Abstract
It is now well established that cancer is a genetic disease and that somatic mutations of oncogenes and tumour suppressor genes are the initiators of the carcinogenic process. The phosphatidylinositol 3-kinase signalling pathway has previously been implicated in tumorigenesis, and evidence over the past year suggests a pivotal role for the phosphatidylinositol 3-kinase catalytic subunit, PIK3CA, in human cancers. In this review, we analyse recent reports describing PIK3CA mutations in a variety of human malignancies, and discuss their possible implications for diagnosis and therapy.
Keywords: PI3K, PIK3CA, p110α, somatic mutations
Recently, somatic mutations in many different human cancers were discovered in the gene encoding for the phosphatidylinositol 3-kinase (PI3K) catalytic subunit, PIK3CA. In this review, we will be analysing and consolidating these findings, and discuss their possible implications in cancer progression and therapy. Due to the focused nature of this review, the PI3K pathway and its biochemical signalling properties will not be discussed in detail. The reader is directed toward several excellent recent reviews for a more comprehensive analysis of this pathway (Hennessy et al, 2005; Wymann and Marone, 2005).
PHOSPHATIDYLINOSITOL 3-KINASE OVERVIEW
The PI3Ks are heterodimeric lipid kinases composed of catalytic and adaptor/regulatory subunit variants encoded by separate genes and alternative splicing. Phosphatidylinositol 3-kinases are important regulators of cellular growth, transformation, adhesion, apoptosis, survival and motility (Volinia et al, 1994; Fruman et al, 1998; Cantley, 2002). The PI3K family of enzymes are organised under three main classes (class I, II and III) and various subgroups have been categorised based on their primary structure, substrate specificity and regulation (Vivanco and Sawyers, 2002).
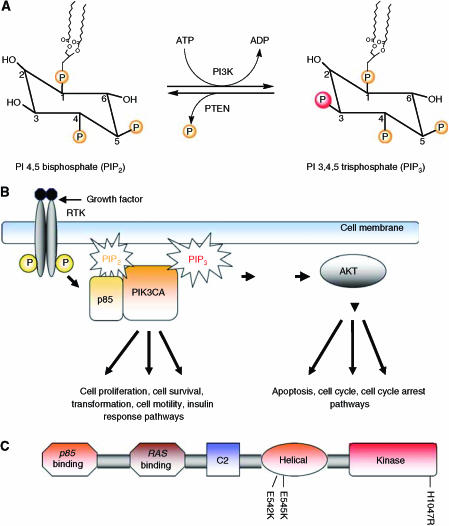
Both the catalytic and regulatory subunits of the human PI3K gene were cloned by Volinia et al (1994) and the overall sequence was found to be highly homologous to the bovine and yeast PI3K genes. The activation of PI3K results in the generation of the second messenger, phosphatidylinositol 3,4,5 trisphosphate (PIP3) from phosphatidylinositol 4,5 bisphosphate (PIP2) (Figure 1A). The activation of PI3K by a growth factor bound (activated) receptor tyrosine kinase (RTK) and subsequent production of PIP3 drives the various downstream pathways that regulate a number of cellular functions including those involved in tumour development and progression (Figure 1B).
Figure 1.
(A) The main reaction catalysed by PI3K: phosphatidylinositol (PI) 4,5 bisphosphate (PIP2) to phosphatidylinositol (PI) 3,4,5-triphosphate (PIP3). (B) PI3K is activated upon ligand binding to a receptor tyrosine kinase (RTK), which then activates the regulatory subunit (p85) to bind the catalytic p110α subunit. This ultimately triggers various downstream signalling cascades resulting in cell survival, apoptosis, transformation, metastasis, and cell migration. (C) Schematic representation of PIK3CA (p110α catalytic subunit of PI3K) and its functional domains with the most common somatic mutations, E542K, E545K and H1047R within the helical and kinase domains indicated.
