cell-produced components of the σK checkpoint, it could provide a check on mother-cell protein synthesis capacity, prior to activation of σK and the ensuing final phase of protein synthesis in the mother cell. We propose that the cascade, by sensing SpoIVB from the forespore via cleavage of SpoIVFA, and sensing CtpB from the mother cell via BofA cleavage, ensures the proper timing and rate of pro-σK RIP, which is important for efficient sporulation (Cutting et al., 1990; Pan et al., 2003).
Components of the σK checkpoint in related bacteria and comparison with other cascades governing RIP
Homologues of the components of the σK checkpoint are found in spore-forming bacteria closely related to B. subtilis, including B. lichenoformis, B. anthracis (causes anthrax), B. cereus (causes food poisoning), B. thuringiensis, B. halodurans, B. clausii, Geobacillus kaustophilus and Oceanobacillus iheyensis (data not shown), so the mechanism governing RIP of pro-σK is likely conserved in close relatives. In contrast, components of the σK checkpoint are missing or significantly different in spore-forming bacteria of the genus Clostridium. SpoIVFA does not appear to be present in the genome of Clostridium perfringens, C. thermocellum, C. acetobutylicum, or C. tetani. SigK homologue are present and appear to have pro sequences (data not shown), but these sequences lack a lysine residue that has been shown to be critical for RIP of B. subtilis pro-σK (Prince et al., 2005). Likewise, potential BofA homologues can be found (with the exception of C. tetani), but lack a histidine residue essential for inhibition of pro-σK RIP in B. subtilis (Zhou and Kroos, 2004). Potential SpoIVFB and CtpB homologues exist, but these could just as well carry out other functions, because they are about as similar to SpoIVFB or CtpB, as to their functionally distinct paralogues, YluC and YydH (paralogues of SpoIVFB) or CtpA (paralogue of CtpB). It remains to be seen how different the mechanisms regulating σK activity are in these Clostridia. In C. difficile, SigK has no pro sequence, and the production of σK is regulated by excision of a prophage-like insertion element in the sigK gene (Haraldsen and Sonenshein, 2003).
Most known RIP events involve a proteolytic cascade in which a substrate is first cleaved in a cytosolic or lumenal/ extracellular domain before the I-CLiP can cleave the substrate in a transmembrane segment. For example, S2P, the human homologue of SpoIVFB, cleaves SREBPs and ATF6 only after initial cleavage by site-1 protease, a serine protease (reviewed in Brown et al., 2000). Similarly, S2P family member RseP (formerly YaeL) of E. coli catalyses intramembrane proteolysis of RseA only after serine protease DegS cleaves RseA’s periplasmic domain (Alba et al., 2002; Kanehara et al., 2002). This theme extends beyond the S2P family. I-CLiPs in the presenilin and signal peptide peptidase families of aspartyl proteases also require prior cleavage of their substrates by another protease (reviewed in Wolfe and Kopan, 2004). In contrast, the Rhomboid family of serine protease I-CLiPs appear to function independently, cleaving their substrates without a prior cleavage. Rhomboid regulation seems to occur mainly at the level of translocation of its substrate from the endoplasmic reticulum to the Golgi (Lee et al., 2001). SpoIVFB is unique among S2P family members in that there is no evidence to suggest that pro-σK is cleaved before RIP by SpoIVFB. Yet, our results provide evidence that regulation of SpoIVFB activity, like that of other S2P family members, is achieved by a proteolytic cascade involving serine proteases. However, in the case of SpoIVFB, our model proposes that the cascade involves two serine proteases from different cell types that target a localization/assembly factor (SpoIVFA) and an I-CLiP inhibitor (BofA), rather than targeting the I-CLiP’s substrate (pro-σK). This elaborate mechanism governing SpoIVFB I-CLiP activity would allow the forespore and mother cell to communicate about the decision to activate pro-σK. Previously, RIP has been shown to initiate inter-cellular communication by generating a signal (reviewed in Brown et al., 2000; Wolfe and Kopan, 2004). Our results provide new insight into how intercellular signalling can culminate in RIP of a transcription factor. It will be interesting to see whether intercellular signalling mechanisms involving proteolytic cascades govern RIP in organisms other than spore-forming bacteria closely related to B. subtilis.
