Abstract
Since apoE allele status is the predominant Alzheimers disease (AD) genetic risk factor, functional single nucleotide polymorphisms (SNP)s in brain apoE receptors represent excellent candidates for association with AD. Recently, we identified a SNP, rs688, as modulating the splicing efficiency of low-density lipoprotein receptor (LDLR) exon 12 in the female human liver and in minigene transfected HepG2 cells. Moreover, the rs688T minor allele associated with significantly higher LDL and total cholesterol in women in the Framingham Offspring Study. Since LDLR is a major apoE receptor in the brain, we hypothesized that rs688 modulates LDLR splicing in neural tissues and associates with AD. To evaluate this hypothesis, we first transfected LDLR minigenes into SH-SY5Y neuroblastoma cells and found that rs688T reduces exon 12 inclusion in this neural model. We then evaluated rs688 association with exon 12 splicing efficiency in vivo by quantifying LDLR splicing in human anterior cingulate tissue obtained at autopsy; the rs688T allele associated with decreased LDLR exon 12 splicing efficiency in aged men but not women. Lastly, we evaluated whether rs688 associates with AD by genotyping DNA from 1,457 men and 2,055 women drawn from three case-control series. The rs688T/T genotype was associated with increased AD odds in males (recessive model, odds ratio (OR) of 1.49, 95% confidence interval (CI) of 1.13−1.97, uncorrected p=0.005), but not in females. In summary, these studies identify a functional apoE receptor SNP that is associated with AD in a sex-dependent fashion.
INTRODUCTION
Genetic variants that alter protein expression or function represent powerful tools to investigate the role of that particular protein in human disease. Single nucleotide polymorphisms (SNP)s that alter exon splicing enhancers (ESE)s and, thereby exon splicing efficiency, are emerging as functional variants capable of modulating disease susceptibility ((1-3), reviewed in (4)). We recently identified the minor T allele of rs688 as neutralizing a putative ESE within exon 12 of the low-density lipoprotein receptor (LDLR). Moreover, rs688T associates with inefficient exon 12 splicing in the human female liver in vivo, causes inefficient LDLR splicing in minigene transfected HepG2 cells, and associates with increased LDL and total cholesterol in women in the Framingham Offspring Study (FOS) (5).
Although the etiology of late onset Alzheimers disease (AD) is unclear, several lines of evidence suggest that LDLR represents an excellent candidate gene for AD association. First, LDLR is a primary brain receptor for apoE, alleles of which are widely accepted to modulate AD risk (6, 7). Second, LDLR deficiency associates with increased amyloid-beta deposition in Tg2576 APP transgenic mice, although not PDAPP transgenic mice, suggesting a possible linkage between LDLR and this AD hallmark (6, 7). Third, murine LDLR deficiency has been associated with learning deficits (7). Fourth, LDLR mutations are a primary cause of familial hypercholesterolemia, suggesting that other LDLR family members are incapable of compensating adequately for insufficient LDLR function (reviewed in (8-10)). Lastly, apoE alleles and rs688 both associate with cholesterol homeostasis, which itself has been implicated in Aß production and risk for AD ((5), reviewed in (11, 12). Here, we report evaluation of rs688 function in neural cells as well as rs688 association with splicing differences in the human brain and risk for AD.
