Abstract
Tumour development is characterised by the increased circulating DNA (cirDNA) concentration and by tumour-related changes in blood plasma DNA. Concentration of cirDNA and methylation of RARβ2, RASSF1A and HIC-1 gene promoters were investigated in cell-free and cell-surface-bound fractions from healthy donors, patients with breast cancer, and patients with breast fibroadenoma. Tumour development was shown to lead to significant changes in the distribution of cirDNA between cell-free and cell-surface-bound fractions. Analysis of RARβ2 and RASSF1A methylation in the total cirDNA provides 95% diagnostic coverage in breast cancer patients, 60% in patients with benign lesions, and is without false-positive results in healthy women. Results of the study indicate that methylation-specific PCR of RARβ2 and RASSF1A genes based on the total cirDNA combined with the quantitative analysis of cirDNA distribution between cell-bound and cell-free fractions in blood provide the sensitive and accurate detection and discrimination of malignant and benign breast tumours.
Keywords: breast cancer, diagnostics, circulating DNA, methylation-specific PCR, DNA quantification
In the last years attempts have been made to develop noninvasive tests for early cancer diagnostics based on analysis of extracellular DNA circulating (cirDNA) in the blood. The increased plasma cirDNA concentration itself can be an indication of tumour development (Anker, 2000). However, this test did not promise to be specific, as far as the comparable increase of DNA concentration in plasma was found in patients with other disorders as systemic lupus erythematosus, rheumatoid arthritis, glomerulonephritis, pancreatitis, hepatitis, etc (Anker, 2000).
Numerous studies have demonstrated tumour-specific alterations, such as aberrant promoter hypermethylation in cirDNA recovered from plasma of patients with different malignancies, and the absence of methylated DNA in healthy persons (Silva et al, 2002). Changes in the status of DNA methylation represent one of the most common molecular alterations in human neoplasia (Egger et al, 2004), including breast cancer (Widschwendter and Jones, 2002). These epigenetic alterations induce neoplastic process by transcriptional silencing of tumour suppressor gene expression and are responsible for initial steps of induction of tumour cell proliferation (Jones and Baylin, 2002). Therefore, analysis of gene methylation patterns in tissues could be of profound significance in the early detection of cancer (Esteller et al, 2001). Expression of more than 40 genes was found to be lost in breast cancer because of promoter hypermethylation (Jones and Baylin, 2002; Widschwendter and Jones, 2002), including RASSF1A, RARβ2, HIC-1 genes. However, the methylated markers are detected with twice lower frequency in plasma samples compared with the frequency of their finding in the tissues of cancer patient (Shao and Nguyen, 2002; Anker P et al, 2003). According to our recent observations, extracellular nucleic acids circulate in blood not only in plasma but are also bound to the surface of blood cells (Rykova et al, 2004). In pilot experiments we evaluated the cell-surface-bound cirDNA as a source of material for the early breast cancer diagnostics based on methylation-specific PCR (MSP) (Rykova et al, 2004). In the present study we investigated the methylation of HIC-1, RASSF1A and RARβ2 genes promoters in cirDNA derived from plasma and cell-surface-bound fractions of patients with breast tumours.
Results of the study evidence that the analysis of the cell-surface-bound cirDNA along with cirDNA from plasma fraction could considerably increase reliability of noninvasive tests for tumour development based on MSP.
MATERIALS AND METHODS
Blood samples
Blood samples of previously untreated breast cancer and nonmalignant patients were obtained from Novosibirsk Regional Oncology Dispensary. The Ethics Committee in Dispensary approved specimen collection procedures. Tumour staging was performed according to the TNM classification. Blood samples of healthy donors, selected for blood transfusion, were obtained from Novosibirsk Central Clinical Hospital. Blood (8 ml) was collected into the tubes containing 2 ml of sterile phosphate-buffered saline (PBS) with 50 mM EDTA. Blood was fractionated into plasma, leucocytes and erythrocytes, and cell-surface-bound DNA fractions were obtained as described earlier (Rykova et al, 2004).