PHOSPHATIDYLINOSITOL 3-KINASE AND HUMAN CANCER
The kinase activity of PI3K was first reported to be associated with viral oncoproteins (Cantley et al, 1991). Subsequent studies employing mouse knockouts of both the regulatory and catalytic subunits of PIK3 resulted in a number of deficits including embryonic lethality, B cell defects, liver necrosis and colorectal cancer (Katso et al, 2001). Other investigations showed that the amplification of the PI3K locus as well as deletions of short nucleotide sequences resulted in elevated lipid kinase activity of the p110α catalytic subunit of PI3K (PIK3CA) in various cancer types with the implication that PI3K was functioning as an oncogene (Volinia et al, 1994; Shayesteh et al, 1999; Ma et al, 2000; Katso et al, 2001; Migozuchi et al, 2004; Pedrero et al, 2005). The PIK3CA p110α catalytic subunit of PI3K will be highlighted in this review due to the recent alterations of this protein found in primary human cancers. PIK3CA is a 34 kb gene located on chromosome 3q26.3 that consists of 20 exons coding for 1068 amino acids yielding a 124 kDa size protein (Figure 1C). Gene amplifications, deletions and more recently, somatic missense mutations in the PIK3CA gene have been reported in many human cancer types including cancers of the colon, breast, brain, liver, stomach and lung. These somatic missense mutations were proposed to increase the kinase activity of PIK3CA contributing to cellular transformation. The first of these mutational reports was published by Samuels et al (2004). In this seminal paper, the authors initially analysed the sequence of eight PI3K and eight PI3K-like genes in a relatively small number of primary colorectal tumours and discovered that PIK3CA was the only gene harbouring somatic mutations. They subsequently expanded their sample size, which included tissues from primary tumours of the colon, brain, breast, stomach and lung. Their results verified their initial observations and demonstrated that somatic mutations were found in all of these tissues at varying frequencies. Notably, colorectal, brain and gastric cancers were found to have a high rate of PIK3CA gene mutation with frequencies of 32, 27 and 25%, respectively. Breast and lung cancers had a relatively low rate of PIK3CA mutations (8 and 4% respectively), although the sample size of all cancer types was relatively small (n=12–24) with the exception of colorectal cancers (n=234). These somatic missense mutations were scattered across most of the exons, but were predominantly found in the kinase and helical domains of the PIK3CA subunit (Table 1). Of note, ‘hotspot’ or frequently recurring mutations were found in exon 9 (G1624A:E542K) and exon 20 (A3140G:H1047R) in this analysis. Based on all sequencing data (Table 1), there now appear to be three hotspots mutations within PIK3CA: H1047R, E542K and E545K. Bachman et al (2004) expanded this report using a larger sample set consisting of primary breast cancers and breast cancer cell lines. Their data demonstrated that on average 25% of breast cancers harbour missense mutations in either the kinase, helical or p85 binding domains, although it should be noted that only the three exons corresponding to these domains were sequenced in their analysis. Many other studies followed, examining PIK3CA mutations in various cancer types (Table 1). Campbell et al (2004) sequenced all of the 20 coding exons of PIK3CA from primary tumour samples of breast, ovarian and colorectal cancers and reported new mutations found in exons 6, 7 and 9, as well as mutations previously reported by others. They reported a PIK3CA mutation frequency of 18.8% in colorectal cancers and among 70 breast cancer samples, they noted a mutation frequency of 40%, which is thus far the highest reported in any cancer type (Table 1). The frequency of ovarian cancers was reported as 6%, but of note, mutations clustered according to the histologic subtype with endometrioid and clear cell variants having a much higher rate than serous and