RESULTS
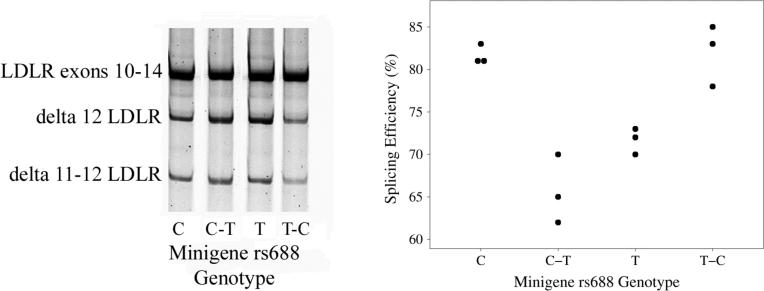
To evaluate whether rs688 modulates LDLR exon 12 splicing in neural cells, we transfected human neuroblastoma SH-SY5Y cells with LDLR minigenes consisting of LDLR exons 9 − 14 as well as the intervening introns. Since the minigenes were cloned from rs688C/C and rs688T/T individuals and varied at additional SNPs (5), we also investigated the role of rs688 specifically, i.e., we used site-directed mutagenesis to convert the rs688 site within the minigenes from rs688C to rs688T (rs688C-T), and, conversely, rs688T to rs688C (rs688T-C). Splicing efficiency resulting from the parent and mutant constructs was analyzed by preparing RNA at 24 hours post-transfection, converting to cDNA, and PCR-amplifying with a sense primer corresponding to sequence derived from the vector and an antisense primer corresponding to sequence within exon 14. This analysis found that the rs688C allele minigene consistently produced a greater proportion of RNA that contained exon 12 relative to the rs688T minigene (Figure 1). Moreover, converting rs688C to the minor allele T reduced the efficiency of exon 12 inclusion. Similarly, when rs688T was converted to rs688C, a greater proportion of the RNA contained exon 12 (Figure 1). In summary, rs688 modulates LDLR exon 12 splicing in neural cells, extending our prior observation that this SNP is functional within HepG2 cells (5).
Figure 1. Rs688 modulates LDLR splicing efficiency in neuroblastoma cells in vitro.
LDLR minigenes containing rs688C and rs688T alleles, or the same haplotypes wherein rs688C and rs688T were specifically converted by site-directed mutagenesis, were transfected into SH-SY5Y cells in parallel and RNA isolated for analyses 24 hours later. These results depict representative images of the splicing results with PCR products corresponding to LDLR exons 10−11−12−13−14, as well as LDLR exons 10−11−13−14 (delta 12 LDLR), and LDLR exons 10−13−14 (delta 11 − 12 LDLR). Quantification of rs688 effects on splicing is also depicted; data points represent separate analyses beginning with cell transfection. The rs688T allele was associated with decreased splicing efficiency regardless of background haplotype, as reflected by Kruskal-Wallis statistical analyses (p=0.026).
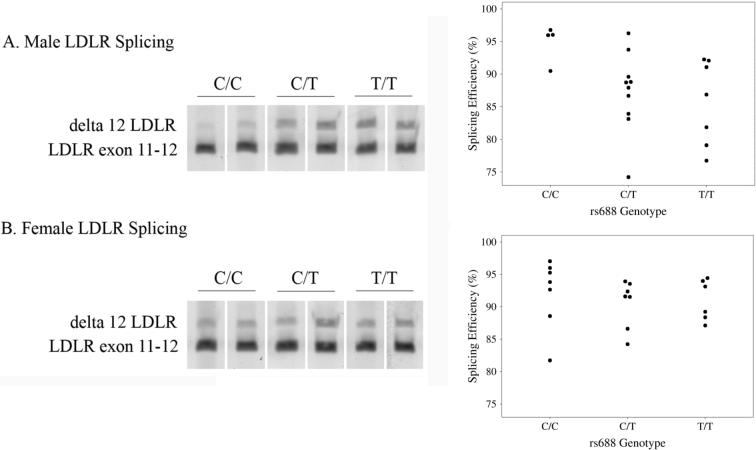
To investigate whether rs688 is associated with exon 12 splicing efficiency in the human brain in vivo, we analyzed 41 RNA samples prepared from the human anterior cingulate, a brain area moderately affected in AD. We focused on exon 12 splicing per se by using appropriate PCR primers in two separate reactions. The first reaction identified the proportion of LDLR mRNA containing exon 12, i.e., the sense primer corresponded to the LDLR exon 11−12 junction while the antisense primer corresponded to exon 12 per se (150 bp PCR product). The second reaction identified the proportion of LDLR mRNA lacking exon 12, i.e., a sense primer corresponding to exon 10 was used in conjunction with an antisense primer specific to the sequence at the junction of exon 11 and exon 13. This latter reaction generated a 170 bp product. This assay revealed that rs688T associated with a significantly lower proportion of exon 12-containing LDLR mRNA in men but not women (Figure 2A-B). Interestingly, these results in the aged brain are the mirror image of our prior findings regarding human liver, where rs688T associated with lower exon 12 splicing efficiency in pre-menopausal women but not in age-matched men (5); we are unclear as to the mechanisms underlying these sex differences in SNP-associated splicing in young adult liver versus aged adult brain. In the brain, the apparent difference in the proportion of exon 12- deficient LDLR between rs688T/T and rs688C/C males was 8%. Since exon 12 deletion causes a frameshift and a premature stop codon, these LDLR isoforms are predicted to encode truncated, non-functional LDLR proteins (5). In summary, rs688 associates with LDLR exon 12 splicing efficiency in males, representing a sex-dependent decrease in the proportion of LDLR mRNA encoding functional LDLR.
