Abstract
Using genome-wide expression profiling of a panel of 27 human mammary cell lines with different mechanisms of E-cadherin inactivation, we evaluated the relationship between E-cadherin status and gene expression levels. Expression profiles of cell lines with E-cadherin (CDH1) promoter methylation were significantly different from those with CDH1 expression or, surprisingly, those with CDH1 truncating mutations. Furthermore, we found no significant differentially expressed genes between cell lines with wild-type and mutated CDH1. The expression profile complied with the fibroblastic morphology of the cell lines with promoter methylation, suggestive of epithelial–mesenchymal transition (EMT). All other lines, also the cases with CDH1 mutations, had epithelial features. Three non-tumorigenic mammary cell lines derived from normal breast epithelium also showed CDH1 promoter methylation, a fibroblastic phenotype and expression profile. We suggest that CDH1 promoter methylation, but not mutational inactivation, is part of an entire programme, resulting in EMT and increased invasiveness in breast cancer. The molecular events that are part of this programme can be inferred from the differentially expressed genes and include genes from the TGFβ pathway, transcription factors involved in CDH1 regulation (i.e. ZFHX1B, SNAI2, but not SNAI1, TWIST), annexins, AP1/2 transcription factors and members of the actin and intermediate filament cytoskeleton organisation.
Keywords: E-cadherin, epithelial–mesenchymal transition, promoter methylation, mammary cell lines, TFGβ pathway
Decreased E-cadherin expression is associated with a more aggressive behaviour of breast cancer (Siitonen et al, 1996). The homotypic cellular adhesion molecule E-cadherin is a transmembrane glycoprotein important for the organisation of epithelial structure (Takeichi, 1995; Huber et al, 1996; Yagi and Takeichi, 2000). E-cadherin can form homophilic interactions with E-cadherin molecules on neighbouring cells in a Ca2+-dependent way and is the main component of adherens junctions. By recruitment of α- and β-catenin, the E-cadherin is anchored to the actin cytoskeleton.
Mutational inactivation of CDH1 has been found in 56% of lobular breast carcinomas (Berx et al, 1996) and 50% of diffuse gastric carcinomas (Becker et al, 1994). In the first tumour type, this is accompanied by loss of the wild-type allele (Berx et al, 1995). Complete loss of E-cadherin protein expression has been found in 84% of lobular breast carcinomas (De Leeuw et al, 1997). Loss of membranous E-cadherin expression results in a reduction of adhesion between epithelial tumour cells and explains the characteristic diffuse growth pattern observed in these tumours (Berx et al, 1998). CDH1 mutations were also identified in lobular carcinoma in situ, the putative precursor of invasive lobular carcinoma (Vos et al, 1997). Thus, in addition to its role as an invasion suppressor, E-cadherin also acts as a classical tumour suppressor gene in pre-invasive lobular breast carcinoma. Besides mutational inactivation of the E-cadherin gene, CDH1 may also be targeted by promoter hypermethylation (Graff et al, 1995; Grady et al, 2000; Tamura et al, 2000), thereby inhibiting CDH1 gene expression. Evidence is accumulating for a prominent role of epithelial-to-mesenchymal transition (EMT) in tumour progression (reviewed by Thiery, 2002). During early embryonic development, E-cadherin is a critical switch in EMT. Upon downregulation of E-cadherin, epithelial cells acquire a fibroblastic phenotype, dissociate from the epithelium and migrate. This process is essential for gastrulation, neural crest formation, kidney development and so on (reviewed by Thiery, 2003). Several proteins have been identified that downregulate E-cadherin expression including SNAI1/SNAIL (Batlle et al, 2000; Cano et al, 2000), ZFHX1B/SIP1 (Comijn et al, 2001), SNAI2/SLUG (Hajra et al, 2002; De Craene et al, 2005), TWIST1 (Yang et al, 2004) and DeltaEF1 (Eger et al, 2005). Altered expression of these transcription factors seems to be also associated with an altered overexpression of transcriptional repressors of E-cadherin in tumour cells (Batlle et al, 2000; Cano et al, 2000; Comijn et al, 2001; Hajra et al, 2002; Yang et al, 2004; Eger et al, 2005). CDH1 gene expression is upregulated by several factors, AML1, p300 and HNF3 (Liu et al, 2005). Also post-transcriptional regulation of E-cadherin has been observed and recently ADAM10 was identified as the cleaving protease (Maretzky et al, 2005).
Given the relevance of E-cadherin in tumour development and progression and the different mechanisms involved in its regulation, we set out to use a genome-wide expression analysis to identify genes or pathways in mammary epithelial cells that are either (in)directly affected by loss of E-cadherin function or are altogether associated with a (epi)genetic programme that determines the biological status of cells: epithelial or mesenchymal. For this purpose, we performed a gene expression profile study on 27 different breast mammary cell lines with a known CDH1 mutation, promoter hypermethylation and expression status. Remarkably, no significant differences in gene expression were identified between breast cancer cell lines with wild-type E-cadherin without promoter methylation and those harbouring CDH1 truncating mutations. The results showed a marked difference in expression profile between cell lines with CDH1 promoter methylation compared to those with CDH1 mutational inactivation, especially for genes involved in EMT and part of the TGFβ pathway.
MATERIALS AND METHODS
Cell lines
The 27 breast and four colon cancer cell lines used in this study are listed in Table 1. MPE600 and SK-BR-5 were provided by Dr F Waldman (California Pacific Medical Centre, San Francisco, CA, USA) and Dr E Stockert (Sloan-Kettering Institute for Cancer Research, New York, NY, USA), respectively. Dr SP Ethier donated SUM44PE (Ethier et al, 1993) and SUM185PE (Forozan et al, 1999). OCUB-F was obtained from the Riken Gene Bank. Other cell lines were obtained from the American Type Culture Collection. We genotyped all cell lines using the Powerplex 1.2 system (Promega, Leiden, The Netherlands) according to the manufacturer's instructions. All cell lines were grown in RPMI1640 medium supplemented with 5 mM glutamine and 10% heat-inactivated fetal calf serum at 37°C under 5% CO2 (culture media from GIBCO Invitrogen, Grand Island, NY, USA).
Table 1. Cell lines used in this study.
| Cell line(source) | Tissue | Tumour type | Genotype confirmation | CDH1 promoter methylation | Protein expression | CDH1 mutation(9) | QPCRa | Flow index MESF |
|---|---|---|---|---|---|---|---|---|
| BT474(1) | Breast | IDC | + | − | + | 192.7 | ||
| BT483(1) | Breast | IDC | NIA | − | 226.6 | |||
| BT549(1) | Breast | PIDC | NIA | ± | − | 3.2 | ||
| CAMA1(1) | Breast | C | +(7) | − | + | + | 23.9 | 17 |
| Du4475(1) | Breast | IDC | + | − | + | − | 24.5 | |
| HBL100(1) | Breast | Normal(8) | NIA(8) | + | − | 0.0 | ||
| Hs578T(1) | Breast | CS | + | ± | − | − | 0.0 | 0 |
| MCF10A(1) | Breast | Normal | ± | 1.0 | ||||
| MCF10F(1) | Breast | Normal | + | ± | − | 0.3 | ||
| MCF12A(1) | Breast | Normal | + | ± | 10.7 | |||
| MCF7(1) | Breast | IDC | + | − | + | − | 201.8 | 47 |
| MDA-MB-134 VI(1) | Breast | IDC | + | − | − | + | 3.8 | |
| MDA-MB-175 VII(1) | Breast | IDC | + | − | + | − | 281.1 | |
| MDA-MB-231(1) | Breast | IDC | + | ± | ± | − | 30.3 | 18 |
| MDA-MB-231*(6) | Breast | IDC | (6) | ± | − | 0.3 | ||
| MDA-MB-330(1) | Breast | IDC | + | − | + | − | 99.1 | |
| MDA-MB-361(1) | Breast | AC | + | − | + | − | 243.6 | |
| MDA-MB-435s(1) | Breast | IDC | + | + | − | 0.0 | 0 | |
| MDA-MB-453(1) | Breast | AC | + | − | − | 6.8 | ||
| MPE600(2) | Breast | C | NIA | − | + | + | 96.2 | 108 |
| OCUB-F(3) | Breast | C | NIA | − | − | + | 75.7 | |
| SK-BR-3(1) | Breast | IDC | + | Del | − | + | 0.1 | 0 |
| SK-BR-5(4) | Breast | C | NIA | − | − | + | 20.1 | 0 |
| SUM185PE(5) | Breast | NIA | − | + | 131.2 | |||
| SUM44PE(5) | Breast | ILC | NIA | − | − | + | 5.4 | |
| T47D(1) | Breast | IDC | + | − | + | − | 97.2 | 121 |
| ZR75-1(1) | Breast | IDC | + | − | + | − | 284.8 | 103 |
| LS180(1) | Colorectal | AC | − | |||||
| LS411N(1) | Colorectal | C | − | |||||
| SW480(1) | Colorectal | AC | − | |||||
| SW837(1) | Rectal | AC | − |
(1)American Tissue Type Collection (ATCC); (2)Dr F Waldman; (3)Riken Gene Bank; (4)Dr E Stockert; (5)Dr S Ethier; (6)a clone spontaneously derived from MDA-MD-231 lacking allele 8 of TPOX; (7)lost the 9.3 allele of THO1; (8)HBL100 was originally isolated from a healthy female; however, it turned out to have a Y chromosome; (9)after van de Wetering et al. (2001).