mucinous ovarian cancers. In both the studies by Bachman et al and Campbell et al, no association was noted between the presence of PIK3CA mutations with other prognostic/clinical features of breast cancer, including histologic subtype, oestrogen/progesterone receptor expression, Her2/neu receptor status, axillary lymph node positivity, grade and/or stage of the tumour. This is in contrast to a more recent analysis by Saal et al (2005) where these authors examined a total of 292 primary breast cancers and found an overall mutation rate of 26%. In this study, the authors described a statistically significant correlation between the presence of PIK3CA mutations and the presence of nodal metastases, oestrogen/progesterone receptor positivity and Her2/neu receptor overexpression/amplification. They also demonstrated a statistically significant correlation between the presence of PIK3CA mutations and the presence of PTEN expression, an intriguing finding given the known roles of these two pathways and similar findings in brain cancers (see below). As described by Saal et al, variations in sample size likely account for the discrepancies between their study and those of Bachman et al and Campbell et al, although regional bias in the tumour samples is still a possibility given that roughly half their samples were from a Swedish cohort that included almost exclusively Stage II breast cancers. Additionally, Levine et al (2005) sequenced PIK3CA exons 9 and 20 in 198 ovarian and 72 breast cancers using primary tissue samples and found an overall mutation rate of 12% for ovarian cancers and 18% for breast cancers, although no correlation with histologic subtypes and/or clinical/prognostic indicators were found for either type of cancer. Finally, Broderick et al (2004) sequenced the PIK3CA gene in 285 brain tumours and found a mutational rate of 5%, which was significantly lower than the rate originally reported by Samuels et al (2005). While the majority of their mutations were in known hotspot regions of the gene, these authors also found that PIK3CA mutations were restricted to certain histologic subtypes. They also showed in a limited analysis that PIK3CA mutations were mutually exclusive with mutations of the tumour suppressor PTEN, suggesting that tumorigenic signalling through this pathway can occur either through activation of PIK3CA or inactivation of PTEN. Given the ubiquitous nature of PIK3CA mutations in human cancers and the conflicting results of the above studies, the association of PIK3CA mutations with other clinical and histologic parameters is still not definitively known.
Table 1. Somatic mutations of PIK3CA in cancer types reported since 10/2005.
| Cancer | % PIK3CA mutationa | Sample source (primary tissue vs cancer cell line) | Exon mutated | Functional domain | Reference |
|---|---|---|---|---|---|
| Liver | 35.6 (26/73) | Primary | 9 and 20 | Helical and kinase | Lee et al (2004) |
| Total liver 36% (26/73) | |||||
| Breast | 33.3 (4/12) | Cell lines | 9 and 20 | Helical and kinase | Bachman et al (2004) |
| Breast | 21.4 (9/42) | Primary | 1, 9 and 20 | p85, helical and kinase | Bachman et al (2004) |
| Breast | 18.1 (13/72) | Primary | 9 and 20 | Helical and kinase | Levine et al (2005) |
| Breast | 40.0 (28/70) | Primary | 6, 7, 9 and 20 | C2, helical and kinase | Campbell et al (2004) |
| Breast | 20.7 (19/92) | Primary | 9 and 20 | Helical and kinase | Wu et al (2005a) |
| Breast | 8.3 (1/12) | Primary | 20 | kinase | Samuels et al (2004) |
| Breast | 33.3 (5/15) | Cell lines | 9 and 20 | Helical and kinase | Wu et al (2005a) |
| Breast | 26.9 (25/93) | Primary | 9 and 20 | Helical and kinase | Lee et al (2004) |
| Breast | 28.0 (14/50) | Cell lines | 1, 9 and 20 | p85, helical and kinase | Saal et al (2005) |
| Breast | 26.4 (77/292) | Primary | 1, 4, 7, 9, 13, 18, 20 | p85, C2, helical and kinase | Saal et al (2005) |
| Total breast 26% (195/750) | |||||
| Colon | 31.6 (74/234) | Primary | 1, 2, 4, 7, 9, 18 and 20 | P85, C2, helical and Kinase | Samuels et al (2004) |