Figure 2. Rs688 is associated with LDLR splicing efficiency in the male human brain in vivo.
This figure depicts representative splicing patterns corresponding to the indicated PCR products as well as quantitation of exon 12 splicing efficiency in the brains of separate males (A) and females (B). Rs688 was associated significantly with splicing in males (p=0.041) but not females (p=0.43) as determined by Jonckheere-Terpstra ranked sum tests.
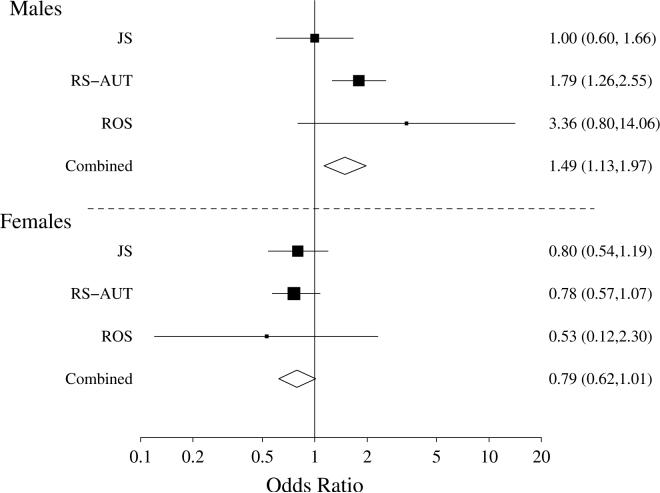
Since LDLR is a primary apoE receptor in the brain (6, 7), we next investigated whether rs688 associates with AD in three AD-case control series; these series were derived from clinically diagnosed subjects at the Mayo Clinic Jacksonville, FL (JS) and the Religious Orders Studies (ROS), as well as clinically diagnosed subjects at the Mayo Clinic Rochester, MN, which were combined with a series of AD autopsies (RS-AUT) (13-15). We genotyped these series and performed analysis of the association of AD with rs688 (recessive model) using logistic regression adjusted for series, presence of apoE4, age and sex. We also considered interaction terms for series and sex with rs688, finding that there was strong evidence of an interaction of rs688 with sex (p=0.001), but no evidence of any heterogeneity in effects of rs688 across series (p=0.46) or of interactions with age (p=0.93) or the presence of apoE4 (p=0.45). Separate logistic regression models were then used to analyze male and female data further. In males, there was strong evidence of increased risk for AD with rs688T/T (OR= 1.49, p=0.005, Figure 3). However, in females, the estimated odds ratio was 0.79 (p=0.056), consistent with the sex-dependent splicing in the brain. There was no significant difference in odds ratios among the three series for either males or females, but the RS-AUT and ROS populations displayed a more robust association in males than the JS series. The statistical significance of this finding in the male series overall was maintained even after correction for multiple testing, i.e., conservative Bonferroni correction for testing allelic, recessive and dominant models would adjust the overall p-value to 0.015. Hence, the rs688T allele that associates with decreased splicing efficiency in males is, when homozygous, also associated with increased risk for AD in males. Since the level of significance in women was marginal, especially after any correction for multiple testing, we interpret the results in women as suggestive evidence that rs688T may be associated with reduced AD risk in women. In summary, rs688T/T is associated with increased risk for AD in males but not in females.
Figure 3. Rs688 is associated with AD in men.
This Forest plot depicts the odds ratio and 95% confidence intervals for the association of rs688 with AD in each series, separately for men and women. The bar width reflects the 95% confidence interval while the symbol size reflects the size of the series. The open diamonds represent the overall odds ratios combining all three series and the width of the diamonds correspond to 95% confidence intervals.