Normalised real-time PCR SQ values for CDH1 expression; IDC=invasive ductal carcinoma; PIDC=papillary invasive ductal carcinoma; C=carcinoma; CS=carcinosarcoma; ILC=invasive lobular carcinoma; AC=adenocarcinoma; NIA=no information available; (blank)=not determined; Del=promoter deletion.
E-cadherin protein expression
Cells were grown until 80% confluence and lysed by adding 1 ml hot lysis solution (1% SDS, 10 mM Tris pH 7.4, 10 mM EDTA, supplemented with complete protease inhibitor (Boehringer Mannheim)). Protein concentration was determined by the Biorad DC Protein Assay (Biorad, Hercules, CA, USA). Western blots of electrophoretically separated proteins (Winter et al, 2003) from whole-cell lysates were probed with HECD-1 (Zymed Laboratories, San Francisco, CA, USA) antibodies to detect E-cadherin (diluted 1 : 1000). Control blots were probed with anti-β-actin (1 : 500) (Sigma, St Louis, MO, USA). Anti-mouse IgG peroxidase conjugates (Transduction Laboratories, Lexington, KY, USA) were used as secondary antibodies, and the blots were developed for 1 min using the enhanced chemiluminescent detection system (Amersham Int., Little Chalfont, UK). Exposure time was 1 min. Membranous E-cadherin expression was analysed using HECD-1 by flow cytometry of viable cells as described previously (Corver et al, 1994). In brief, 1 μg of HECD1 was used to label the cells. For each sample, measurements from 20 000 single cells were collected using a standard FACSCalibur™ flow cytometer (BD Biosciences, San Diego, CA, USA). Data were analysed using WinList 5.0 software (Verity Software House Inc., Topsham, ME, USA).
Methylation-specific PCR
CDH1 promoter methylation was determined as described by Herman et al (1996). Modification of DNA before PCR was carried out with the EZ Methylation Kit™ (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions. Methylation-specific fragments were sequenced to determine methylation of CpGs between the primer binding sites. As a control for efficient modification, we used primers for fragments of CDH3 and TERF2, flanking CDH1, which do not contain CpGs and exclusively amplified modified DNA (primer sequences and PCR conditions are available upon request).
RNA isolation
When cell cultures reached 70–80% confluence, RNA was isolated using TRIzol (Invitrogen Life Technologies Breda, The Netherlands) and purified using Qiagen RNeasy mini kit columns (Qiagen Sciences, Germantown, MD, USA). Samples were DNase treated using the Qiagen RNase-free DNase kit (Qiagen). The isolation and purification were carried out according to the manufacturer's instructions.
cDNA microarrays
cDNA clones were amplified to generate PCR products for the cDNA microarray from a sequence verified clone collection (Research Genetics, Invitrogen, Huntsville, AL, USA). Apart from these clones, cDNAs related to adhesion, cytoskeleton and carcinogenesis were selected. These additional clones were obtained from the Deutsches Krebsforschungszentrum (Deutsches Ressourcenzentrum für Genomforschung, Berlin, Germany) or created by performing an RT–PCR (reverse transcriptase) reaction on RNA from mammary cell lines. Subsequently, these PCR products were cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA). All plasmid inserts were PCR amplified using plasmid-specific primers.
Purified PCR products, in 3 × SSC buffer, were spotted in duplicate on MicroMax SuperChip™ I slides (Perkin-Elmer Life Science, Boston, MA, USA) at the Leiden Genome Technology Center (http://www.lgtc.nl/) using the OmniGrid 100 robot (GeneMachines, San Carlos, CA, USA). In total, 9216 PCR products, including ‘landing marks’, consisting of biotin- and fluorescein-labelled PCR products to facilitate spot identification, were spotted in duplicate on each slide.
cDNA labelling and hybridisation
For cDNA labelling and signal amplification, we used the tyramide signal amplification (TSA) kit (Perkin-Elmer Life Science, Boston, MA, USA) according to the manufacturer's instructions with minor modification. A 1 μg portion of RNA was used to generate biotin- or fluorescein-labelled cDNA. In general, cell line cDNA was labelled with fluorescein (Cy3) whereas reference cDNA was labelled with biotin (Cy5). This reference RNA consists of RNA isolated from several human tumour cell lines (HL-60, K562, NCI-H226, COLO205, SNB-19, LOX-IMVI, OVCAR-3, OVCAR-4, CAK-IPC-3, MCF7, Hs578 T, MCF10F, MCF12A, OUMS27 and SW1353), analogous to the panel described by Ross et al (2000). Labelled cDNA was purified on YM-30 Microcon columns (Millipore Corporation, Bedford, MA, USA), dissolved in the hybridisation buffer and applied to cDNA arrays. Slides were hybridised overnight at 65°C in Corning Hybridization Chambers (Corning, NY, USA). The TSA reaction and washing of the slides was carried out using ThermoShandon Coverplates (http://www.thermo.com/).
Data analysis
Microarrays were scanned using the GeneTAC LSIV laser scanner (Genomic Solutions, Ann Arbor, MI, USA). Each slide was scanned at two different gain settings, namely, a low gain to avoid detector saturation by high-amplitude signals and high gain to improve signal detection from weakly expressed genes. This approach provides a larger dynamic range of signal detection. Fluorescent spots were detected and quantified using GenePix Pro 3.0 software (Axon Instruments Inc., Union City, CA, USA). An MS-Excel macro was created for automated spot selection. This enables exclusion of saturated spots and spots with a signal below the threshold. The remaining spots were corrected for local background noise. The intensity for both dyes of each spot was normalised to the median of all spots on the array. For each spot, the ratio of the sample to the reference was calculated. For spots measured both at high gain and low gain, ratios were averaged. Finally, ratios were log 10-transformed.
Unsupervised cluster analysis (using the options ‘Complete linkage’ and ‘Correlation’) was made on log 10-transformed ratios with Cluster 2.12 (Eisen et al, 1998) and visualised using Treeview 1.6 (MB Eisen, http://rana.lbl.gov/index.htm). For the identification of differentially expressed genes, R version 1.9.0 (http://www.R-project.org/) (R Development Core Team, 2004) using the Limma (linear models for microarray data) package of Bioconductor (http://www.bioconductor.org/) was applied. Limma is a moderated T-statistic that detects differentially expressed genes between groups given the natural variance within these groups, corrected for false discovery rate due to multiple testing (Wettenhall and Smyth, 2004). Cluster analysis was made for genes yielding a signal in at least 90% of the samples. Independent hybridisations including dye-swaps of the same sample generally clustered together; therefore, we averaged experimental data for every cell line. Furthermore, we averaged ratios of duplicate spots on the array to improve significance. Differentially expressed genes were determined for cDNAs that gave a signal in at least 80% of the samples.