Experimental procedures
General methods
The plasmids used in this study are described briefly in Table 1. A complete description of plasmid construction and polymerase chain reaction (PCR) primers is available in Tables S1 and S2. DNA sequencing of all cloned PCR products and all genes subjected to mutagenesis (QuikChange kit, Stratagene) confirmed the presence of the desired sequences. All B. subtilis strains were derived from the prototrophic strain PY79 and are listed in Table 2. ZR264, ZR266 and ZR368 were constructed by transformation of BK754, ZR264 and BDR954 with DNA from BDR88, BDR954 and QPB161, respectively, selecting with appropriate antibiotics. Sporulation was induced by resuspending growing cells in SM medium (Harwood and Cutting, 1990). The time of resuspension is considered the onset of sporulation.
Table 1.
Plasmids used in this study.
| Plasmid | Description |
|---|---|
| pZR2 | ApR, T7-H10SpoIVFB-GFPa |
| pZR12 | KmR, T7-pro-σK(1-126)H6 |
| pZR13 | ApR, T7-H10SpoIVFB-GFP |
| pZR33 | ApR, T7-H10SpoIVFB-GFP/SpoIVFA |
| pZR42 | ApR, T7-H10SpoIVFB-GFP/T7-BofA/SpoIVFA |
| pZR50 | KmR, T7-pro-σK(1-126)H6/T7-SpoIVB |
| pZR53 | ApR, T7-SpoIVB-H6 |
| pZR57 | KmR, T7-pro-σK(1-126)H6/T7-SpoIVB-H6 |
| pZR62 | ApR, T7-GFPΔ27BofA |
| pZR63 | KmR, T7-pro-σK(1-126)H6/T7-GFPΔ27BofA |
| pZR67 | ApR, T7-H10SpoIVFB-GFP/T7-GFPΔ27BofA |
| pZR69 | ApR, T7-H10SpoIVFB-GFP/T7-GFPΔ27BofA/SpoIVFA |
| pZR71 | KmR, T7-pro-σK(1-126)H6/T7-SpoIVB-H6 S378A |
| pZR73 | ApR, T7-SpoIVFA |
| pZR74 | KmR, T7-H6SpoIVFA |
| pZR114 | KmR, T7-CtpB-H6 |
| pZR115 | KmR, T7-H6GFPΔ27BofA |
| pZR119 | KmR, T7-CtpB-H6 S309A |
The T7 RNA polymerase promoter (T7) plus the optimal E. coli translation initiation sequence and DNA coding for 10 histidine (H10) residues in pET-16b was fused to the B. subtilis gene spoIVFB, which had been fused to a gene coding for the green fluorescent protein (GFP). The descriptions of other plasmids are abbreviated in a similar fashion and details of their construction are in Tables S1 and S2 or have been described previously (Zhou and Kroos, 2004).
ApR, ampicillin-resistant; KmR, kanamycin-resistant.
Table 2.
Bacillus subtilis strains used in this study.
| Strain | Relevant characteristics | Reference or construction |
|---|---|---|
| PY79 | Spo+ prototroph | Youngman et al. (1984) |
| LK1 | spoIVBΔ::spc | Kroos et al. (2002) |
| BK754 | spoIVB165 | Cutting et al. (1991a) |
| BDR88 | bofA::erm | Rudner and Losick (2002) |
| BDR954 | bofA::erm, amyE::PspoIID-gfp(mut.2)−Δ27bofA (spc) | Rudner and Losick (2002) |
| QPB161 | ctpB::tet | Pan et al. (2003) |
| ZR264 | spoIVB165 bofA::erm | This study |
| ZR266 | spoIVB165 bofA::erm, amyE::PspoIID-gfp(mut.2)−Δ27bofA (spc) | This study |
| ZR368 | ctpB::tet, bofA::erm, amyE::PspoIID-gfp(mut.2)−Δ27bofA (spc) | This study |
Cotransformation of plasmids into E. coli and induction of B. subtilis proteins
Two plasmids bearing different antibiotic resistance genes and different B. subtilis genes fused to a T7 RNA polymerase promoter were cotransformed into E. coli BL21(DE3), and expression of T7 RNA polymerase and B. subtilis proteins was induced as described previously (Zhou and Kroos, 2004).