DISCUSSION
The primary findings of this report are that rs688 is a functional SNP within neural cells, associates with LDLR exon 12 splicing efficiency only in the male brain, and associates with significantly increased risk for AD in men. Hence, the overall significance of this study is twofold. First, these results support the hypothesis that a functional SNP in LDLR, a brain apoE receptor, is associated with AD. Second, the unusual sex-dependent association of rs688 with AD may be relevant to other genetic variants, suggesting that AD genetic studies may benefit by analyzing men and women separately.
Given that rs688 is a common SNP within an excellent AD candidate gene, several prior studies have evaluated the association between AD and rs688 or linked SNPs (16-20). When these studies were undertaken, the investigators were not aware of the sex dependence of rs688 function and therefore did not parse their series by sex. Importantly, when we analyzed males and females together by logistic regression adjusting for series, presence of apoE4 and gender, the associations within each sex balanced one another and no association was detected between rs688 T/T and AD; the overall OR for the recessive model was 1.04 (95% CI of 0.86−1.25, p=0.69). Hence, stratifying for sex was critical to detecting the association described here. Additional case control series must be analyzed to confirm the association of rs688 T/T with increased risk for AD in male but not female subjects. The correlation between this association and functional alteration in splicing, which also occurs in male but not female subjects, strongly suggests, however, that replication is likely in confirmatory series of sufficient size.
Although apoE and LDLR SNPs associate with both cholesterol and AD, we do not interpret our results as supporting a positive correlation between peripheral cholesterol and risk for AD generally. Although apoE4 and rs688T both associate with increased LDL-cholesterol and increased risk for AD, the apoE4 and rs688T alleles have clear differences in their sex dependence. The presence of an apoE4 allele in women is associated with a 5 mg/dl increase in LDL-cholesterol (21) and an increased risk of AD with an odds ratio of 5.15 (95% CI of 4.23−6.27, data from this study). Males are similar in that apoE4 is associated with a 3 mg/dl increase in LDL-cholesterol (21) and a 3.81 AD odds ratio (95% CI 3.05−4.75). In contrast, rs688T is associated with a 6 mg/dl increase in LDL-cholesterol in women (5) but an AD odds ratio of 0.79 (95% CI 0.62−1.01). Moreover, rs688T in men was associated with an insignificant trend towards decreased cholesterol (5) but had an odds ratio of 1.49 (95% CI 1.13−1.97) for AD risk. Thus the effects of apoE4 and rs688T on peripheral cholesterol do not correlate with their effects on AD risk. For this reason, we interpret our data as suggesting that rs688 modulates LDLR function with respect to cholesterol in the periphery and to apoE metabolism in the CNS. In the former, the relationship between LDLR and cholesterol has been well-substantiated; individuals with impaired LDLR function have less LDL-cholesterol removed from the extracellular space, resulting in increased intracellular cholesterol synthesis, further exacerbating plasma cholesterol levels (reviewed in (10)). Hence, LDLR in the periphery is intimately linked with cholesterol homeostasis per se. In contrast, LDLR deficiency in the brain does not alter brain cholesterol, at least as modeled by the LDLR deficient murine brain (6). Rather, LDLR deficiency increases murine brain apoE levels, as well as levels of apoE3 and apoE4 in mice expressing these human alleles (6, 7). Since increased apoE expression associates with increased Aß pathology in the mouse (22), the most parsimonious interpretation of our data in men is a model wherein rs688T decreases exon 12 splicing efficiency, leading to reduced functional LDLR and reduced apoE clearance, and thereby, increased amyloid pathology and risk for AD. This hypothesis is supported by several additional lines of reasoning. First, the rs688T allele increases the proportion of LDLR that lacks exon 12, which shifts the LDLR reading frame, leading to a premature stop codon in exon 13 and a LDLR isoform lacking the transmembrane domain encoded by exons 16−17 (5); consistent with this possibility, a similarly truncated LDLR produced by a nonsense mutation in exon 13 is retained within the endoplasmic reticulum and causes familial hypercholesterolemia (23). Second, LDLR deficiency in murine models of amyloidogenesis may exacerbate amyloid accumulation and memory deficits (6, 7). Third, the association between rs688 and increased risk for AD achieved statistical significance only when considered as a recessive model, which is suggestive that rs688T acts in a loss-of-function fashion. In summary, while experimentation to evaluate this model is underway, the parsimonious interpretation of these results is a recessive model wherein LDLR protein encoded by the exon 12-deficient LDLR isoform is not functional, representing a loss of apoE receptor, reduced apoE clearance and increased risk for AD in males.