To test reproducibility of hybridisation and data acquisition, duplicate or triplicate microarray hybridisations were performed for 13 of the 31 cell lines, and seven dye-swap experiments. As the results of the duplicate or triplicate array hybridisations of the same cell line are highly similar, these are averaged as well as duplicate spots.
Real-time PCR
Quantitative real-time PCR (qPCR) was performed to verify results found by the cDNA microarray analysis and examine the expression profile of candidate genes (Table 4). Primers were developed with the Beacon Designer 3 software (Premier Biosoft International, Palo Alto, CA, USA). Primer sequences and PCR conditions are available upon request. Reactions were performed using qPCR Corekits for SybrGreen or TAQman probes (Eurogentec, Seraing, Belgium). Cycle threshold (Ct) and starting quantities (SQ) were determined using the Biorad iCycler software (Biorad, Hercules, CA, USA). Ct and SQ values were normalised to the expression levels of three housekeeping genes, HNRPM, CPSF6 and TBP, selected from the microarray results as being stably expressed (Van Wezel et al, 2005), using the geNorm program (Vandesompele et al, 2002). Statistical analysis (ANOVA) was carried out using SPSS 10.0 (SPSS Inc., Chicago, IL, USA).
Table 4. Quantitative RT–PCR data.
| Symbol | Name | Locus ID | Fold change | P-value | Up or down | Keyword |
|---|---|---|---|---|---|---|
| CPSF6 | Cleavage and polyadenylation specific factor 6 | 11052 | 0.98 | 7.15E−01 | NS | Housekeeping gene |
| HNRPM | Heterogeneous nuclear ribonucleoprotein M | 4670 | 1.0 | 1.00E+00 | NS | Housekeeping gene |
| TBP | TATA box binding protein | 6908 | 1.0 | 1.00E+00 | NS | Housekeeping gene |
| CTNNB1 | Catenin beta 1 | 1499 | 1.1 | 1.00E+00 | NS | Adhesion and transcription factor |
| SNAI1 | Snail homolog 1 | 6515 | 1.3 | 1.00E+00 | NS | Transcription repressor |
| TWIST1 | Twist homolog 1 | 7291 | 1.7 | 1.00E+00 | NS | Transcription repressor |
| ILK | Integrin-linked kinase | 3611 | 1.26 | 9.50E−02 | NS | TGFbeta |
| TFGB1 | Transforming growth factor beta | 7040 | 2.0 | 1.50E−02 | Up | TGFbeta |
| SERPINE1 | Serine or cysteine proteinase inhibitor | 5054 | 5.1 | 3.00E−03 | Up | Matrix degradation |
| FN1 | Fibronectin 1 | 2335 | 2.8 | 2.00E−03 | Up | Mesenchymal marker |
| FOSL1 | FOS-like antigen 1 | 8061 | 2.4 | 0.00E+00 | Up | Transcription factor |
| SNAI2* | Snail homolog 2, slug | 6591 | 12 | 0.00E+00 | Up | Transcription repressor |
| VIM* | Vimentin | 7431 | 12 | 0.00E+00 | Up | Mesenchymal marker |
| ZFHX1B | Zinc finger homeobox 1b, sip1 | 9839 | 8.9 | 0.00E+00 | Up | Transcription repressor |
| CDH1 | E-cadherin | 999 | 0.15 | 0.00E+00 | Down | Cytoskeleton/adhesion |
| ELF3 | E74-like factor 3 | 1999 | 0.24 | 0.00E+00 | Down | Transcription factor |
Up indicates upregulated in the ‘fibroblastic’ cell lines. NS=not significant.
Genes also identified by Lacroix and Leclerq.
RESULTS
Cell line characterisation
All breast cell lines were genotyped to verify their identity. For 18 cell lines, results could be verified by data available from the ATCC (http://www.atcc.org). Only for CAMA1, we were unable to identify the 9.3 allele of THO1 on chromosome 11p15.5. Although for six cell lines no genotypes of the loci tested with the Powerplex kit were available, all had a unique profile. The MDA-MB-231* cell line is a derivative of MDA-MB-231 that spontaneously arose in our laboratory. Both genotypes are identical except the loss of allele 8 of TPOX in MDA-MD-231*. Mutations in the DNA sequence of the CDH1 gene have been reported previously (van de Wetering et al, 2001).
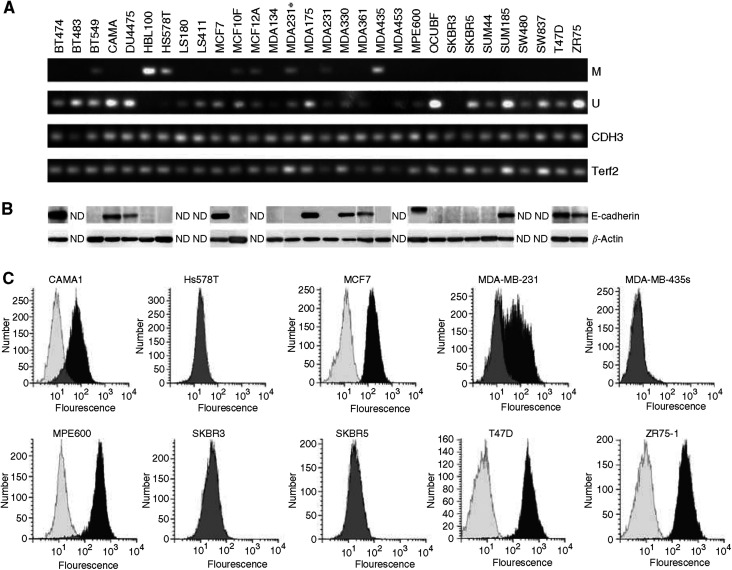
CDH1 promoter methylation status was verified by methylation-specific PCR (MSP) (Figure 1A) (Graff et al, 1995; Herman et al, 1996; Hiraguri et al, 1998; Paz et al, 2003). Three different patterns were identified: (i) Complete promoter hypermethylation (cell lines HBL100 and MDA-MB-435s); (ii) partial promoter methylation (cell lines BT549, Hs578T, MDA-MB-231, MDA-MB-231*, MCF10F and MCF12A) and (iii) no promoter methylation in all other cell lines. Partial methylation indicates that not all CpGs in a promoter region are methylated, as reported previously (Lombaerts et al, 2004). This has been established by sequencing of the PCR products. The results are in agreement with published data for cell lines showing either complete or no promoter methylation. Cell lines showing partial promoter methylation (BT549 and MDA-MB-231) were previously reported as ‘methylated’ (Graff et al, 1995; Herman et al, 1996; Paz et al, 2003) without further specification. The partial CDH1 promoter methylation of MCF10F and MCF12A was not published previously.
Figure 1.
Verification of CDH1 status in mammary cell lines. (A) Methylation-specific PCR. M=MSP specific for methylated CDH1 promoter, U=MSP specific for unmethylated CDH1 promoter, CDH3 and Terf2=control PCR fragments for integrity and modification of template DNA. (B) Western blot analysis for E-cadherin protein expression; β-actin is a loading control. (C) Fluorescence-activated cell sorting analysis for E-cadherin protein expression: overlay of control (white), without antibody and test (black), grey indicates overlap between control and test. The y-axis shows the number of cells and the x-axis the fluorescence of cells.
Western blotting of lysates of 28 cell lines confirmed the published presence or absence of E-cadherin protein expression (Sommers et al, 1994b; Hiraguri et al, 1998; Oberst et al, 2001; van de Wetering et al, 2001) (Figure 1B and Table 1).