Western blot analysis
Equivalent amounts of E. coli cells from different cultures were collected and whole-cell extracts were prepared and subjected to Western blot analysis as described previously (Zhou and Kroos, 2004). For B. subtilis, samples (1 ml) were collected at the indicated times after the onset of sporulation, cells were collected by centrifugation (12 000 g), and the cell pellet was stored at −80°C. Whole-cell extracts were prepared and subjected to Western blot analysis as described previously (Zhou and Kroos, 2004). Antibodies against pentahistidine (penta-His) (Qiagen), GFP and SpoIVFA (Kroos et al., 2002) and pro-σK (Lu et al., 1990), were used at a 1:5000 dilution, except the antipenta-His antibodies were used at a 1:10 000 dilution.
Purification of proteins
To purify the membrane proteins H6SpoIVFA and H6GFPΔ27BofA for use as substrates, E. coli from cultures (4 l) induced with isopropyl β-D-thiogalactopyranoside (IPTG) as described previously (Zhou and Kroos, 2004) were harvested by centrifugation and resuspended in 40 ml phosphate buffered saline (PBS) pH 7.2 (Invitrogen) containing 1 mM Pefabloc, 10 μg ml−1 DNase I and 10 μg ml−1 RNase A, then passed twice through a French pressure cell at 14 000 lb in−2. The lysate was centrifuged at 4°C (12 000 g, 10 min). The supernatant was further centrifuged at 4°C for 1.5 h at 200 000 g in a Ti 42.1 rotor (Beckman). The pellet, representing a crude membrane fraction, was solubilized with 6 ml of 1% digitonin buffer (PBS pH 7.2, 150 mM NaCl, 10% glycerol, 1 mM Pefabloc) by rotating for 2 h at 4°C. The sample was centrifuged at 4°C for 1 h at 100 000 g in a Ti 42.1 rotor (Beckman). The supernatant was incubated with cobalt resin (Clontech) and purification of H6-tagged proteins was performed according to the manufacturer’s instructions except elution was with 0.1% digitonin buffer (PBS pH 7.2, 200 mM imidazole, 150 mM NaCl). Purified proteins were dialysed in 0.05% digitonin buffer (PBS pH 7.2, 50 mM NaCl) at 4°C for 6 h before use as substrates for in vitro proteolysis.
To purify CtpB-H6, E. coli from a culture (2 l) induced with IPTG as described previously (Zhou and Kroos, 2004) were harvested by centrifugation and resuspended in 30 ml PBS pH 7.2 containing 1 mM Pefabloc, 10 μg ml−1 DNase I and 10 μg ml−1 RNase A, then lysed as described above. The lysate was centrifuged at 4°C for 1.5 h at 200 000 g in a Ti 42.1 rotor (Beckman). The supernatant was incubated with cobalt resin (Clontech) and purification of H6-tagged proteins was performed according to the manufacturer’s instructions. The purified protein was dialysed in PBS pH 7.2 containing 50 mM NaCl at 4°C for 6 h prior to use in assays for in vitro proteolysis.
Pulse-chase and immunoprecipitation
Pulse-labelling of cells was performed as described previously (Zhang and Kroos, 1997). The chase involved addition of a 2000-fold molar excess of unlabelled methionine. Immunoprecipitation was performed as described previously (Kroos et al., 2002). Polyclonal anti-SpoIVFA antibodies (6 μl) or anti-GFP antibodies (14 μl) were used and these amounts were sufficient to quantitatively precipitate SpoIVFA or GFPΔ27BofA, respectively, from 1 ml culture lysates in control experiments. Immunoprecipitates were separated on sodium dodecyl sulphate – 14% Prosieve polyacrylamide gels (FMC) with Tris-Tricine electrode buffer (0.1 M Tris, 0.1 M Tricine, 0.1% SDS). SpoIVFA or GFPΔ27BofA bands were visualized by fluorography with En3Hance (NEN) as enhancing fluors and quantified with a Storm 820 PhosphorImager (Molecular Dynamics), with the background of each lane subtracted from the band intensity.