| Colon | 13.6 (14/103) | Primary | 9 and 20 | Helical and kinase | Velho et al (2005) |
| Colon | 18.8 (6/32) | Primary | 9 and 20 | Helical and kinase | Campbell et al (2004) |
| Total colon 25% (94/369) | |||||
| Ovarian | 12.1 (24/198) | Primary | 9 and 20 | Helical and kinase | Levine et al (2005) |
| Ovarian | 6.0 (11/182) | Primary | 9 and 20 | Helical and kinase | Campbell et al (2004) |
| Total ovarian 9% (35/380) | |||||
| Gastric | 25.0 (3/12) | Primary | 18 and 20 | Kinase | Samuels et al (2004) |
| Gastric | 10.6 (5/47) | Primary | 9 and 20 | Helical and kinase | Velho et al (2005) |
| Gastric | 6.5 (12/185) | Primary | 9 and 20 | Helical and kinase | Lee et al (2004) |
| Gastric | 4.3 (4/94) | Primary | 9 and 20 | Helical and kinase | Li et al (2005) |
| Total gastric 7% (24/338) | |||||
| Brain | 26.7 (4/15) | Primary | 4, 5 and 13 | C2 and helical | Samuels et al (2004) |
| Brain | 4.6 (13/285) | Primary | 9 and 20 | Helical and kinase | Broderick et al (2004) |
| Total brain 6% (17/300) | |||||
| Lung | 1.3 (3/229) | Primary | 9 and 20 | Helical and kinase | Lee et al (2004) |
| Lung | 4.2 (1/24) | Primary | 9 | Helical | Samuels et al (2004) |
| Total lung 2% (4/253) | |||||
| Leukaemia | 1.1 (1/88) | Primary | 9 | Helical | Lee et al (2004) |
| Total leukaemia 1% (1/88) | |||||
| Total cancers reported 15% (382/2551) | |||||
The majority of PIK3CA documented mutations being somatic missense mutations, this table does not include other genetic changes (i.e. gene amplifications, deletions, insertions, etc.).
Another recent study (Lee et al, 2004) demonstrated a very high rate (36%) of PIK3CA somatic mutations in liver cancer. In the same study, these authors also analysed tissues from breast, gastric and lung cancers using a relatively high sample size and found mutation rates similar to other studies (Table 1). Interestingly, the authors also found one PIK3CA mutation out of 88 acute leukaemias (mutation rate=1.1%) that were analysed in this study, suggesting that PIK3CA mutations are not limited to solid tumours of epithelial origin. An analysis of PIK3CA somatic mutations and amplifications in thyroid cancers (Wu et al, 2005b) did not reveal any PIK3CA mutations; however, this group did find PIK3CA gene amplification in 12% of thyroid adenomas, 5% of papillary thyroid cancers, 24% of follicular thyroid cancers and 71% of thyroid cancer cell lines. More recently, there has been a report of somatic mutations in genes (i.e. PDK1, AKT2 and PAK4) downstream of the PI3K signalling pathway (Parsons et al, 2005).
Although the frequency of mutations and the discovery of hotspot heterozygous mutations strongly argue for the importance of PIK3CA in the carcinogenic process, functional analysis of these mutations has also been performed to confirm this supposition. Overexpression of common hotspot PIK3CA mutations, as well as gene deletion experiments using somatic cell knockouts, has demonstrated that these mutations are in fact oncogenic (Ikenoue et al, 2005; Kang et al, 2005; Samuels et al, 2005). Kang et al (2005) overexpressed cDNAs containing the common PIK3CA mutations, E542K, E545K, and H1047R, in chicken embryo fibroblasts. Their study demonstrated that overexpression of these mutant PIK3CA proteins led to cellular transformation with concomitant phosphorylation of proteins in the AKT pathway. Through the use of somatic cell knockouts, Samuels et al (2005) reported that mutation of the PIK3CA kinase domain in the HCT116 colon cancer cell line, and mutation of the helical domain in the DLD1 colon cancer cell line, resulted in increased activity of the PIK3CA enzyme as manifested by increased cell signalling, cell growth and invasion. Another functional study examining the E542K, E545K and H1047R hotspots was reported by Ikenoue et al (2005). These authors found that an increase in PIK3CA kinase activity and cellular transformation occurred when the above-mentioned mutant PIK3CA sequences were introduced into mouse NIH 3T3 cells.