Using flow cytometry on a subset of 11 cell lines, we detected membranous E-cadherin expression in five cell lines whereas six cell lines were negative (Figure 1C and Table 1). Highest expression was found for ZR75-1, MPE600 and T47D. These results were in agreement with those from Western blotting. The MDA-MB-231 cell line showed no E-cadherin expression by Western blotting (Figure 1B); however, the flow cytometric results on MDA-MB-231 indicated the presence of two subpopulations, one lacking E-cadherin protein and a weakly positive one (MESF value 18) (Figure 1C), apparently too weak to be detected by Western blot. Methylation-specific PCR analysis suggested partial promoter methylation (Figure 1A). Possibly, promoter methylation in this cell line is dynamic and reversible. This is illustrated by the lower mRNA level in a derivative of this cell line, MDA-MB-231* (Table 4), and it is corroborated by recent findings (Kang et al, 2003) where subpopulations were selected from MDA-MB-231 cells with different metastatic propensities. This plasticity of the CDH1 promoter has been observed previously (Graff et al, 2000).
The results on methylation status were in agreement with the protein expression data. Cell lines showing partial or complete promoter methylation lacked E-cadherin protein expression, with the exception of MDA-MB-231. Although the methylation profile was similar to that of other cell lines showing partial methylation (BT549, MCF10F and MCF12A), flow cytometry showed weak membranous protein expression in a subpopulation of the cells. For SK-BR-3, we could not detect a fragment in the MSP analysis. In order to verify the quality of DNA modification, we performed a PCR on modified DNA with primers specific for two fragments flanking CDH1. As PCR fragments were obtained for both flanking genes, a homozygous deletion of the DNA containing the CDH1 promoter is likely. This result is in discordance with the reported loss of only exons 2–12 (van de Wetering et al, 2001).
Cluster analysis
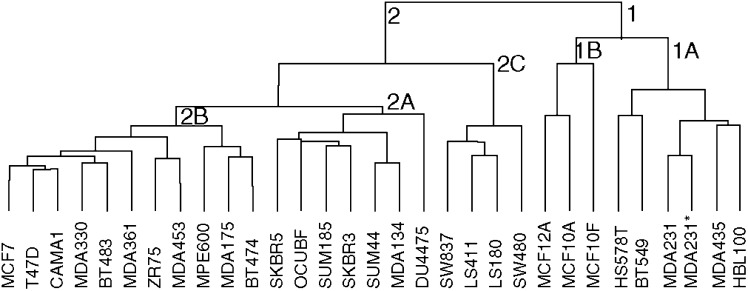
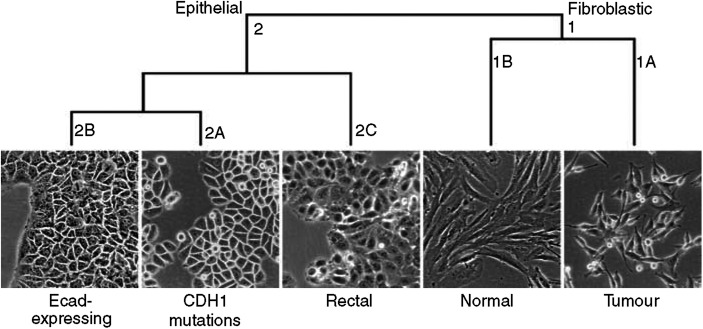
Unsupervised cluster analysis of all cell lines identified two main clusters (Figure 2). Cluster 1 contains cell lines with a fibroblastic morphology, whereas the cluster 2 includes cell lines with a more or less epithelial appearance (Figure 3). The ‘Fibroblastic’ cluster includes two subclusters: 1A includes the breast cancer cell lines BT549, HBL100, Hs578T, MDA-MB-231, MDA-MB-231* and MDA-MB-435s, which all show CDH1 promoter methylation and are oestrogen receptor negative (ER−). Remarkably, cluster 1B (‘Fibroblastic-Normal’) contains three cell lines derived from normal breast tissue (MCF10A, MCF10F and MCF12A). Cluster 2, containing cell lines with an epithelial morphology, is divided into three subclusters. 2A (‘Epithelial-Ecad-expressing’) contains the cell lines BT483, BT474, MCF7, MDA-MB-175VII, MDA-MB-330, MDA-MB-361, MDA-MB-453, T47D and ZR75-1, with wild type CDH1 and two cell lines with CDH1 mutations, CAMA1 and MPE600. Cluster 2A includes eight ER+ cell lines and two ER− cell lines. Interestingly, CAMA1 and MPE600 carry in-frame CDH1 exon deletions (van de Wetering et al, 2001) and show membrane-bound E-cadherin protein expression in our flow cytometry analysis (Figure 1C). Thus, all cell lines in this cluster express E-cadherin protein, but interestingly, the size of the altered protein of MPE600 is larger than normal E-cadherin, suggesting a failure in the removal of the signal peptide. Cluster 2B (‘Epithelial-CDH1-mutated’) includes all breast cancer cell lines harbouring inactivating CDH1 mutations (MDA-MB-134VI, SK-BR-3, SK-BR-5, SUM44PE and OCUB-F) and two cell lines with wild-type CDH1, SUM185PE and Du4475. 2B includes two ER− and one ER+ cell line. Both Ocub-F and Du4475 grow in suspension, but no deviating growth pattern was observed for SUM185PE. Cluster 2C (‘Epithelial-Rectal’) contains cell lines derived from (colo-) rectal tumours (LS180, LS411N, SW480 and SW837). The separation of the colorectal cell lines validates the resolving power of the microarray method.
Figure 2.
Hierarchical cluster analysis of cDNA microarray data for all cell lines reveals an ‘epithelial’ cluster (2) with wild-type CDH1 (2A), mutated CDH1 (2B) and colorectal cell lines (2C) and a ‘fibroblastic’ cluster (1), including tumour (1A) and ‘normal’ mammary cell lines (1B).
Figure 3.
Morphology of representative cell lines in the different clusters. Cluster 1A is represented by MDA-MB-435, 1B by MCF10A, 2A by SKBR3, 2B by MCF7 and 2C by SW480.
Differentially expressed genes
To identify differentially expressed genes associated with differences in E-cadherin expression, we first compared the seven breast tumour cell lines in the ‘Epithelial’ cluster with CDH1 mutations with 12 harbouring wild-type CDH1 from the same cluster. Remarkably, we did not identify any significant differentially expressed genes using the Limma package. As CAMA1 and MPE600 harbour a mutation in the CDH1 gene but still express E-cadherin, we next removed CAMA1 and MPE600 from this analysis. Also, this comparison did not yield any differentially expressed genes (data not shown).
Next, we compared the 18 breast tumour cell lines from the ‘Epithelial’ cluster with those from the ‘Fibroblastic’ cluster. We identified 121 clones showing a highly significant difference in expression (false discovery rate (FDR) <0.01), whereas an additional 187 clones showed differential expression at a lower level of significance (0.01⩽FDR<0.05). Twenty-eight genes were represented by two or more clones in this list. In total, we identified 273 genes that were significantly up- or downregulated in cell lines with a CDH1 promoter methylation vs cell lines without promoter methylation (FDR<0.05). Table 2 shows the top 10 up- and downregulated genes based on the false discovery rate. Table 3 shows a subset of genes that are of particular interest because they are involved in the TGFβ pathway, EMT control or cytoskeletal (re)organisation.