In conclusion, rs688 is a common and functional LDLR SNP that modulates LDLR exon splicing efficiency in vitro in neural cells and in vivo in the male brain. Moreover, rs688 associates significantly with increased AD risk in men. Since LDLR expression in the CNS could be manipulated, e.g., by statins that penetrate the blood-brain barrier, these results suggest that LDLR modulation may represent a therapeutic target in at-risk populations. Overall, we anticipate these studies may prove useful for understanding the role of sex and apoE receptors in AD. Moreover, these studies provide a model for identifying AD susceptibility alleles by focusing on well-characterized functional SNPs in genes likely to influence risk for AD.
MATERIALS AND METHODS
Evaluation of LDLR mini-gene splicing in vitro
The role of rs688 in LDLR exon 12 splicing efficiency in neural cells was evaluated by transfecting SH-SY5Y cells with LDLR minigenes consisting of LDLR exons 9−14 along with their intervening introns. These minigenes were cloned from rs688C/C and rs688T/T individuals into a pcDNA3.1 backbone (5). The specific effects of rs688 were evaluated by comparing splicing efficiency in clones wherein site-directed mutagenesis (Quik-Change, Stratagene) was used to specifically mutate rs688C to rs688T, and vice versa. LDLR splicing efficiency was evaluated by transfecting the clones into SH-SY5Y cells by using Nucleofection as directed by the manufacturer (Amaxa, Inc., Gaithersberg, MD). Twenty-four hours post-transfection, total RNA was isolated and analyzed for LDLR splicing patterns by RT- PCR as described (5). Briefly, RNA was converted to cDNA (SuperScript II, Invitrogen) and sequences corresponding to LDLR minigene splice products were PCR-amplified (Platinum Taq, Invitrogen) by using a sense primer corresponding to vector sequence at the beginning of the transcription product (5’ACTAGTCCAGTGTGGTGGAATTGCC 3’) and an antisense primer corresponding to sequence within LDLR exon 14 (5’- CATCGTGGTGGATCCTGTTC). PCR profiles consisted of pre-incubation at 94°C for 60 sec, followed by cycles of 94°C for 30s, 60°C for 45s, and 72°C for 90s (Perkin Elmer 9600). The minimal number of PCR cycles necessary to discern products were performed, i.e., 30 cycles. PCR products were separated by polyacrylamide gel electrophoresis (PAGE) and visualized by SYBR-gold fluorescence on a fluorescence imager (Fuji FLA-2000). The identities of the PCR products were determined by gel purification and direct sequencing (Davis Sequencing). The amounts of full length and inefficiently spliced LDLR isoforms were quantified by fluorescence intensity. For each sample, fluorescence values were corrected for background and normalized for length differences among amplicons. Sample splicing efficiency was then quantified as the amount of LDLR PCR product containing exons 10, 11, 12, 13 and 14 divided by the total LDLR PCR product for that sample. Statistical significance of the results were analyzed by Kruskal-Wallis non-parametric tests.
