Abstract
Changes in DNA methylation during tobacco pollen development have been studied by confocal fluorescence microscopy using a monoclonal anti-5-methylcytosine (anti-m5C) antibody and a polyclonal anti-histone H1 (anti-histone) antibody as an internal standard. The specificity of the anti-m5C antibody was demonstrated by a titration series against both single-stranded DNA and double-stranded DNA substrates in either the methylated or unmethylated forms. The antibody was found to show similar kinetics against both double- and single-stranded DNA, and the fluorescence was proportional to the amount of DNA used. No signal was observed with unmethylated substrates. The extent of methylation of the two pollen nuclei remained approximately constant after the mitotic division that gave rise to the vegetative and generative nuclei. However, during the subsequent development of the pollen, the staining of the generative nucleus decreased until it reached a normalized value of  of that of the vegetative nucleus. The use of a confocal microscope makes these data independent of possible focusing artefacts. The anti-histone antibody was used as a control to show that, while the antibody staining directed against 5-methylcytosine changed dramatically during pollen maturation, the histone signal did not. We observed the existence of structural dimorphism amongst tobacco pollen grains, the majority having three pollen apertures and the rest with four. However, the methylation changes observed occurred to the same extent in both subclasses.
of that of the vegetative nucleus. The use of a confocal microscope makes these data independent of possible focusing artefacts. The anti-histone antibody was used as a control to show that, while the antibody staining directed against 5-methylcytosine changed dramatically during pollen maturation, the histone signal did not. We observed the existence of structural dimorphism amongst tobacco pollen grains, the majority having three pollen apertures and the rest with four. However, the methylation changes observed occurred to the same extent in both subclasses.
Keywords: fluorescence microscopy, Nicotiana tabacum
Tobacco pollen undergoes a series of well defined stages during its development. Four amphihaploid sporocytes are produced by means of a meiotic division from an amphidiploid pollen mother cell. The cells then each form a highly resilient cell wall composed primarily of callose and cellulose. The nucleus then migrates to one of the poles of the pollen grain, where it undergoes a mitotic division to give two amphihaploid daughter cells. This division is not an equal one, and the cell closest to the pollen pole contains minimal cytoplasm and is known as the generative cell because it is destined to divide and form the gametes (1). The larger cell is called the vegetative cell, and its nucleus is responsible for controlling the development of the pollen tube. The generative cell rapidly detaches from the pole and floats freely within the cytoplasm of the vegetative cell (2). The two cells are separated only by plasma membranes (2). When mature pollen germinates, it produces a pollen tube that grows chemotropically toward the ovaries (3–5). The vegetative nucleus of tobacco always enters the pollen tube first and is then followed by the generative nucleus. Shortly after this, the vegetative nucleus disintegrates. The generative cell then undergoes a second mitotic division in the pollen tube to give two amphihaploid sperm cells that can fertilize the egg and the polar nuclei in the ovary (6), resulting in the embryo and endosperm, respectively.
Previous studies, using genomic sequencing (7), have indicated the existence of methylation differences between the vegetative and generative nuclei of mature tobacco pollen. However, these studies were restricted to a short region of the genome, and the significance and origins of these differences remained elusive. The major difficulty in studying methylation differences by genomic sequencing is that the starting material originates from heterogeneous cellular populations. It is far from trivial to obtain pure samples of each of the two nuclei from ungerminated pollen grains and then to chemically identify any nucleus-specific differences between the two. Genomic sequencing is also of limited value when studying genome-wide changes. Therefore we decided to adopt a simpler, histochemical, approach, which will be discussed in this paper.
MATERIALS AND METHODS
Pollen Collection.
Pollen was collected, from Nicotiana tabacum var. SR1 plants, from the first appearance of flower buds to the opening of the anthers and subsequent dehiscence of the pollen. The pollen was always harvested between 2 and 3 hr after sunrise. (In our experience, pollen could be isolated only at the mitotic stage of development, early in the morning. When it was collected later in the day we never observed mitosis.)