Table 2. Top 10 upregulated (>1) and downregulated (<1) genes in the ‘Fibroblastic-Tumour’ cluster when compared with breast tumour cell lines in the ‘Epithelial’ cluster.
| Symbol | Name | Locus ID | Fold Change | P-value | Keyword |
|---|---|---|---|---|---|
| FOSL1 | FOS-like antigen 1 | 8061 | 692 | 1.03E−07 | Transcription factor |
| GBE1 | Glucan (1,4-alpha-) branching enzyme 1 | 2632 | 9.77 | 6.09E−07 | Sucrose metabolism |
| MMP15 | Matrix metalloproteinase 15 | 4324 | 9.55 | 2.83E−06 | Matrix degradation |
| AMPD1 | Adenosine monophosphate deaminase 1 | 270 | 7.94 | 3.27E−06 | Purine metabolism |
| COL4A2 | Collagen type IV alpha 2 | 1284 | 36.3 | 1.31E−05 | Matrix |
| ACADL | Acyl-coenzyme A dehydrogenase long chain | 33 | 72.4 | 1.32E−05 | Fatty acid metabolism |
| PLAUR | Plasminogen activator, urokinase receptor | 5329 | 43.6 | 2.99E−05 | Matrix degradation |
| AXL | AXL receptor tyrosine kinase | 558 | 11.8 | 2.99E−05 | |
| PHLDA1 | Pleckstrin homology-like domain family A member 1 | 22822 | 13.2 | 3.56E−05 | |
| ANXA5 | Annexin A5 | 308 | 5.62 | 3.65E−05 | |
| IGFBP2* | Insulin-like growth factor binding protein 2 | 3485 | 0.00371 | 1.27E−05 | IGFBP |
| ST14* | Suppression of tumorigenicity 14 | 6768 | 0.0110 | 1.27E−05 | Serine protease |
| TFDP2 | Transcription factor Dp-2 | 7029 | 0.0355 | 2.58E−05 | Transcription factor |
| XBP1 | X-box binding protein 1 | 7494 | 0.0302 | 2.61E−05 | Transcription factor |
| KRT13 | Keratin 13 | 3860 | 0.0195 | 3.65E−05 | Cytoskeleton |
| KIAA0089 | KIAA0089 | 23171 | 0.170 | 7.60E−05 | unknown |
| ELF3 | E74-like factor 3 | 1999 | 0.0263 | 1.20E−04 | Transcription factor |
| FXYD3 | FXYD domain containing ion transport regulator 3 | 5349 | 0.0107 | 1.24E−04 | Ion transport |
| KRT14 | Keratin 14 | 3861 | 0.0195 | 1.24E−04 | Cytoskeleton |
| IL1RN | Interleukin 1 receptor antagonist | 3557 | 0.0933 | 1.24E−04 |
Genes also identified by Lacroix and Leclerq.
Table 3. Selection of differentially expressed genes involved in TGFβ, matrix remodelling and cytoskeleton.
| Symbol | Name | Locus ID | Fold change | P | Up or down | Group |
|---|---|---|---|---|---|---|
| VEGFC | Vascular endothelial growth factor C | 7424 | 30.9 | 2.42E−02 | Up | Angiogenesis |
| PLOD2 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 5352 | 6.17 | 2.96E−02 | Up | Angiogenesis |
| STC1 | Stanniocalcin 1 | 6781 | 12.0 | 2.35E−03 | Up | Angiogenesis |
| SPARC | Secreted protein, acidic, cysteine-rich | 6678 | 28.2 | 2.57E−02 | Up | AP1 target |
| CSF1* | Colony stimulating factor 1 | 1435 | 102 | 6.36E−05 | Up | Cytokine |
| ITGB1 | Integrin, beta 1 | 3688 | 5.13 | 2.03E−02 | Up | Cytoskeleton |
| RAC2 | Ras-related C3 botulinum toxin substrate 2 | 5880 | 6.76 | 1.59E−02 | Up | Cytoskeleton |
| KRT7 | Keratin 7 | 3855 | 0.0602 | 1.97E−02 | Down | Cytoskeleton |
| KRT8 | Keratin 8 | 3856 | 0.0251 | 5.38E−04 | Down | Cytoskeleton |
| KRT19 | Keratin 19 | 3880 | 0.00891 | 3.10E−04 | Down | Cytoskeleton |
| RHOB | Ras homolog gene family, member B | 388 | 0.0589 | 9.87E−03 | Down | Cytoskeleton |
| ARHD | Ras homolog gene family, member D | 29984 | 0.0871 | 3.42E−02 | Down | Cytoskeleton |
| DSP* | Desmoplakin | 1832 | 0.0631 | 1.22E−03 | Down | Cytoskeleton/adhesion |
| CAV1 | Caveolin 1 | 857 | 126 | 3.65E−05 | Up | Cytoskeleton/adhesion |
| CAV2 | Caveolin 2 | 858 | 10.7 | 1.50E−03 | Up | Cytoskeleton/adhesion |
| CD44 | CD44 antigen | 960 | 51.3 | 5.27E−05 | Up | Cytoskeleton/adhesion |
| FN1 | Fibronectin 1 | 2335 | 17.4 | 2.36E−03 | Up | Cytoskeleton/adhesion |
| MSN | Moesin | 4478 | 115 | 1.26E−04 | Up | Cytoskeleton/adhesion |
| S100A2 | S100 calcium binding protein A2 | 6273 | 5.50 | 4.72E−02 | Up | Cytoskeleton/adhesion |
| S100A3 | S100 calcium binding protein A3 | 6274 | 7.41 | 1.08E−02 | Up | Cytoskeleton/adhesion |
| CDH1* | E-cadherin | 999 | 0.0589 | 2.82E−02 | Down | Cytoskeleton/adhesion |
| MAPK1 | Mitogen-activated protein kinase 1 | 5594 | 54.9 | 1.69E−03 | Up | MAPK |
| COL5A1 | Collagen type V alpha 1 | 1289 | 8.32 | 1.58E−02 | Up | Matrix |
| COL15A1 | Collagen type XV alpha 1 | 1306 | 3.31 | 3.94E−02 | Up | Matrix |
| BMP1 | Bone morphogenetic protein 1 | 649 | 9.55 | 1.85E−02 | Up | Matrix degradation |
| MMP2 | Matrix metalloproteinase 2 | 4313 | 27.5 | 9.29E−03 | Up | Matrix degradation |
| MMP3 | Matrix metalloproteinase 3 | 4314 | 2.45 | 4.81E−02 | Up | Matrix degradation |
| MMP14* | Matrix metalloproteinase 14 | 4323 | 53.7 | 5.27E−05 | Up | Matrix degradation |
| PLAU* | Plasminogen activator, urokinase | 5328 | 14.45 | 3.32E−03 | Up | Matrix degradation |
| SERPINE1* | Serine or cysteine proteinase inhibitor member 1 | 5054 | 19.1 | 1.44E−02 | Up | Matrix degradation |
| SERPINE2 | Serine or cysteine proteinase inhibitor member 2 | 5270 | 759 | 1.40E−04 | Up | Matrix degradation |
| TPD52* | Tumor protein D52 | 7163 | 0.25 | 1.86E−02 | Down | Morphogenesis |
| ANXA1 | Annexin A1 | 301 | 6.31 | 3.82E−05 | Up | Phospho-lipid binding |
| ANXA5 | Annexin A5 | 244 | 5.75 | 3.66E−05 | Up | Phospho-lipid binding |
| ANXA8 | Annexin A8 | 244 | 3.09 | 3.24E−02 | Up | Phospho-lipid binding |
| TGFB1 | Transforming growth factor beta 1 | 7040 | 11.8 | 3.76E−02 | Up | TGFbeta |
| TGFB2 | Transforming growth factor beta 2 | 7042 | 23.4 | 1.50E−03 | Up | TGFbeta |
| TGFBR2 | Transforming growth factor beta receptor II | 7048 | 11.2 | 9.56E−04 | Up | TGFbeta |
| FST | Follistatin | 10468 | 11.5 | 6.41E−04 | Up | TGFbeta |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 2354 | 2.24 | 4.47E−02 | Up | Transcription factor |
| TFAP2A | Transcription factor AP-2 alpha | 7020 | 0.224 | 2.29E−03 | Down | Transcription factor |
| TFAP2C* | Transcription factor AP-2 gamma | 7022 | 0.144 | 3.58E−02 | Down | Transcription factor |
| MDM2* | Mouse double minute 2 homolog isoform | 4193 | 0.380 | 5.59E−03 | Down | Ubiquitination |
Up indicates upregulated in the ‘fibroblastic’ cell lines.