By combining the copious amount of sequencing data over the past year, we find that the PIK3CA gene is mutated on average in 15% of human cancers, although there is obviously great variability in the tissue type, that is, colon vs breast vs lung (Table 1). In most tissue types, mutations predominantly cluster within the three aforementioned hotspots: E542K, E545K and H1047R (Figure 1C). It is now evident that cancers of the liver, colon and breast harbour the most PIK3CA mutations with average mutational frequencies (across the reported studies) of 36, 26 and 25%, respectively (Table 1). The mutational studies that are summarised in Table 1 do reveal some conflicting results, however, and as previously mentioned these are likely due to a number of factors including geographical variation/influence, sample source preservation and methods used for DNA isolation. However, despite these discrepancies, the high frequency of PIK3CA mutation and the discovery of hotspot mutations have important clinical implications for diagnosis, prognosis and therapy. For example, using PCR and sequencing of hotspot mutations, increased diagnostic sensitivity of cancer may be possible in situations of histologic ambiguity. As a case in point, detection of disease positive nodes in breast cancer may benefit from this type of molecular diagnostic test. The detection and prognostic significance of micrometastatic nodal disease in breast cancer has yielded conflicting and controversial results, and so far, no definitive data have been presented (Sakorafas et al, 2004; Colleoni et al, 2005; Kuijt et al, 2005). This may be due in part to the lack of specificity used in these studies to detect cancerous cells within normal appearing lymph nodes. One could envision that if a woman's primary breast cancer harboured a PIK3CA mutation, then that same mutation could be screened for in her axillary lymph nodes that were otherwise histologically normal, using recently developed technologies that allow for the detection of minute amounts of mutant DNA molecules (Dressman et al, 2003; Diehl et al, 2005). From a prognostic standpoint, long-term prospective, blinded randomised trials could be performed to determine if the presence or absence of PIK3CA mutations have any correlation with clinical outcome in various cancer types. This would then allow for the clinician to predict with a fair amount of certainty whether or not cancers harbouring these mutations would be more or less aggressive and could therefore influence decisions regarding additional systemic therapies. Finally, targeted therapies such as Imatinib mesylate (anti-BCR/ABL and cKIT), Gefitinib and Erlotinib (anti-EGFR) that appear to impart a high degree of specificity for translocated/mutated oncogenes give hope that therapies targeted specifically against mutant PIK3CA can be developed (Druker et al, 2001; Lynch et al, 2004; Paez et al, 2004). Given the high degree of PIK3CA mutations in human cancers, this could have a tremendous impact on eliminating the morbidity and mortality of malignant diseases.
Acknowledgments
We thank Abde Abukhdeir for assistance and thoughtful discussions. This work was supported by The Flight Attendant's Medical Research Institute (FAMRI), The American Cancer Society (#IRG-58-005-41), NIH Breast SPORE Grant P50 CA88843, the Maryland Cigarette Restitution Fund, The Entertainment Industry Foundation, The Department of Defense Breast Cancer Research Program (DAMD17-03-1-0241), and the Avon Foundation. BHP is an Avon Scholar for Breast Cancer Research and also receives generous support from The V Foundation for Cancer Research.