Dot-Blot Analysis of Antibody Specificity.
Four 21-bp-long oligonucleotides were synthesized as a specificity control for the antibody to 5-methylcytosine (m5C). Each oligonucleotide contained five cytosines, and their compositions were as follows: oligo 1, 5′-AGCTCAGACTTTGAGGCCTAA-3′; oligo 2, 5′-TTAGGCCTCAAAGTCTGAGCT-3′; oligo 3, as oligo 1 except that all of the Cs were replaced with m5C; and oligo 4, as oligo 2 except that all of the Cs were replaced with m5C. Oligonucleotides 1 and 2 were annealed by heating an equimolar solution to 95°C for 2 min in 100 mM NaCl and allowing the solution to cool to room temperature, on the bench, in a 100-ml beaker full of hot water. The annealed oligonucleotides were then precipitated with ethanol and used as the double-stranded unmethylated control. The same procedure was used to anneal oligonucleotides 3 and 4 to give the double-stranded methylated control. Oligonucleotides 1 and 3 were used for the single-stranded unmethylated and methylated controls, respectively.
The test oligonucleotides were spotted onto a DEAE membrane (Schleicher & Schuell, NA 85) in three different amounts (5 μg, 2.5 μg, and 1 μg). They were then incubated with a 1:1000 dilution of the monoclonal anti-m5C antibody for 16 hr at 4°C. The membrane was washed in DEAE wash buffer (50 mM NaCl/10 mM Tris⋅HCl/1 mM EDTA, pH 7.5) + 1% Triton X-100 for 4 hr at room temperature (22°C) and then incubated overnight with a fluorescein isothiocyanate (FITC)-linked goat anti-mouse polyclonal IgG (1:1000 dilution) secondary antibody (Sigma). The signals were detected by using a Storm PhosphorImager (Molecular Dynamics). After the detection procedure, the membrane was stained for 30 sec in DEAE wash buffer + 500 μg⋅ml−1 ethidium bromide and then washed for half an hour in DEAE wash buffer to destain it. The DNA was then visualized on a UV-transilluminator.
Permeabilization of Tobacco Pollen.
Tobacco pollen grains were rendered permeable to the antibodies by incubating them for 2 hr at 30°C in 200 mg⋅ml−1 cellulase (Fluka) in 0.6 M mannitol/25 mM Mes, pH 4.8. This was followed by two washes in phosphate-buffered saline (PBS). The sample was then heated in 10% dimethyl sulfoxide/10 mM EGTA at 96°C for 10 min to heat-inactivate any residual cellulase and to denature the genomic DNA.
Antibody Staining.
The permeabilized pollen grains were coincubated, in 1× PBS + 1% Triton X-100, with the anti-m5C (1:50 dilution) and anti-histone (1:10 dilution) antibodies for 20 hr at 4°C. The grains were then washed by two centrifugations (2,000 rpm, 10 min, in a microcentrifuge) and resuspension (1 ml of 1× PBS + 1% Triton X-100) steps followed by 6 hr at 4°C in 1 ml of 1× PBS + 1% Triton X-100. The primary antibodies were detected by incubation with FITC-labeled anti-mouse IgG (1:100 dilution) and Texas red isothiocyanate (TRITC)-labeled anti-rabbit IgG (1:100 dilution) (both from Sigma) for 20 hr at 4°C followed by wash steps as described above.
Confocal Microscopy.
The antibody signals were quantified by taking 40 optical sections through each nucleus, using a confocal microscope (Leika) connected to a Silicon Graphics workstation. The resulting images were used to calculate the volume and total luminance under FITC emission. The background fluorescence from the cytoplasmic region around each nucleus was also sampled and subtracted from the data. Once the luminance had been normalized to take nuclear volume and background fluorescence into account, the process was repeated for the TRITC emission.