Genes also identified by Lacroix and Leclerq.
Real-time PCR
Quantitative PCR was performed to validate cDNA microarray expression data for six differentially expressed genes (CTNNB1, CDH1, ELF3, FN1, FOSL1 and TGFB1; see Table 4). The correlation between results from the microarray and qPCR was highly significant (Figure 4). The microarray expression data led us to hypothesise that breast cell lines with CDH1 promoter methylation all have undergone EMT. To verify this, we also performed qPCR for genes that are involved in the regulation of CDH1 expression (TWIST1, ZFHX1B, SNAI1 and SNAI2) or (a marker for) EMT (ILK, VIM and SERPINE1), but could not be evaluated on the array because of poor hybridisation results or absence.
Figure 4.
Correlation between cDNA microarray data and real-time qPCR for six genes.
As determined by one-way ANOVA, the normalised starting quantity, a measure for the amount of mRNA in the sample, differed significantly between tumour cell lines with CDH1 promoter methylation with a fibroblastic morphology (‘Fibroblastic-Tumour’ cluster) and epithelial breast cancer cell lines (‘Epithelial-Ecad-Expressing’ and ‘Epithelial-CDH1-Mutated’ clusters) for several genes (Table 4). CDH1 and ELF3 were expressed at a significantly higher level in cells with an epithelial morphology, whereas FN1, FOSL1, VIM, ZFHX1B, SNAI2, SERPINE1 and TFGB1 showed a higher expression in fibroblastic breast tumour cells (Table 4 and Figure 5).
Figure 5.
Three examples of qPCR data showing the following: CDH1, high expression in the ‘epithelial’ cluster; SNAI1, no significant difference; SNAI2, high expression in the ‘fibroblastic’ cluster. The y-axis represents the relative expression levels as determined by qPCR. Black bars=epithelial with wild-type CDH1, dark grey=epithelial, CDH1 mutation, light grey=fibroblastic tumour, CDH1 methylation, white=fibroblastic ‘normal’ lines, CDH1 methylation.
Cell lines in the ‘Epithelial-Ecad-Expressing’ subcluster showed considerable variation in CDH1 expression, with higher expression levels predominantly in the ER+ cell lines. Remarkably, the highest levels of ELF3 expression were identified in both SK-BR-3 and SK-BR-5 cell lines.
Several cell lines within the ‘Fibroblastic-Tumour’ subcluster show high expression of TWIST1, particularly BT549 and MDA-MB-435s, but for the whole group the results did not differ significantly from those of the epithelial cluster. Of the ‘Epithelial’ cluster, SUM44PE, which was originally derived from a lobular carcinoma (Ethier et al, 1993; van de Wetering et al, 2001), showed the highest expression. Remarkably, cell lines derived from normal breast epithelial cells, MCF10A, MCF10F and MCF12A, showed very high expression levels of FN1, SERPINE1, ZFHX1B, SNAI2, TGFB1, TWIST1 and VIM and low values for CDH1 and ELF3, which is comparable to expression data from cell lines in the ‘Fibroblastic-Tumour’ subcluster.
DISCUSSION
Alterations in E-cadherin are an important event in carcinogenesis; however, there is controversy about the corollary of the type of E-cadherin inactivation (mutation or promoter hypermethylation) and the aggressiveness of tumour cells. In view of its function in adhesion, it is considered as an invasion suppressor, which is indeed corroborated by in vitro experiments (Vleminckx et al, 1991). Nevertheless, mutational inactivation is already identified in pre-invasive lobular carcinoma in situ (Vos et al, 1997), thereby supporting a role in early carcinogenesis instead of invasive capacity. In order to identify pathways that are affected by E-cadherin inactivation, we performed a genome-wide expression study on 27 human mammary cell lines, which are well characterised on E-cadherin RNA and protein expression status.
Cluster analysis of the microarray data identified two main clusters that coincide with the epithelial and fibroblastic phenotype of the cells, respectively. Importantly, the ‘Fibroblastic’ cluster included only cell lines with either partial or complete CDH1 promoter methylation. This contrasts with the ‘Epithelial’ cluster that included cell lines with wild-type as well as cell lines with mutant CDH1 status. Based on published data on in vitro invasion assays, cellular phenotype and gene expression profiles, Lacroix and Leclercq, (2004) classified breast cancer cell lines into three groups: luminal epithelial, weakly luminal epithelial and ‘mesenchymal’ or ‘stromal’ like. In our analyses, all cell lines belonging to both the luminal and weakly epithelial luminal type group into the ‘Epithelial’ cluster. The position of colorectal tumour cell lines as ‘epithelial’ subcluster rather than forming an out-group suggests that the origin of these cells is subordinate to the phenotype (epithelial or fibroblastic), underscoring the large phenotypic differences induced by EMT.
The fibroblastic phenotype of the cell lines in the latter cluster is strongly indicative of EMT. This is also supported by the increased invasiveness of these cell lines (BT549, Hs578T, MDA-MB-231 and MDA-MB-435s) in vitro and their metastatic potential in mouse models (Price et al, 1990; Thompson et al, 1992; Sommers et al, 1994a). Lacroix and Leclercq (2004) identified 72 differentially expressed genes between the (weakly) luminal epithelial and mesenchymal cell lines of which 15 genes (21%) coincide with differentially expressed genes in our ‘Epithelial’ vs ‘Fibroblastic’ cluster and are indicated in Tables 2, 3 and 4 with an asterisk. The finding that the three non-tumorigenic mammary cell lines derived from normal epithelium form a cluster close to the fibroblastic tumour cell lines is remarkable. However, given their fibroblastic morphology and the CDH1 promoter methylation, this is not unexpected.
Our observation that EMT only occurs in breast cancer cell lines with CDH1 promoter hypermethylation and not with a CDH1 mutational inactivation questions the presumed central role of E-cadherin loss as the initial or primary cause for EMT. This is furthermore illustrated by the surprising lack of significantly differentially expressed genes when comparing cell lines with wild-type and mutant CDH1. It strongly suggests that E-cadherin transcriptional inactivation is an epi-phenomenon and part of an entire programme, with much more severe effects than loss of E-cadherin expression alone. The genes that are involved in this programme can be inferred from the significantly differentially expressed genes when comparing ‘Fibroblastic’ and ‘Epithelial’ cell lines. Two of the identified upregulated genes are upstream repressors of CDH1 transcription, thereby emphasising that E-cadherin itself is not the initiating event in this programme.
We identified 273 differentially expressed genes between breast cancer cell lines in the ‘Epithelial’ vs the ‘Fibroblastic’ cluster, underscoring that these two phenotypes are highly different (Lacroix and Leclercq, 2004). We hypothesise that mutational inactivation is selected for early in carcinogenesis and results in increased growth. In contrast, the transcriptional inactivation by promoter methylation seems part of a larger programme directed towards EMT, thereby increasing invasive and tumorigenic capacity or providing normal epithelial cells with the propensity to divide infinitely in culture as can be inferred from cluster 1B. The TGFβ pathway and furthermore transcription factors that regulate E-cadherin (ZFHX1B and SNAI2), FOSL1 and other AP1/AP2 transcription factors, members of cytoskeleton organisation, IGFBPs, caveolae components, annexins and the AXL receptor tyrosine kinase seem part of such a programme. Further below we will discuss how the major groups of gene products that differentiate the ‘Fibroblastic’ and ‘Epithelial’ breast tumour cell line fit in the this EMT paradigm.