Supplementary Material
The following supplementary material is available for this article online:
Table S1. Construction of plasmids used in this study.
Table S2. Primers used in this study.
This material is available as part of the online article from http://www.blackwell-synergy.com
Acknowledgments
We thank B. Kunkel, D. Rudner, Q. Pan and R. Losick for providing strains BK754, BDR88, BDR954 and QPB161; and L. Gu for art work on the figures. This work was supported by NIH Grant GM43585 to L.K. and by the Michigan Agricultural Experiment Station.
References
- Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry. 2000;39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-σK processing in Bacillus subtilis. Genes Dev. 1991a;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991b;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Dong TC, Cutting SM. SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of pro-σK in Bacillus subtilis. Mol Microbiol. 2003;49:1425–1434. doi: 10.1046/j.1365-2958.2003.03651.x. [DOI] [PubMed] [Google Scholar]
- Dong TC, Cutting SM. The PDZ domain of the SpoIVB transmembrane signalling protein enables cistrans interactions involving multiple partners leading to the activation of the pro-σK processing complex in Bacillus subtilis. J Biol Chem. 2004;279:43468–43478. doi: 10.1074/jbc.M407048200. [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Green D, Cutting S. Membrane topology of the Bacillus subtilis pro-σK processing complex. J Bacteriol. 2000;182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsen JD, Sonenshein AL. Efficient sporulation in Clostridium difficile requires disruption of the σK gene. Mol Microbiol. 2003;48:811–821. doi: 10.1046/j.1365-2958.2003.03471.x. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Chichester, UK: John Wiley and Sons; 1990. [Google Scholar]
- Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σE, RseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Sauer RT. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J Biol Chem. 1996;271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- Kroos L, Yu YT. Regulation of sigma factor activity during Bacillus subtilis development. Curr Opin Microbiol. 2000;3:553–560. doi: 10.1016/s1369-5274(00)00140-5. [DOI] [PubMed] [Google Scholar]
- Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- Kroos L, Yu YT, Mills D, Ferguson-Miller S. Forespore signaling is necessary for pro-σK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest. J Bacteriol. 2002;184:5393–5401. doi: 10.1128/JB.184.19.5393-5401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Lu S, Halberg R, Kroos L. Processing of the mother-cell σ factor σK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Losick R, Rudner DZ. A second PDZ-containing serine protease contributes to activation of the sporulation transcription factor σK in Bacillus subtilis. J Bacteriol. 2003;185:6051–6056. doi: 10.1128/JB.185.20.6051-6056.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Losick R. Sporulation genes and intercompartmental regulation. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Its Closest Relatives: From Genes to Cells. Washington, DC: American Society for Microbiology Press; 2002. pp. 483–518. [Google Scholar]
- Prince H, Zhou R, Kroos L. Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-σK. J Bacteriol. 2005;187:961–971. doi: 10.1128/JB.187.3.961-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-σK processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner D, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci USA. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varcamonti M, Marasco R, De Felice M, Sacco M. Membrane topology analysis of the Bacillus subtilis BofA protein involved in pro-σK processing. Microbiology. 1997;143:1053–1058. doi: 10.1099/00221287-143-4-1053. [DOI] [PubMed] [Google Scholar]
- Wakeley PR, Dorazi R, Hoa NT, Bowyer JR, Cutting SM. Proteolysis of SpoIVB is a critical determinant in signalling of pro-σK processing in Bacillus subtilis. Mol Microbiol. 2000;36:1336–1348. doi: 10.1046/j.1365-2958.2000.01946.x. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposonborne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Yu YTN, Kroos L. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J Bacteriol. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kroos L. A feedback loop regulates the switch from one sigma factor to the next in the cascade controlling Bacillus subtilis mother cell gene expression. J Bacteriol. 1997;179:6138–6144. doi: 10.1128/jb.179.19.6138-6144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Hofmeister A, Kroos L. The pro-sequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Kroos L. BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 2004;101:6385–6390. doi: 10.1073/pnas.0307709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supplementary material is available for this article online:
Table S1. Construction of plasmids used in this study.
Table S2. Primers used in this study.
This material is available as part of the online article from http://www.blackwell-synergy.com