RESULTS
DNA methylation was analyzed by using monoclonal antibodies directed against m5C (8), visualized with a FITC-labeled anti-mouse IgG, and polyclonal antibodies directed against mammalian histone H1 proteins, visualized with a TRITC-labeled anti-rabbit IgG.
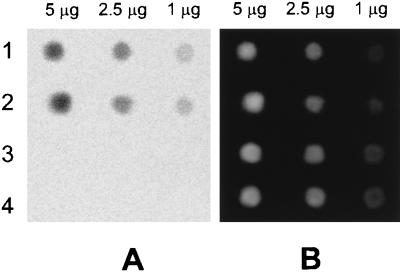
The specificity of the anti-m5C antibody was tested by using a dot-blot of methylated and unmethylated oligonucleotides. Single-stranded methylated, double-stranded methylated, single-stranded unmethylated, and double-stranded unmethylated oligonucleotides were spotted onto a DEAE membrane, and the ability of the monoclonal antibody to detect the different DNA samples was determined as described in Materials and Methods. As a control the membrane was stained with ethidium bromide, after quantification, to ensure even loading of the DNA. Fig. 1A shows that signals were obtained only in the tracks containing the methylated substrates, indicating that the antibody does not cross-react with unmethylated cytosine in either single-stranded or double-stranded DNA. The intensities of each spot were obtained from three experimental repetitions using the PhosphorImager (Table 1), and a linear relationship (r > 0.99) was found between mass of DNA and fluorescence (Table 1). Fig. 1B shows that the amount of each DNA type bound to the membrane was approximately equal. A more detailed examination of the specificity of our antibody may be found in ref. 8.
Figure 1.
Immunofluorescence of DNA standards. (A) PhosphorImager scan of a dot-blot of DNA detected with the anti-m5C antibody. Lane 1 is methylated single-stranded DNA, 2 is methylated double-stranded DNA, 3 is unmethylated single-stranded DNA, and 4 is unmethylated double-stranded DNA. (B) Same blot stained with ethidium bromide.
The development of the pollen falls into approximately five stages (1): (i) primary postmeiotic pollen grain with a single amphihaploid nucleus; (ii) mitotic division of the primary pollen cell to give the vegetative cell and the generative cell; (iii) reformation of the generative nuclear envelope, followed by; (iv) reformation of the vegetative nuclear envelope; and (v) pollen dehydration and dehiscence. The development of the pollen grains was approximately synchronized so that those samples collected during the mitotic division showed all of the stages of mitosis from mononuclear pollen, through metaphase chromosomal condensation, up to early binuclear pollen.
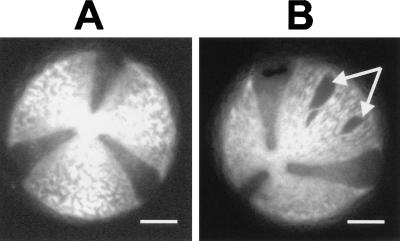
The pollen wall is an extremely resistant structure that is not easily permeabilized by chemical treatments (1). In this experiment we rendered the pollen grains permeable to the antibodies by the procedure outlined in Materials and Methods. The effect of the permeabilization regime was visualized by viewing the unstained pollen grains under a fluorescence microscope using the 4′,6-diamidino-2-phenylindole (DAPI) excitation filter to induce autofluorescence of the pollen wall. This showed that the permeabilization treatment resulted in some ruptures in the pollen wall (Fig. 2B, arrows) but did not cause the physical lysis of the pollen grain.
Figure 2.
Permeabilization of tobacco pollen grains. (A) Autofluorescence micrograph of tobacco pollen prior to permeabilization. (B) Pollen after permeabilization. The arrows indicate holes within the exine coat of the pollen. The dark sectors between the bright plates are not holes but are simply less fluorescent. (Bars = 10 μm.)