DNA extraction and concentration measurement
DNA was extracted from 1 ml of plasma samples, 2 ml of PBS/EDTA samples and 1 ml of trypsin samples by glass milk-based protocol providing quantitative isolation of DNA (Tamkovich et al, 2004). Concentration of DNA was measured using Hoechst 33258 assay (Labarca and Paigen, 1980). Detection limit for DNA calculated to the initial blood volume was 8 ng ml−1 in plasma, 20 ng ml−1 in trypsin eluates and 40 ng ml−1 in PBS/EDTA eluates.
Methylation-specific PCR
Aberrant methylation of RASSF1A, RARβ2 and HIC-1 gene promoters was determined by MSP according to modified protocol (Herman et al, 1996). DNA was extracted from 1 ml of plasma samples, 6 ml of PBS-EDTA samples and 1 ml of trypsin samples by glass milk-based protocol (Tamkovich et al, 2004). Isolated DNA was modified by sodium bisulfite, purified using glass milk protocol, eluted into 30 μl of water and stored in aliquots at −40°C. A volume of 3 μl of the bisulfite-modified DNA was used in MSP reaction. The primers used are listed in Table 1. Polymerase chain reaction products were visualised on 6% PAAG stained with ethidium bromide.
Table 1. Sequences of primers used in MSP.
| Gene | Primer sequence (5′–3′) | Genbank accession no. | Amplicon location (Genebank numbering) | T (in °C) | Length (in bp) |
|---|---|---|---|---|---|
| RASSF1A | Mf: GGG TTT TGC GAG AGC GCG | 169 | |||
| Mr: GCT AAC AAA CGC GAA CCG | AC002481 | 17881–18115 | 64 | ||
| Uf: GGT TTT GTG AGA GTG TGT TTAG | 59 | 169 | |||
| Ur: CAC TAA CAA ACA CAA ACC AAG | |||||
| RARβ2 | Mf: GCT TAG TAG TTC GGG TAG GGT TTA TC | 235 | |||
| Mr: CCG AAT CCT ACC CCG ACG | X56849 | 773–1007 | 64 | ||
| Uf: TTA GTA GTT TGG GTA GGG TTT ATT | 55 | 233 | |||
| Ur: CCA AAT CCT ACC CCA ACA | |||||
| HIC-1 | Mf: TCG GTT TTC GCG TTT TGT TCG T | 95 | |||
| Mr: AAC CGA AAA CTA TCA ACC CTC G | L41919 | 26–120 | 64 | ||
| Uf: TTG GGT TTG GTT TTT GTG TTT TG | 64 | 118 | |||
| Ur: CAC CCT AAC ACC ACC CTA AC |
RESULTS
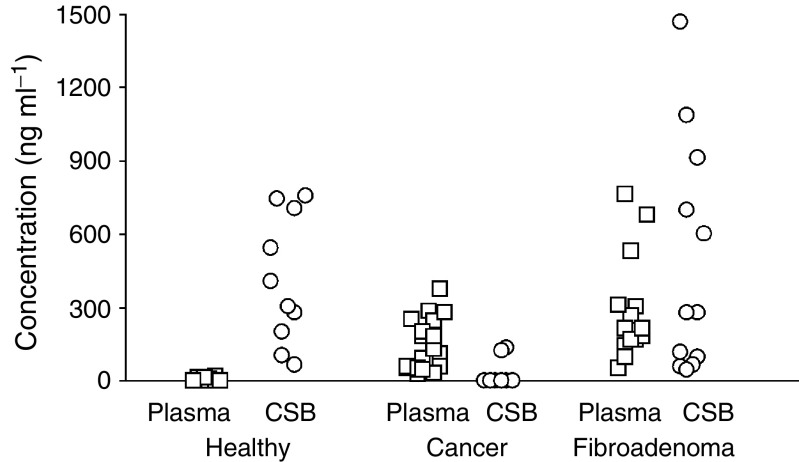
We analysed concentrations of cell-surface-bound and cell-free cirDNA in the blood samples of 20 breast cancer patients, 15 patients with nonmalignant tumour (fibroadenoma) and 10 healthy women. Analysis of the distribution of cirDNA in blood of breast cancer patients supports our earlier data (Laktionov et al, 2004) that the main part of the cirDNA in the blood of healthy donors is absorbed at the surface of blood cells (Figure 1). A strong decrease of the cell-surface-bound cirDNA amount, as well as an increase of cell-free cirDNA was found in cancer relative to health (Figure 1). In the blood of 95% of breast cancer patients, increased amounts of cirDNA were found in plasma and only negligible amounts of cell-surface-bound cirDNA were found in few patients of this group. Patients with fibroadenoma were characterised by increased level of circulating DNA in plasma similar to cancer patients, albeit cirDNA were found also at the surface of blood cells similar to what was found in healthy women (Figure 1).