The increased expression of several genes involved in the TGFβ pathway in the ‘Fibroblastic’ cluster, that is, TGFβ1, TGFβ2 and their receptor TGFβR2, is in agreement with the important role of this pathway in the induction of EMT (Thiery, 2003 and references therein). Furthermore, one of the known downstream targets of the TGFβ pathway is ZFHX1B/SIP1, which is a direct repressor of CDH1 (Comijn et al, 2001). Another transcriptional repressor of CDH1, SNAI2/SLUG, a downstream target of the cKIT pathway (Perez-Losada et al, 2002), is also significantly upregulated in the ‘Fibroblastic’ cluster, suggesting that other pathways might also be involved in EMT-related E-cadherin downregulation. For two other transcription factors that are well known to regulate E-cadherin expression in relation to EMT, TWIST (Yang et al, 2004) and SNAI1 (Cano et al, 2000), no significantly altered expression was observed in ‘Fibroblastic’ cells. Together, this suggests that ZFHX1B and SNAI2 are the predominant transcriptional regulators of CDH1 accounting for the EMT phenotype of breast tumour cell lines. Remarkably, TWIST upregulation was reported in lobular breast cancer as an alternative for inactivating mutations of CDH1 (Yang et al, 2004) and, moreover, SUM44PE, the only breast cancer cell line of lobular origin in our panel, showed the highest expression of TWIST. As TWIST is not significantly differentially expressed in the fibroblastic cell lines, we suggest that its protein product has a direct effect on CDH1 and results in a similar phenotype as CDH1 mutations, thereby contributing to the typical phenotype of lobular breast cancer. The lack of upregulation of SNAI1 is unexpected, especially given the recently identified role of this gene in breast cancer recurrence (Moody et al, 2005).
FOSL1 (also called FRA1) and FOSB, albeit to a lesser extent, were significantly upregulated genes in cell lines with CDH1 promoter methylation. These FOS family members form heterodimers with JUN family members (mainly, c-Jun, JunB and JunD) thereby forming the AP1 transcription factor (Karin et al, 1997). Various studies have shown that alterations of the composition of AP1 are related to changes in proliferation, malignant transformation and aggressiveness of cells (Mechta et al, 1997; Smith et al, 1999). Detectable FOSL1 protein expression in mammary carcinomas was demonstrated to be associated with poor differentiation, Ki67 and cyclin E expression and an oestrogen receptor-negative phenotype (Milde-Langosch et al, 2000). As an AP1 site has been identified in the TGFBR2 promoter, upregulation of FOSL1 and FOSB may stimulate TGFBR2 transcription. Interestingly, RHOB binds to the promoter of TGFBR2 and in this way prevents AP1-dependent transcription creating a negative feedback loop that regulates TGFβ signal transduction (Adnane et al, 2002). RHOB was significantly downregulated in the ‘Fibroblastic’ cluster. Other targets of AP1 are extracellular matrix modulating enzymes that on their turn may contribute to an increased migratory and metastatic phenotype. Indeed, genes encoding metalloproteinases MMP 2, 3, 14 and 15 as well as PLAUR, encoding the urokinase receptor, showed increased expression in the CDH1 methylated cells.
Besides downregulation of E-cadherin and loss of cell–cell interactions, EMT is accompanied by extensive reorganisation of actin as well as intermediate filamental cytoskeleton. Therefore, it is not surprising that we observed a differential expression of various genes that encode parts of the intermediate filaments, including KRT7, -8, -13, -14, -19 and vimentin, a fibroblastic marker. Also genes that regulate the organisation and turnover of the F-actin filaments such as RAC, MSN as well as RHOB were differentially expressed (Table 2). The balance between RHO and RAC is shifted towards RAC in cell lines with fibroblastic morphology. The upregulation of RAC in these latter cells fits with increased protrusions and lamellopodia that are required for cell migration.
Also, annexin gene family members, of which ANXA1, ANXA5 and ANXA8 show increased expression in fibroblastic cells, may indirectly affect the cytoskeleton. AXL is a member of a family of receptor tyrosine kinases characterised by an extracellular domain resembling cell adhesion molecules and an intracellular conserved tyrosine kinase domain. Its upregulation in the ‘Fibroblastic-Tumour’ cluster is in agreement with the reported elevated expression in highly metastatic osteosarcoma cell lines (Nakano et al, 2003) and metastatic tumours including colon cancer, gastric cancer and melanoma (Quong et al, 1994; Craven et al, 1995; Wu et al, 2002).
We are aware that this study requires translation to tissue samples. Unfortunately, such a study is hampered by infiltrating lymphocytes that confound a reliable detection of CDH1 promoter hypermethylation by MSP (Lombaerts et al, 2004).
In conclusion, our data indicate that CDH1 promoter hypermethylation but not CDH1 mutational inactivation is a part of an entire EMT programme resulting in breast tumour cells with a more aggressive phenotype, thus enabling metastasis formation. At this moment, we do not know the initial steps for this epigenetically controlled EMT. Nevertheless, it has become generally accepted that metastasis is facilitated by EMT and thus interfering with this process in breast cancer might prevent tumour dissemination. Hence, targeting of abnormal TGFβ signalling could be one of the main priorities in preventing EMT and its adverse effects on the prognosis of patients with breast cancer (Sokol et al, 2005). Future investigations will therefore be directed at verification of this transcriptional programme associated with CDH1 methylation in primary breast tumour samples and an association with disease outcome.
Acknowledgments
We thank S Uljee, HJ Baelde and Dr ASK de Hooge for providing SERPINE1 and FN1 primers and the anti-β-actin antibody, respectively. We also thank IH Briaire-de Bruijn and Dr W Corver for technical assistance on Western blotting and flow cytometry. JN Kloth, E van Roon, Dr S Romeo and Dr E de Heer are acknowledged for their advice and useful discussions. We thank Dr M Schutte and Dr A Hollestelle for their help on cell cultures. This study was financially supported by the Association for International Cancer Research (AICR) (ML) grant number 00-258 and by the Dutch Cancer Foundation (KWF) grant number 2000-2205 (TvW and KP).
References
- Adnane J, Seijo E, Chen Z, Bizouarn F, Leal M, Sebti SM, Munoz-Antonia T (2002) RhoB, not RhoA, represses the transcription of the transforming growth factor beta type II receptor by a mechanism involving activator protein 1. J Biol Chem 277: 8500–8507 [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia DH (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2: 84–89 [DOI] [PubMed] [Google Scholar]
- Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Hofler H (1994) E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res 54: 3845–3852 [PubMed] [Google Scholar]
- Berx G, Becker KF, Hofler H, van Roy F (1998) Mutations of the human E-cadherin (CDH1) gene. Hum Mutat 12: 226–237 [DOI] [PubMed] [Google Scholar]
- Berx G, Cleton-Jansen AM, Nollet F, De Leeuw WJ, Van de Vijver MJ, Cornelisse C, van Roy F (1995) E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J 14: 6107–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx G, Cleton-Jansen AM, Strumane K, De Leeuw WJ, Nollet F, van Roy F, Cornelisse C (1996) E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene 13: 1919–1925 [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2: 76–83 [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F (2001) The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell 7: 1267–1278 [DOI] [PubMed] [Google Scholar]
- Corver WE, Cornelisse CJ, Fleuren GJ (1994) Simultaneous measurement of two cellular antigens and DNA using fluorescein-isothiocyanate, R-phycoerythrin, and propidium iodide on a standard FACScan. Cytometry 15: 117–128 [DOI] [PubMed] [Google Scholar]
- Craven RJ, Xu LH, Weiner TM, Fridell YW, Dent GA, Srivastava S, Varnum B, Liu ET, Cance WG (1995) Receptor tyrosine kinases expressed in metastatic colon cancer. Int J Cancer 60: 791–797 [DOI] [PubMed] [Google Scholar]
- De Craene B, Gilbert B, Stove C, Bruyneel E, van Roy F, Berx G (2005) The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res 65: 6237–6244 [DOI] [PubMed] [Google Scholar]