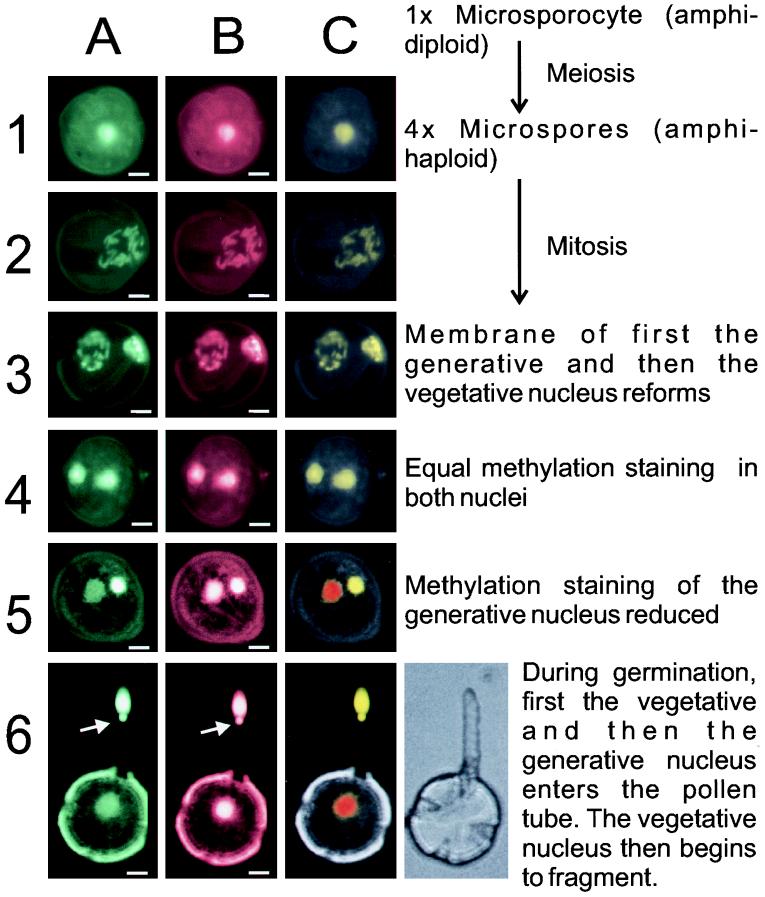
The permeabilized pollen grains were coincubated with the anti-m5C and anti-histone antibodies, and these primary antibodies were detected by coincubation with FITC-anti-mouse and TRITC-anti-rabbit IgGs (Sigma), following the procedure outlined in Materials and Methods. Incubation with the preimmune serum for the anti-histone antibodies, in place of the primary antibodies, did not give any specific staining (data not shown). Fig. 3 shows the results from the staining of cells at each stage of pollen development. It is clear from Fig. 2A that all of the metaphase chromosomes show similar immunostaining levels, indicating that there was no major methylation difference between the newly replicated chromosomes at this stage. Row 3 in Fig. 3 shows the rapid reformation of the generative nucleus. This nucleus may be positively identified because at this stage it is known that the generative cell, in all plants, is located in contact with the pollen wall at one pole of the pollen grain (9). Once the two nuclei have reformed (row 4) the generative cell detaches from the pollen wall and enters the cytoplasm of the vegetative cell. At this stage both nuclei show approximately even immunofluorescence (panel 4A). Over the next 1–3 days of floral development the anti-m5C fluorescence from the generative nucleus decreased relative to that of the vegetative nucleus (panel 5A), whereas the fluorescence from the anti-histone antibody remained constant (panel 5B). The generative nucleus fluorescence decreased until it stabilized at about  that of the vegetative nucleus (Table 2). The positive identification of the two nuclei came from the observation that the intensely immunostained nucleus was the first to enter the pollen tube (Fig. 3, panel 6A). The arrow on row 6 indicates the beginning of the nuclear fragmentation of the vegetative nucleus upon pollen germination.
that of the vegetative nucleus (Table 2). The positive identification of the two nuclei came from the observation that the intensely immunostained nucleus was the first to enter the pollen tube (Fig. 3, panel 6A). The arrow on row 6 indicates the beginning of the nuclear fragmentation of the vegetative nucleus upon pollen germination.