Figure 1.
Concentrations of circulating DNA in plasma (Plasma) and cell-surface-bound DNA (CSB) in blood of healthy women (Health), breast cancer patients (Cancer) and patients with breast fibroadenoma (Fibroadenoma).
Methylation of RASSF1A, RARβ2 and HIC-1 gene promoters were studied in healthy donors and patients with breast tumours. These genes had been reported to be frequently methylated in breast cancer (Esteller et al, 2001; Jones and Baylin, 2002; Widschwendter and Jones, 2002). CirDNA isolated from the plasma of 10 healthy women were found to be negative for methylation forms of all three genes. Methylation of RARβ2 and RASSF1A promoters was not found in the cirDNA isolated from cell-surface-bound fractions, whereas five healthy women out of ten were positive for methylated HIC-1 in different cell-associated cirDNA fractions (Table 2). Methylated form of RASSF1A gene was detected with the same frequency (60%) in the cell-bound cirDNA from patients with cancer and nonmalignant breast tumours (Table 2). In contrast, methylated RARβ2 form was detected three times more frequently in breast cancer patients than in fibroadenoma patients: in 60% of cell-associated cirDNA in cancer and 20% in nonmalignant cases (Table 2). The methylation of all three genes was detected considerably more frequently in the cell-bound cirDNA than in the plasma cirDNA of the tumour-bearing patients (Table 2). The methylation pattern of the different genes differ among patients; some patients demonstrated simultaneous methylation of RARβ2 and RASSF1A, while some of them were positive only in either RARβ2 or RASSF1A regions. To summarise, two methylated markers RARβ2 and RASSF1A were not found in healthy women but were detected in 95% of the cancer patients and in 60% of the patients with fibroadenoma when total circulating DNA were used for MSP reaction (Table 2).
Table 2. Frequency of methylation of RASSF1A, RARβ and HIC-1 genes in plasma and cell-surface-bound DNA in blood of breast cancer patients, patients with fibroadenoma and healthy donors.
|
Erythrocytes
|
Leucocytes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Diagnosis | Gene | Plasma | PBS eluate | Trypsin eluate | PBS eluate | Trypsin eluate | Total cell-surf- bound | Plasma and total cell-surf-bound |
| Breast cancer (n=20) | HIC-1 | 55* | 35 | 45 | 70 | 45 | 90 | 90 |
| RASSF1A | 15 | 35 | 0 | 35 | 30 | 65 | 75 | |
| RARβ2 | 15 | 5 | 25 | 20 | 15 | 60 | 65 | |
| RASSF1A or RARβ2 | 30** | 95 | ||||||
| Fibroadenoma (n=15) | HIC-1 | 40 | 27 | 73 | 67 | 53 | 93 | 93 |
| RASSF1A | 7 | 27 | 33 | 40 | 27 | 53 | 53 | |
| RARβ2 | 13 | 0 | 7 | 0 | 13 | 20 | 33 | |
| RASSF1A or RARβ2 | 13 | 60 | ||||||
| Healthy donors (n=10) | HIC-1 | 0 | 10 | 10 | 50 | 20 | 50 | 50 |
| RASSF1A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| RARβ2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| RASSF1A or RARβ2 | 0 | 0 | ||||||
Percentage of cases with RASSF1A, RARβ2, HIC-1 hypermethylation in plasma DNA, cell-surface bound DNA and total circulating DNA.
Percentage of cases with RASSF1A or RARβ2 hypermethylation in plasma DNA and total circulating DNA.
