Abstract
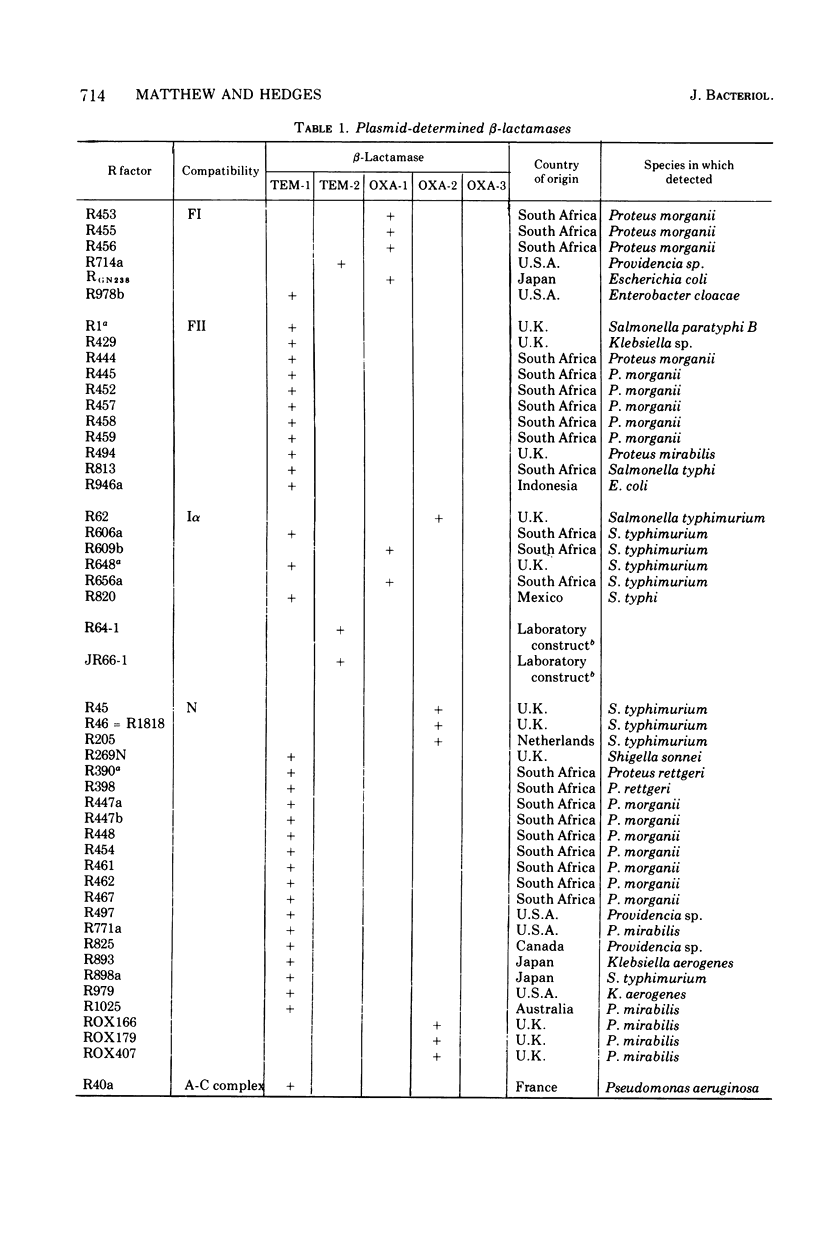
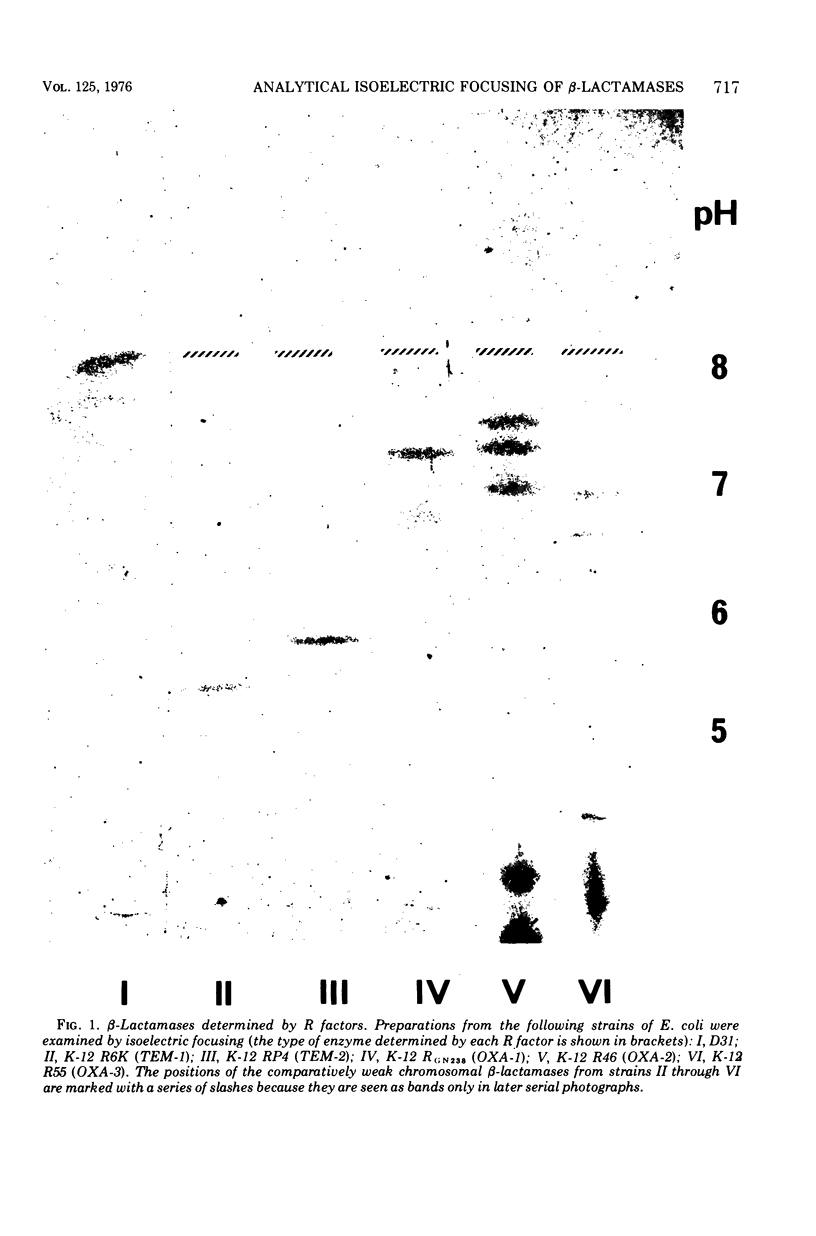
R factor-determined beta-lactamases have been investigated by analytical isoelectric focusing. The enzymes such as those specified by the R6K and RP4 plasmids (TEM-type enzymes) are notably homogenous in biochemical tests (Hedges et al., 1974), but two subclasses can be distinguished by isoelectric focusing. Three subclasses can be distinguished among the oxacillin-hydrolyzing enzymes, in good agreement with the classification based upon biochemical characteristics (Dale and Smith, 1974). The TEM-type beta-lactamases are promiscuously distributed among plasmids of a wide variety of compatibility groups, whereas the various oxacillin-hydrolyzing enzymes show some degree of correlation with compatibility.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burman L. G., Nordström K., Boman H. G. Resistance of Escherichia coli to penicillins. V. Physiological comparison of two isogenic strains, one with chromosomally and one with episomally mediated ampicillin resistance. J Bacteriol. 1968 Aug;96(2):438–446. doi: 10.1128/jb.96.2.438-446.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee J. N., Datta N., Hedges R. W. R factors from Proteus rettgeri. J Gen Microbiol. 1972 Oct;72(3):543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- Dale J. W., Smith J. T. R-factor-mediated beta-lactamases that hydrolyze oxacillin: evidence for two distinct groups. J Bacteriol. 1974 Aug;119(2):351–356. doi: 10.1128/jb.119.2.351-356.