Figure 3.
Nuclear methylation changes in pollen during different stages of its development. Column A shows the FITC fluorescence from the anti-m5C antibody. Column B shows the TRITC fluorescence from the anti-histone antibody. Column C shows a false-color image composed of the FITC image (green channel), the TRITC image (red channel), and pollen autofluorescence (blue channel). The nuclear color shows the ratio of antibody staining: the more green it is the more hypermethylated the nucleus, the more red it is the more hypomethylated the nucleus. (Bars = 10 μm.) The terms amphihaploid and amphidiploid refer to the fact that tobacco is a stable hybrid of two different species, Nicotiana tomentosiformis and Nicotiana sylvestris. Although this means that it is technically tetraploid, it behaves as if it were a diploid organism. Row 6 also shows a phase-contrast micrograph of a germinating pollen grain to indicate the position of the pollen tube (which is not fluorescent). The arrow in row 6 indicates the beginning of the vegetative nuclear fragmentation.
The fluorescence of the two nuclei was calculated, using a confocal microscope to scan through the pollen grains. The TRITC emission was used as a control for the accessibility of the antibodies to the two nuclei. Nuclear fluorescence could then be normalized by taking the ratio of the FITC to TRITC staining for each nucleus. These normalized data are presented in Table 2. The purpose of this control was to establish whether or not any differences observed were due to changes in chromatin packing, or other accessibility differences, that might render one nucleus or the other more or less susceptible to antibody binding. The histone control shows that, while there is a difference in the relative binding affinities of the histone antibody during pollen maturation, this difference is trivial compared with that observed for the methylcytosine signal (Table 2). This internal standard also allows the comparison of signal strength from one experiment to another to be more tightly controlled. Fig. 3 column C shows false-color images made by combining the signals from the FITC, TRITC, and autofluorescent images as the green, red, and blue channels respectively. In Fig. 3 panel 4C it is clear from the colors that the FITC to TRITC ratio is constant between the two nuclei, indicating constant methylation. In panels 5C and 6C the demethylation of the generative nucleus is clear because its staining becomes red-shifted (i.e., less green in the picture).
There are two distinct subclasses of pollen within SR1 tobacco. The majority of the pollen grains had three large epifluorescent sectors running from one pole to the other with the nonfluorescent pollen apertures in between them (Fig. 2A—fluorescent micrograph). The rest of the pollen was essentially identical except that it had four sectors rather than three (Fig. 3, row 6, phase-contrast micrograph). The changes in methylation observed during pollen development did not appear to be different between these two subclasses (data not shown). It should be stated that both classes could be readily observed in pollen isolated from a single flower of a single tobacco plant, indicating that this structural dimorphism is not predetermined during floral development. This pollen dimorphism shows striking similarities to that reported by Dajoz et al. (10), who observed pollen aperture polymorphism in Viola diversifolia. In Viola there are also two subtypes of pollen with either three or four potential pollen apertures. Both forms were able to germinate, also true for tobacco, but the four-aperture pollen germinated faster than the three- in vitro. Once germinated, the pollen tube from a three-aperture pollen grew faster (10). It is interesting to speculate whether a similar explanation might also be true for tobacco.
DISCUSSION
Specific methylation changes occur after mitosis between the generative and the vegetative nuclei in the developing pollen cell. The methylation levels of the two nuclei, immediately after they had reformed, were the same. During subsequent development, the methylation of the generative nucleus decreased until it reached a stable level of about  of that of the vegetative nucleus. The histone staining showed that these changes were not due to differences in antibody accessibility to the nucleus. The observed methylation change is particularly interesting when it is compared with the observation that the regeneration of haploid plants is possible from pollen grains in anther cultures taken either at the single-cell stage or immediately after mitosis (11), but it becomes increasingly difficult as the pollen matures (12). Androgenic reproduction from binuclear pollen is believed to occur only from the vegetative nucleus, as the generative nucleus degenerates during tobacco anther culture (13). It may be that early methylation changes in the generative nucleus render it unable to regenerate, whereas the less severe changes to the vegetative nucleus permit its regeneration over a longer time scale. These genome-wide methylation changes may also play a crucial role in the resetting of epigenetic phenomena often observed in primary transgenic plants after tissue culture.