DISCUSSION
CirDNA is present in blood plasma in health and is increased in cancer and other malignancies (Anker, 2000). These findings have attracted much attention to the potential use of elevated concentration of cirDNA as a tumour marker. It is known that nucleic acids can bind with cell-surface DNA-binding proteins (Bennett et al, 1985; Laktionov et al, 1999), as well as with phospholipids of cellular membrane through bivalent ions (Beliaev et al, 1988). These interactions together with large amount of cells in the bloodstream provide an opportunity for circulation of DNA in blood being absorbed at cell surface. Earlier we have found that a fraction of the bound nucleic acids could be detached from the cell surface by PBS/EDTA treatment suggesting involvement of bivalent ions in the complex formation. Further removal of the surface-bound cirDNA can be achieved by mild trypsin treatment, which destroys cell surface proteins binding with cirDNA. In accordance with previous data (Laktionov et al, 2004) results of the present study indicate that the main part of the cirDNA in the blood of healthy donors is mainly absorbed at the surface of blood cells (Table 2). Our data demonstrate redistribution of extracellular DNA in blood between cell surface and plasma in breast tumour-bearing patients, which suggest a possibility of the development of simple noninvasive tumour-screening assay.
Mechanisms regulating appearance and distribution of extracellular DNA in blood are not clear to date. In the bloodstream extracellular DNA are under the pressure of factors influencing its circulation and clearance, including hydrolysing enzymes. It is known that tumour invasion in cancer patients is accompanied by increased level of proteases (Farias et al, 2000). Damage of the cell-surface DNA-binding proteins by these enzymes can result in detachment of cell-bound DNA. Cancer development is characterised by the change of DNase activity in blood plasma. It was shown recently, that the decreased DNase activity in plasma of patients with gastrointestinal cancer correlates with the increased integrity of cirDNA and their concentration in blood plasma (Tamkovich et al, 2006). Other factors should be further investigated, which could influence extracellular DNA distribution in the circulation in health and in the process of tumour development.
Tumour-related methylated forms of three genes (RARβ2, RASSF1A and HIC-1) were much more frequently evaluated in the cell-surface-bound cirDNA compared with cell-free cirDNA in tumour-bearing patients (Table 2). The data obtained clearly indicate that cell-surface-bound DNA provides valuable source of material for MSP diagnostics along with cirDNA isolated from the plasma. According to our data RARβ2 gene methylation possesses the most cancer-specific propensity. In contrast, methylation of HIC-1 promoter region, which was examined here, is not tumour specific.
Analysis of methylated DNA sequences in the cirDNA demonstrated no regularities in their fraction distribution (Table 2). Phosphate-buffered saline and trypsin treatments both provide cell-bound DNA, which should be used combined as a template for the MSP-based assay. There is no significant difference in the methylation frequencies for the leucocyte-bound cirDNA compared with erythrocyte-bound cirDNA, when any of three genes was analysed. However, as long as there are 1000 times less leucocytes than erythrocytes in blood, we propose that specific content of methylated DNA at the surface of a single leucocyte is higher than at the surface of a single erythrocyte.
Whatever might be the mechanism for tumour-related DNA binding to blood cells, we can now ask whether this phenomenon has potentially important implications in cancer patients. Experiments with human fibroblasts provided evidences that tumour DNA may be horizontally transferred by the uptake of apoptotic bodies (Holmgren et al, 1999). Garcia-Olmo et al (1999)suggested that metastases might develop as a result of transfection of susceptible cells in distant target organs with dominant oncogenes that circulate in plasma and are derived from the primary tumour. Monocyte cells from leucocyte fraction, which can migrate into the tissues from the blood vessels, could play the role of ‘carriers’ of the surface-bound tumour DNA to healthy cells.
Our data indicate the presence of methylated RASSF1A and RARβ2 in the cirDNA of all the detected patients bearing the Ist tumour stage. This sounds optimistic for the development of tumour-specific screening test, taking into account the early stage of these changes to occur. To summarise, assay based on the methylated RARβ2 or RASSF1A detection in the total circulating DNA in blood provides 95% sensitivity for breast cancer detection. Additional quantitative analysis of the distribution of extracellular DNA between cell-bound and cell-free fractions in blood is helpful to differentiate healthy women, patients with breast fibroadenoma and breast cancer patients.
Acknowledgments
We gratefully acknowledge Dr Philippe Anker for very helpful discussions and Valentina Permjakova for the medical assistance. The present work was supported by Russian Academy of Sciences Program ‘Science to medicine’, Russian Foundation for Basic Research Grant 06-04-49732, Interdisciplinary Project from Siberian Division of Russian Academy of Sciences No. 13, Scientific Schools Grant SS-1384.2003.4.