Evaluation of LDLR splicing in vivo
Human anterior cingulate brain samples were generously provided by the Sanders-Brown Alzheimers Disease Center Neuropathology Core. The samples were from deceased individuals with an average age at death for women of 82 ± 8 years (mean ± SD, n=20) and for men of 81 ± 8 (n=21). The average post-mortem interval (PMI) for women was 3.0 ± 0.8 hours (mean ± SD, n=20) while the PMI for men was 3.0 ± 0.9 hours (n=21). Available samples included four rs688C/C, ten rs688C/T, and seven rs688T/T males, as well as seven rs688C/C, seven rs688C/T, and six rs688T/T females. Total RNA was prepared and converted to cDNA in one microgram aliquots with random hexamers and reverse transcriptase (Invitrogen, SuperScriptII) as we described previously (5, 24, 25). LDLR mRNAs containing exon 12 were identified by subjecting 1/60th of the cDNA to PCR with a sense primer corresponding to the LDLR exon 11−12 junction (5’ AATGGCATCACCCTAGATCTC) and an antisense primer that recognized exon 12 (5’ GGTGGGCCAGCCTCTTTTCATC). LDLR isoforms lacking exon 12 were identified by using a sense primer that recognized exon 10 (5’ CATCGTGGTGGATCCTGTTC ) and an antisense primer specific to the LDLR exon 11−13 junction (5’ CAAAATACTTTGTCCTAGGGTGA). PCR products directed by these two pairs of primers were amplified separately under conditions of 94°C for 4 min followed by 31 cycles of 94°C for 30 sec; 60°C for 30 sec; and 72°C for 30 sec. Post-amplification, the PCR products from each individual were pooled, separated by PAGE, and visualized and quantified by SYBR-gold fluorescence as described above. Data for men and women were analyzed separately, with statistical differences in splicing efficiency analyzed by two-tailed, non-parametric tests for 3 ordered groups, i.e., Jonckheere-Terpstra tests (SPSS).
Genetics Studies
Three different series (JS, RS-AUT, and ROS) were analyzed. The JS series was collected through the Mayo Clinic Jacksonville memory disorders clinic (13). This study included 215 AD men (mean age ± standard deviation: 79 ± 8) and 227 non-AD men (age: 78 ± 7) as well as 362 women with AD (age: 78 ± 6) and 339 non-AD women (age: 78 ± 8). The RS series was collected through the prospective, community-based Mayo Clinic AD registry, as well as the Mayo Clinic Rochester memory disorders clinic (13). For this cohort, we obtained data for 188 men with AD (age at diagnosis: 81 ± 8, and 519 non-AD men (age: 78 ± 6), as well as 330 women with AD (age: 81 ± 8), and 607 non-AD women (age: 79 ± 6). The AD cases in the AUT series, which were combined with the RS series, were obtained from brains with neuropathologically confirmed AD that were collected at autopsy from Caucasians in Jacksonville, FL, Rochester, MN and many additional sites, and included 235 men (age at death: 81 ± 6) as well as 337 women (age: 85 ± 7) The ROS series represents the deceased members of the larger, prospective, community-based ROS series and have been described elsewhere (20). For this project, we used DNA from 37 men with AD (age: 86 ± 7), and 36 non-AD men (age: 80 ± 7), as well as 47 women with AD (age: 89 ± 6), and 33 non-AD women (age: 83 ± 6). Clinical AD diagnoses on all series were made with NINCDS-ADRDA criteria (26). Genomic DNA was extracted from peripheral blood leukocytes or autopsied tissue by using routine methods. Samples were genotyped for rs688 and/or rs5925, a surrogate which is in near perfect linkage disequilibrium with rs688, by using unlabeled PCR primers and TaqMan MGB probes (FAM and VIC dye labeled) obtained via the ‘Assays-by-Design’ service from Applied Biosystems (Foster City, CA, USA) on an ABI-7000 or ABI-7900 (Applied Biosystems). The association of AD with rs688 was assessed by using logistic regression as adjusted for series, age and presence of apoE4. Models were fit separately for males and females after strong evidence of an interaction of sex with the effect of rs688 was found in a single model. Tests of interaction of rs688 by series were also conducted to check that there was no evidence of heterogeneity across series. For apoE and LDLR, the genotype frequencies in males and females from each series were consistent with Hardy Weinberg equilibrium.
ACKNOWLEDGEMENTS
The authors acknowledge tissue supplied by the University of Kentucky Alzheimers Disease Center, which is supported by P30AG028383, as well as NIH for the following grant support: R01AG026147 (SE), P30AG10161, R01AG15819 (DAB) and AG16574 (RCP, SGY).
ABBREVIATIONS
- AD
Alzheimers disease
- SNPs
single nucleotide polymorphisms
- LDLR
low-density lipoprotein receptor
- OR
odds ratio
- CI
95% confidence interval
- ESEs
exon splicing enhancers
- FOS
Framingham Offspring Study
- PMI
post-mortem interval
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest regarding this manuscript.