of that of the vegetative nucleus. The histone staining showed that these changes were not due to differences in antibody accessibility to the nucleus. The observed methylation change is particularly interesting when it is compared with the observation that the regeneration of haploid plants is possible from pollen grains in anther cultures taken either at the single-cell stage or immediately after mitosis (11), but it becomes increasingly difficult as the pollen matures (12). Androgenic reproduction from binuclear pollen is believed to occur only from the vegetative nucleus, as the generative nucleus degenerates during tobacco anther culture (13). It may be that early methylation changes in the generative nucleus render it unable to regenerate, whereas the less severe changes to the vegetative nucleus permit its regeneration over a longer time scale. These genome-wide methylation changes may also play a crucial role in the resetting of epigenetic phenomena often observed in primary transgenic plants after tissue culture.
Genome-wide changes in DNA methylation patterns have been well studied during gamete production in animal cells. It is believed that premeiotic mammalian sperm cells are hypomethylated and that high levels of DNA methyltransferase (14) result in the de novo methylation of sperm DNA during meiosis to give hypermethylated sperm nuclei (15). This change is exactly the opposite direction to that observed by us in tobacco pollen. However, CpG islands in mammalian oocytes have been shown to be actively demethylated during development (16). For a general review on active demethylation see ref. 17. It would be interesting to examine tobacco oocytes and test changes in their methylation levels during floral development. However, such experiments have proved difficult to date because of the high activity of polyphenol oxidase in tobacco flowers, which makes fluorescent analysis more complicated (personal observation).
We conclude that pollen is an interesting model system for studying methylation-dependent developmental changes and possibly the mechanisms underlying the active demethylation of DNA in plants. These changes may have important consequences for the resetting of gene imprinting and for the control of methylation in transgenic tobacco plants.
Table 1.
Immunofluorescence of DNA standards
| DNA, μg | Methylated
|
Unmethylated
|
||
|---|---|---|---|---|
| ssDNA | dsDNA | ssDNA | dsDNA | |
| 5 | 7,852 ± 305 | 6,985 ± 255 | 135 ± 25 | 122 ± 15 |
| 2.5 | 3,751 ± 214 | 3,577 ± 189 | 112 ± 15 | 128 ± 3 |
| 1 | 1,349 ± 301 | 1,152 ± 298 | 121 ± 10 | 131 ± 7 |
ssDNA, single-stranded DNA; dsDNA, double-stranded DNA. Results are mean intensity ± SD.
Table 2.
Changes in nuclear methylation between the vegetative and generative pollen cells in developing tobacco
| Immature pollen
|
Mature pollen
|
||||||
|---|---|---|---|---|---|---|---|
| Pollen | Ratio of vegetative to generative nuclei
|
Pollen | Ratio of vegetative to generative nuclei
|
||||
| m5C F | Histone F | m5C/histone | m5C F | Histone F | m5C/histone | ||
| A | 1.50 | 1.26 | 1.19 | K | 5.48 | 0.92 | 5.96 |
| B | 1.42 | 1.52 | 0.93 | L | 5.54 | 1.06 | 5.23 |
| C | 1.32 | 1.36 | 0.97 | M | 5.19 | 0.97 | 5.35 |
| D | 1.27 | 1.26 | 1.01 | N | 5.32 | 1.10 | 4.84 |
| E | 1.26 | 1.28 | 0.98 | O | 4.79 | 0.93 | 5.15 |
| F | 1.36 | 1.51 | 0.90 | P | 5.42 | 1.09 | 4.97 |
| G | 1.25 | 1.22 | 1.02 | Q | 6.05 | 1.37 | 4.42 |
| H | 1.28 | 1.44 | 0.89 | R | 5.47 | 0.93 | 5.88 |
| I | 1.23 | 1.18 | 1.04 | S | 6.49 | 1.44 | 4.51 |
| J | 1.14 | 1.25 | 0.91 | T | 5.57 | 1.04 | 5.36 |
| Mean | 0.98 | 5.17 | |||||
| SD | 0.09 | 0.51 | |||||
“Immature pollen” shows the corrected relative fluorescence values (F) for the generative and vegetative nuclei in postmitotic pollen. “Mature pollen” shows the relative fluorescence values for the generative and vegetative nuclei in mature pollen immediately prior to another dehiscence.