- De Leeuw WJ, Berx G, Vos CB, Peterse JL, Van de Vijver MJ, Litvinov S, van Roy F, Cornelisse CJ, Cleton-Jansen AM (1997) Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. J Pathol 183: 404–411 [DOI] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R (2005) DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 24: 2375–2385 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier SP, Mahacek ML, Gullick WJ, Frank TS, Weber BL (1993) Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res 53: 627–635 [PubMed] [Google Scholar]
- Forozan F, Veldman R, Ammerman CA, Parsa NZ, Kallioniemi A, Kallioniemi OP, Ethier SP (1999) Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br J Cancer 81: 1328–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady WM, Willis J, Guilford PJ, Dunbier AK, Toro TT, Lynch H, Wiesner G, Ferguson K, Eng C, Park JG, Kim SJ, Markowitz S (2000) Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet 26: 16–17 [DOI] [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG (2000) Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem 275: 2727–2732 [DOI] [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB (1995) E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res 55: 5195–5199 [PubMed] [Google Scholar]
- Hajra KM, Chen DY, Fearon ER (2002) The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res 62: 1613–1618 [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri S, Godfrey T, Nakamura H, Graff J, Collins C, Shayesteh L, Doggett N, Johnson K, Wheelock M, Herman J, Baylin S, Pinkel D, Gray J (1998) Mechanisms of inactivation of E-cadherin in breast cancer cell lines. Cancer Res 58: 1972–1977 [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R (1996) Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev 59: 3–10 [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3: 537–549 [DOI] [PubMed] [Google Scholar]
- Karin M, Liu Z, Zandi E (1997) AP-1 function and regulation. Curr Opin Cell Biol 9: 240–246 [DOI] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G (2004) Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat 83: 249–289 [DOI] [PubMed] [Google Scholar]
- Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH (2005) Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 24: 8277–8290 [DOI] [PubMed] [Google Scholar]
- Lombaerts M, Middeldorp JW, van der WE, Philippo K, van Wezel T, Smit VT, Cornelisse CJ, Cleton-Jansen AM (2004) Infiltrating leukocytes confound the detection of E-cadherin promoter methylation in tumors. Biochem Biophys Res Commun 319: 697–704 [DOI] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P (2005) ADAM10 mediates E-cadherin shedding and regulates epithelial cell–cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci USA 102: 9182–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechta F, Lallemand D, Pfarr CM, Yaniv M (1997) Transformation by ras modifies AP1 composition and activity. Oncogene 14: 837–847 [DOI] [PubMed] [Google Scholar]
- Milde-Langosch K, Bamberger AM, Methner C, Rieck G, Loning T (2000) Expression of cell cycle-regulatory proteins rb, p16/MTS1, p27/KIP1, p21/WAF1, cyclin D1 and cyclin E in breast cancer: correlations with expression of activating protein-1 family members. Int J Cancer 87: 468–472 [PubMed] [Google Scholar]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA (2005) The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8: 197–209 [DOI] [PubMed] [Google Scholar]
- Nakano T, Tani M, Ishibashi Y, Kimura K, Park YB, Imaizumi N, Tsuda H, Aoyagi K, Sasaki H, Ohwada S, Yokota J (2003) Biological properties and gene expression associated with metastatic potential of human osteosarcoma. Clin Exp Metast 20: 665–674 [DOI] [PubMed] [Google Scholar]
- Oberst M, Anders J, Xie B, Singh B, Ossandon M, Johnson M, Dickson RB, Lin CY (2001) Matriptase and HAI-1 are expressed by normal and malignant epithelial cells in vitro and in vivo. Am J Pathol 158: 1301–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M (2003) A systematic profile of DNA methylation in human cancer cell lines. Cancer Res 63: 1114–1121 [PubMed] [Google Scholar]
- Perez-Losada J, Sanchez-Martin M, Rodriguez-Garcia A, Sanchez ML, Orfao A, Flores T, Sanchez-Garcia I (2002) Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood 100: 1274–1286 [PubMed] [Google Scholar]
- Price JE, Polyzos A, Zhang RD, Daniels LM (1990) Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res 50: 717–721 [PubMed] [Google Scholar]
- Quong RY, Bickford ST, Ing YL, Terman B, Herlyn M, Lassam NJ (1994) Protein kinases in normal and transformed melanocytes. Melanoma Res 4: 313–319 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2004) R Foundation for Statistical Computing. Vienna, Austria. ISBN 3-900051-00-3, URL http://www.R-project.org Ref Type: Generic [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de RM, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 24: 227–235 [DOI] [PubMed] [Google Scholar]
- Siitonen SM, Kononen JT, Helin HJ, Rantala IS, Holli KA, Isola JJ (1996) Reduced E-cadherin expression is associated with invasiveness and unfavorable prognosis in breast cancer. Am J Clin Pathol 105: 394–402 [DOI] [PubMed] [Google Scholar]
- Smith LM, Wise SC, Hendricks DT, Sabichi AL, Bos T, Reddy P, Brown PH, Birrer MJ (1999) cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene 18: 6063–6070 [DOI] [PubMed] [Google Scholar]
- Sokol JP, Neil JR, Schiemann BJ, Schiemann WP (2005) The use of cystatin C to inhibit epithelial–mesenchymal transition and morphological transformation stimulated by transforming growth factor-beta. Breast Cancer Res 7: R844–R853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers CL, Byers SW, Thompson EW, Torri JA, Gelmann EP (1994a) Differentiation state and invasiveness of human breast cancer cell lines. Breast Cancer Res Treat 31: 325–335 [DOI] [PubMed] [Google Scholar]
- Sommers CL, Gelmann EP, Kemler R, Cowin P, Byers SW (1994b) Alterations in beta-catenin phosphorylation and plakoglobin expression in human breast cancer cells. Cancer Res 54: 3544–3552 [PubMed] [Google Scholar]
- Takeichi M (1995) Morphogenetic roles of classic cadherins. Curr Opin Cell Biol 7: 619–627 [DOI] [PubMed] [Google Scholar]
- Tamura G, Yin J, Wang S, Fleisher AS, Zou T, Abraham JM, Kong D, Smolinski KN, Wilson KT, James SP, Silverberg SG, Nishizuka S, Terashima M, Motoyama T, Meltzer SJ (2000) E-cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst 92: 569–573 [DOI] [PubMed] [Google Scholar]
- Thiery JP (2002) Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer 2: 442–454 [DOI] [PubMed] [Google Scholar]
- Thiery JP (2003) Epithelial–mesenchymal transitions in development and pathologies. Curr Opin Cell Biol 15: 740–746 [DOI] [PubMed] [Google Scholar]
- Thompson EW, Paik S, Brunner N, Sommers CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME (1992) Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol 150: 534–544 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Barker N, Harkes IC, van der HM, Dijk NJ, Hollestelle A, Klijn JG, Clevers H, Schutte M (2001) Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Res 61: 278–284 [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel T, Lombaerts M, van Roon EH, Philippo K, Baelde HJ, Szuhai K, Cornelisse CJ, Cleton-Jansen AM (2005) Expression analysis of candidate breast tumour suppressor genes on chromosome 16q. Breast Cancer Res 7: R998–R1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleminckx K, Vakaet L, Mareel M, Fiers W, van Roy F (1991) Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 66: 107–119 [DOI] [PubMed] [Google Scholar]
- Vos CB, Cleton-Jansen AM, Berx G, De Leeuw WJ, ter Haar NT, van Roy F, Cornelisse CJ, Peterse JL, Van de Vijver MJ (1997) E-cadherin inactivation in lobular carcinoma in situ of the breast: an early event in tumorigenesis. Br J Cancer 76: 1131–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK (2004) limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20: 3705–3706 [DOI] [PubMed] [Google Scholar]
- Winter MJ, Nagelkerken B, Mertens AE, Rees-Bakker HA, Briaire-de Bruijn IH, Litvinov SV (2003) Expression of Ep-CAM shifts the state of cadherin-mediated adhesions from strong to weak. Exp Cell Res 285: 50–58 [DOI] [PubMed] [Google Scholar]
- Wu CW, Li AF, Chi CW, Lai CH, Huang CL, Lo SS, Lui WY, Lin WC (2002) Clinical significance of AXL kinase family in gastric cancer. Anticancer Res 22: 1071–1078 [PubMed] [Google Scholar]
- Yagi T, Takeichi M (2000) Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev 14: 1169–1180 [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117: 927–939 [DOI] [PubMed] [Google Scholar]