References
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters AB, Velculescu VE, Park BH (2004) The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 3: 772–775 [DOI] [PubMed] [Google Scholar]
- Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H (2004) Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res 64: 5048–5050 [DOI] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DYH, Montgomery KG, Ciavarella ML, Hooi CSF, Cristiano BE, Pearson RB, Phillips WA (2004) Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 64: 7678–7681 [DOI] [PubMed] [Google Scholar]
- Cantley LC (2002) The phosphoinositide 3-kinase pathway. Science 296: 1655–1657 [DOI] [PubMed] [Google Scholar]
- Cantley LC, Auger KR, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S (1991) Oncogenes and signal transduction. Cell 64: 281–302 [DOI] [PubMed] [Google Scholar]
- Colleoni M, Rotmensz N, Peruzzotti G, Maisonneuve P, Mazzarol G, Pruneri G, Luini A, Intra M, Veronesi P, Galimberti V, Torrisi R, Cardillo A, Goldhirsch A, Viale G (2005) Size of breast cancer metastases in axillary lymph nodes: clinical relevance of minimal lymph node involvement. J Clin Oncol 23: 1379–1389 [DOI] [PubMed] [Google Scholar]
- Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz Jr LA, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B (2005) Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 102: 16368–16373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B (2003) Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic varitatons. Proc Natl Acad Sci USA 100: 8817–8822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL (2001) Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 344: 1031–1037 [DOI] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC (1998) Phosphoinositide kinases. Annu Rev Biochem 67: 481–507 [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB (2005) Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov 4: 988–1004 [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, Ohta M, Jazag A, Guleng B, Tateishi K, Asaoka Y, Matsumura M, Kawabe T, Omata M (2005) Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res 65: 4562–4567 [DOI] [PubMed] [Google Scholar]
- Kang S, Bader AG, Vogt PK (2005) Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA 102: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD (2001) Cellular function of phosphoinositide 3-kinases: implications for development, immunity, homeostasis, and cancer. Annu Rev Cell Dev Biol 17: 615–675 [DOI] [PubMed] [Google Scholar]
- Kuijt GP, Voogd AC, van de Poll-Franse LV, Scheijmans LJ, van Beek MW, Roumen RM (2005) The prognostic significance of axillary lymph-node micrometastases in breast cancer patients. Eur J Surg Oncol 31: 500–505 [DOI] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH (2004) PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 24: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, Boyd J (2005) Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res 11: 2875–2878 [DOI] [PubMed] [Google Scholar]
- Li V, Wong C, Chan T, Chan A, Zhao W, Chu KM, So S, Chen X, Yuen S, Leung S (2005) Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer 5: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, Liu JM, Yang DM, Yang WK, Shen CY (2000) PIK3CA as an oncogene in cervical cancer. Oncogene 19: 2739–2744 [DOI] [PubMed] [Google Scholar]
- Migozuchi M, Nutt CL, Mohapatra G, Louis DN (2004) Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol 14: 372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M (2004) EGFR mutations in lung cancer: corr-elation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JKV, Markowitz S, Kinzler KW, Vogelstein B, Lengauer C, Velculescu VE (2005) Colorectal cancer mutations in a signalling pathway. Nature 436: 792. [DOI] [PubMed] [Google Scholar]
- Pedrero JMG, Carracedo DG, Pinto CM, Zapatero AH, Rodrigo JP, Nieto CS, Gonzales MV (2005) Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer 114: 242–248 [DOI] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R (2005) PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 65: 2554–2559 [DOI] [PubMed] [Google Scholar]
- Sakorafas GH, Geraghty J, Pavlakis G (2004) The clinical significance of axillary lymph node micrometastases in breast cancer. Eur J Surg Oncol 30: 807–816 [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304: 554. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz J, Schmidt-Kittler O, Cummins JM, DeLong L, Cheong I, Rago C, Huso DL, Lengauer C (2005) Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7: 561–573 [DOI] [PubMed] [Google Scholar]
- Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW (1999) PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet 21: 99–102 [DOI] [PubMed] [Google Scholar]
- Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz J, Duval A, Carneiro F, Machado JC (2005) The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer 41: 1649–1654 [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL (2002) The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer 2: 489–501 [DOI] [PubMed] [Google Scholar]
- Volinia S, Hiles I, Ormondroyd E, Nizetic D, Antonacci R, Rocchi M, Waterfield MO (1994) Molecular cloning, cDNA sequence, and chromosomal localization of the human phosphatidylinositol 3-kinase p110[alpha] (PIK3CA) Gene. Genomics 24: 472–477 [DOI] [PubMed] [Google Scholar]
- Wu G, Xing M, Mabbo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, Trink B, Sidransky D (2005a) Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res 7: R609–R616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Mambo E, Guo Z, Hu S, Huang X, Gollin SM, Trink B, Ladenson PW, Sidransky D, Xing M (2005b) Uncommon mutation, but common amplifications, of the PIK3CA gene in thyroid tumors. J Clin Endocrinol Metab 90: 4688–4693 [DOI] [PubMed] [Google Scholar]
- Wymann MP, Marone R (2005) Phosphoinositide 3-kinase in disease: timing, location, and scaffolding. Curr Opin Cell Biol 17: 141–149 [DOI] [PubMed] [Google Scholar]