Acknowledgments
We thank Dr. Beat Ludin and Dr. Ortrun Scheid for critical reading of this manuscript. We also thank Dr. Beat Ludin, Dr. Stefanie Käch, and Dr. Jörg Hagmann for help with the fluorescence and confocal microscopy.
ABBREVIATIONS
- m5C
5-methylcytosine
- FITC
fluorescein isothiocyanate
- TRITC
Texas red isothiocyanate
References
- 1.Bhojwani S S, Bhatnager S P. The Embryology of Angiosperms. Delhi, India: Vikas; 1975. [Google Scholar]
- 2.Noguchi T, Ueda K. Cell Struct Funct. 1990;15:379–384. doi: 10.1247/csf.15.379. [DOI] [PubMed] [Google Scholar]
- 3.Cai G, Moscatelli A, Cresti M. Trends Plant Sci. 1997;2:86–91. [Google Scholar]
- 4.Cecich R A. Forest Sci. 1997;43:140–146. [Google Scholar]
- 5.Cheung A Y. Sexual Plant Rep. 1996;9:330–336. [Google Scholar]
- 6.Liu B, Palevitz B A. Protoplasma. 1996;195:78–89. [Google Scholar]
- 7.Oakeley E J, Jost J P. Plant Mol Biol. 1996;31:927–930. doi: 10.1007/BF00019481. [DOI] [PubMed] [Google Scholar]
- 8.Podestà A, Castiglione M A, Avanzi S, Montagnoli G. Int J Biochem. 1993;25:929–933. doi: 10.1016/0020-711x(93)90249-e. [DOI] [PubMed] [Google Scholar]
- 9.Heslop-Harrison J. In: Plant Physiology. Steward F C, editor; Steward F C, editor. Vol. 6. New York: Academic; 1972. pp. 133–289. [Google Scholar]
- 10.Dajoz I, Tillbottraud I, Gouyon P H. Evolution. 1993;47:1080–1093. doi: 10.1111/j.1558-5646.1993.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 11.Raghavan V. Embryogenesis in Angiosperms: A Developmental and Experimental Study. New York: Cambridge Univ. Press; 1986. [Google Scholar]
- 12.Reynolds T L. Plant Mol Biol. 1997;33:1–10. doi: 10.1023/a:1005748614261. [DOI] [PubMed] [Google Scholar]
- 13.Sunderland N, Wicks F. J Exp Bot. 1973;22:213–226. [Google Scholar]
- 14.Singer-Sam J, Robinson M O, Bellvé A R, Simon M I, Riggs A D. Nucleic Acids Res. 1990;18:1255–1259. doi: 10.1093/nar/18.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer-Sam J, Riggs A D. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Salutz H P, editors; Jost J P, Salutz H P, editors. Basel: Birkhäuser; 1993. pp. 358–384. [Google Scholar]
- 16.Frank D, Keshet I, Shani M, Levine A, Razin A, Cedar H. Nature (London) 1991;351:239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- 17.Jost J P. In: Epigenetic Mechanisms of Gene Regulation. Russo V E A, Martienssen R A, Riggs A D, editors; Russo V E A, Martienssen R A, Riggs A D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 109–125. [Google Scholar]