Supplementary Material
REFERENCES
- 1.Zatkova A, Messiaen L, Vandenbroucke I, Wieser R, Fonatsch C, Krainer AR, Wimmer K. Disruption of Exonic Splicing Enhancer Elements Is the Principal Cause of Exon Skipping Associated with Seven Nonsense or Missense Alleles of Nf1. Hum. Mutat. 2004;24:491–501. doi: 10.1002/humu.20103. [DOI] [PubMed] [Google Scholar]
- 2.Pfarr N, Prawitt D, Kirschfink M, Schroff C, Knuf M, Habermehl P, Mannhardt W, Zepp F, Fairbrother W, Loos M, et al. Linking C5 Deficiency to an Exonic Splicing Enhancer Mutation. J. Immunol. 2005;174:4172–4177. doi: 10.4049/jimmunol.174.7.4172. [DOI] [PubMed] [Google Scholar]
- 3.Steiner B, Truninger K, Sanz J, Schaller A, Gallati S. The Role of Common Single-Nucleotide Polymorphisms on Exon 9 and Exon 12 Skipping in Nonmutated Cftr Alleles. Hum. Mutat. 2004;24:120–129. doi: 10.1002/humu.20064. [DOI] [PubMed] [Google Scholar]
- 4.Faustino NA, Cooper TA. Pre-Mrna Splicing and Human Disease. Genes & Development. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, Cupples LA, Estus S. A Common Polymorphism Decreases Low-Density Lipoprotein Receptor Exon 12 Splicing Efficiency and Associates with Increased Cholesterol. Hum. Mol. Genet. 2007;16:1765–1772. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fryer JD, Demattos RB, McCormick LM, O'Dell MA, Spinner ML, Bales KR, Paul SM, Sullivan PM, Parsadanian M, Bu G, et al. The Low Density Lipoprotein Receptor Regulates the Level of Central Nervous System Human and Murine Apolipoprotein E but Does Not Modify Amyloid Plaque Pathology in Pdapp Mice. The J. Biol. Chem. 2005;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- 7.Cao D, Fukuchi KI, Wan H, Kim H, Li L. Lack of Ldl Receptor Aggravates Learning Deficits and Amyloid Deposits in Alzheimer Transgenic Mice. Neurobiol. Aging. 2005;27:1632–1643. doi: 10.1016/j.neurobiolaging.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Pullinger CR, Kane JP, Malloy MJ. Primary Hypercholesterolemia: Genetic Causes and Treatment of Five Monogenic Disorders. Expert Rev. Cardiovasc. Ther. 2003;1:107–119. doi: 10.1586/14779072.1.1.107. [DOI] [PubMed] [Google Scholar]
- 9.Anderson RG. Joe Goldstein and Mike Brown: From Cholesterol Homeostasis to New Paradigms in Membrane Biology. Trends Cell Biol. 2003;13:534–539. doi: 10.1016/j.tcb.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Hobbs HH, Brown MS, Goldstein JL. Molecular Genetics of the Ldl Receptor Gene in Familial Hypercholesterolemia. Hum. Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 11.Puglielli L, Tanzi RE, Kovacs DM. Alzheimer's Disease: The Cholesterol Connection. Nat. Neurosci. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- 12.Michikawa M. Cholesterol Paradox: Is High Total or Low Hdl Cholesterol Level a Risk for Alzheimer's Disease? J. Neurosci. Res. 2003;72:141–146. doi: 10.1002/jnr.10585. [DOI] [PubMed] [Google Scholar]
- 13.Ertekin-Taner N, Ronald J, Feuk L, Prince J, Tucker M, Younkin L, Hella M, Jain S, Hackett A, Scanlin L, et al. Elevated Amyloid Beta Protein (Abeta42) and Late Onset Alzheimer's Disease Are Associated with Single Nucleotide Polymorphisms in the Urokinase-Type Plasminogen Activator Gene. Hum. Mol. Genet. 2005;14:447–460. doi: 10.1093/hmg/ddi041. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Taylor JW, Bennett DA, Younkin SG, Estus S. Lack of Association of Hepatic Lipase Polymorphisms with Late-Onset Alzheimer's Disease. Neurobiol Aging. 