References
- Anker P (2000) Quantitative aspects of plasma/serum DNA in cancer patients. Ann NY Acad Sci 906: 5–7 [DOI] [PubMed] [Google Scholar]
- Anker P, Mulcahy H, Stroun M (2003) Circulating nucleic acids in plasma and serum as a noninvasive investigation for cancer: time for large-scale clinical studies? Int J Cancer 103: 149–152 [DOI] [PubMed] [Google Scholar]
- Beliaev ND, Budker VG, Gorokhova OE, Sokolov AV (1988) Mg2+-dependent interaction of DNA with eukaryotic cells. Mol Biol (Moskovskogo) 22: 1667–1672 [PubMed] [Google Scholar]
- Bennett RM, Gabor GT, Merritt MM (1985) DNA binding to human leukocytes. Evidence for a receptor-mediated association, internalization, and degradation of DNA. J Clin Invest 76: 2182–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463 [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG (2001) A gene hypermethylation profile of human cancer. Cancer Res 61: 3225–3229 [PubMed] [Google Scholar]
- Farias E, Ranuncolo S, Cresta C, Specterman S, Armanasco E, Varela M, Lastiri J, Pallotta MG, Bal de Kier Joffe E, Puricelli L (2000). Plasma metalloproteinase is enhanced in the euglobulin fraction of breast and lung cancer patients. Int J Cancer 89: 389–394 [DOI] [PubMed] [Google Scholar]
- Garcia-Olmo D, Garcia-Olmo DC, Ontanon J, Martinez E, Vallejo M (1999) Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol Histopathol 4: 1159–1164 [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren L, Szeles A, Rajnavolgyi E, Folkman J, Klein G, Ernberg I, Falk KI (1999) Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood 93: 3956–3963 [PubMed] [Google Scholar]
- Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3: 415–428 [DOI] [PubMed] [Google Scholar]
- Labarca C, Paigen K (1980) A simple, rapid, and sensitive DNA assay procedure. Anal Biochem 102: 344–352 [DOI] [PubMed] [Google Scholar]
- Laktionov PP, Dazard JE, Vives E, Rykova EY, Piette J, Vlassov VV, Lebleu B (1999) Characterisation of membrane oligonucleotide-binding proteins and oligonucleotide uptake in keratinocytes. Nucleic Acids Res 27: 2315–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laktionov PP, Tamkovich SN, Rykova EY, Bryzgunova OE, Starikov AV, Kuznetsova NP, Valssov VV (2004) Cell-surface-bound nucleic acids: Free and cell-surface-bound nucleic acids in blood of healthy donors and breast cancer patients. Ann NY Acad Sci 1022: 221–227 [DOI] [PubMed] [Google Scholar]
- Rykova EY, Laktionov PP, Skvortsova TE, Starikov AV, Kuznetsova NP, Vlassov VV (2004) Extracellular DNA in breast cancer: cell-surface-bound, tumor-derived extracellular DNA in blood of patients with breast cancer and nonmalignant tumors. Ann NY Acad Sci 1022: 217–221 [DOI] [PubMed] [Google Scholar]
- Silva JM, Garcia JM, Dominguez G, Silva J, Miralles C, Cantos B, Coca S, Provencio M, Espana P, Bonilla F (2002) Persistence of tumor DNA in plasma of breast cancer patients after mastectomy. Ann Surg Oncol 9: 71–76 [DOI] [PubMed] [Google Scholar]
- Shao ZM, Nguyen M (2002) Tumor-specific DNA in plasma of breast cancer patients. Anticancer Drugs 13: 353–357 [DOI] [PubMed] [Google Scholar]
- Tamkovich SN, Cherepanova AV, Rykova EY, Pyshnyi DV, Vlassov VV, Laktionov PP (2006) Circulating DNA and DNAse activity in human blood. Ann NY Acad Sci 1080, (in press) [DOI] [PubMed] [Google Scholar]
- Tamkovich SN, Laktionov PP, Rykova EY, Vlassov VV (2004) Simple and rapid procedure suitable for quantitative isolation of low and high molecular weight extracellular nucleic acids. Nucleosides Nucleotides Nucleic Acids 23: 873–877 [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Jones PA (2002) DNA methylation and breast carcinogenesis. Oncogene 21: 5462–5482 [DOI] [PubMed] [Google Scholar]