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965 Oct 16;208(5007):239–241. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- Guerry P., Falkow S., Datta N. R62, a naturally occurring hybrid R plasmid. J Bacteriol. 1974 Jul;119(1):144–151. doi: 10.1128/jb.119.1.144-151.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Kontomichalou P., Smith J. T. Molecular specificities of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1974 Jan;117(1):56–62. doi: 10.1128/jb.117.1.56-62.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Mobilization of plasmid-borne drug resistance determinants for transfer from Pseudomonas aeruginosa to Escherichia coli. Mol Gen Genet. 1975 Sep 15;140(1):69–79. doi: 10.1007/BF00268990. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Hedges R. W. R factors from Providence. J Gen Microbiol. 1974 Mar;81(1):171–181. doi: 10.1099/00221287-81-1-171. [DOI] [PubMed] [Google Scholar]
- Heffron F., Sublett R., Hedges R. W., Jacob A., Falkow S. Origin of the TEM-beta-lactamase gene found on plasmids. J Bacteriol. 1975 Apr;122(1):250–256. doi: 10.1128/jb.122.1.250-256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A., Harris A. M., Marshall M. J., Ross G. W. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J Gen Microbiol. 1975 May;88(1):169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- Sawai T., Takahashi K., Yamagishi S., Mitsuhashi S. Variant of penicillinase mediated by an R factor in Escherichia coli. J Bacteriol. 1970 Nov;104(2):620–629. doi: 10.1128/jb.104.2.620-629.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]