2006 doi: 10.1016/j.neurobiolaging.2006.11.015. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, et al. The Neuronal Sortilin-Related Receptor Sorl1 Is Genetically Associated with Alzheimer Disease. Nat. Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertram L, Hsiao M, McQueen MB, Parkinson M, Mullin K, Blacker D, Tanzi RE. The Ldlr Locus in Alzheimer's Disease: A Family-Based Study and Meta-Analysis of Case-Control Data. Neurobiol. Aging. 2005;28:18.e11–14. doi: 10.1016/j.neurobiolaging.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Cheng D, Huang R, Lanham IS, Cathcart HM, Howard M, Corder EH, Poduslo SE. Functional Interaction between Apoe4 and Ldl Receptor Isoforms in Alzheimer's Disease. J. Med. Genet. 2005;42:129–131. doi: 10.1136/jmg.2004.024968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez E, Mateo I, Llorca J, Sanchez-Quintana C, Infante J, Berciano J, Combarros O. No Association between Low Density Lipoprotein Receptor Genetic Variants and Alzheimer's Disease Risk. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141:541–543. doi: 10.1002/ajmg.b.30341. [DOI] [PubMed] [Google Scholar]
- 19.Retz W, Thome J, Durany N, Harsanyi A, Retz-Junginger P, Kornhuber J, Riederer P, Rosler M. Potential Genetic Markers of Sporadic Alzheimer's Dementia. Psych. Genet. 2001;11:115–122. doi: 10.1097/00041444-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Gopalraj RK, Zhu H, Kelly JF, Mendiondo M, Pulliam JF, Bennett DA, Estus S. Genetic Association of Low Density Lipoprotein Receptor and Alzheimer's Disease. Neurobiol. Aging. 2005;26:1–7. doi: 10.1016/j.neurobiolaging.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer EJ, Lamon-Fava S, Johnson S, Ordovas JM, Schaefer MM, Castelli WP, Wilson PW. Effects of Gender and Menopausal Status on the Association of Apolipoprotein E Phenotype with Plasma Lipoprotein Levels. Results from the Framingham Offspring Study. Arterioscler. Thromb. 1994;14:1105–1113. doi: 10.1161/01.atv.14.7.1105. [DOI] [PubMed] [Google Scholar]
- 22.Holtzman DM, Fagan AM, Mackey B, Tenkova T, Sartorius L, Paul SM, Bales K, Ashe KH, Irizarry MC, Hyman BT. Apolipoprotein E Facilitates Neuritic and Cerebrovascular Plaque Formation in an Alzheimer's Disease Model. Ann. Neurol. 2000;47:739–747. [PubMed] [Google Scholar]
- 23.Lehrman MA, Schneider WJ, Brown MS, Davis CG, Elhammer A, Russell DW, Goldstein JL. The Lebanese Allele at the Low Density Lipoprotein Receptor Locus. Nonsense Mutation Produces Truncated Receptor That Is Retained in Endoplasmic Reticulum. J. Biol. Chem. 1987;262:401–410. [PubMed] [Google Scholar]
- 24.Chomczynski P, Sacchi N. Single-Step Method of Rna Isolation by Acid Guanidinium Thiocynate-Phenol-Chloroform Extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 25.Aksenov MY, Tucker HM, Nair P, Aksenova MV, Butterfield DA, Estus S, Markesbery WR. The Expression of Several Mitochondrial and Nuclear Genes Encoding the Subunits of Electron Transport Chain Enzyme Complexes, Cytochrome C Oxidase, and Nadh Dehydrogenase, in Different Brain Regions in Alzheimer's Disease. Neurochem. Res. 1999;24:767–774. doi: 10.1023/a:1020783614031. [DOI] [PubMed] [Google Scholar]
- 26.McKhann G, Drachman D, Folstein M, Katzman R, Price DL, Stadlan EM. Clinical Diagnosis of Alzheimers Disease: Report of the Nincds-Adrda Work Group under the Auspices of Department of Health and Human Services Task Force on Alzheimers Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.