Abstract
We describe a plant protoplast transformation method that provides transformants with a simple pattern of integration of a foreign gene. The approach is to deliver into plant protoplasts by direct gene transfer the Agrobacterium virulence genes virD1 and virD2 with or without virE2, together with a target plasmid containing a gene of interest flanked by Agrobacterium T-DNA border repeat sequences of 25 bp. We present evidence of T-DNA formation in maize protoplasts and its integration into the maize genome. The frequency of VirD1-VirD2-mediated integration events was about 20–35% of the total number of transformants. The addition of virE2 doubled the transformation efficiency. The method described here is of sufficient efficiency and simplicity to be useful for the production of transgenic plants with single-copy well-defined transgenic inserts.
Plant cell transformation technologies are an important tool for the genetic manipulation and improvement of crop species (1, 2). Various transformation methods allow the direct transfer of foreign genetic material into plant cells and are capable of giving rise to fertile plants. These include the delivery of DNA to protoplasts by electroporation or direct DNA uptake, microinjection, and gene transfer by bombardment with DNA-coated microprojectiles (3). In each of these delivery systems, the foreign genes tend to integrate as multiple copies, which are often fragmented and rearranged (4, 5). Such complex insertion events tend to be unstable. In contrast, Agrobacterium-mediated transformation often results in single-copy or low-copy integration of full-length T-strands carrying intact copies of the transgenes (6).
Agrobacterium has evolved an elaborate mechanism to achieve this orderly and predictable arrangement of transgenes in the plant chromosomal DNA. The bacterium transfers a segment of the Ti plasmid, called T-DNA (transferred DNA), to the nucleus of the plant cell, where it is integrated into the plant nuclear genome (7, 8). T-DNA is defined by 25-bp direct repeats (“border sequences”). DNA situated between such border repeats will be transferred to the plant cell. The processing and transfer of T-DNA are mediated by products encoded by the virulence (vir) region of the Ti plasmid. VirD1 and VirD2 are essential for the generation of the T-strand, the single-stranded copy of T-DNA (7). The gene products of virD1 and virD2 recognize the 25-bp border sequence and produce an endonucleolytic cleavage in the bottom strand between the third and fourth nucleotides of each border sequence (9–11). After nicking, VirD2 remains covalently joined to the 5′ end of the T-strand (12). It has been proposed that VirD2 may also be involved in precise ligation of the 5′ end of the T-strand to plant chromosomal DNA (13). Another protein, VirE2, a single-stranded-DNA-binding protein, is proposed to protect the T-strand against nucleases (14). Both VirD2 and VirE2 contain short peptide sequences that act as nuclear localization signals (NLSs) (15).
While Agrobacterium offers many advantages for plant transformation, its host range limits transformation efficiency in many monocots (16). An ideal transformation system would result in a simple pattern of foreign gene integration without integration of extraneous DNA, and with no host range limitations.
We have previously described an alternative procedure for generating simple insertions of foreign DNA into plant cells. We have termed the procedure “agrolistic.” Agrolistic transformation involves biolistic delivery of plant-expressible forms of virD1 and virD2 genes together with a substrate containing a gene of interest flanked by T-DNA border sequences (17). In the present report, we employ direct gene transfer to protoplasts for introduction of these elements into maize protoplasts. We find that virD1 and virD2 gene products produce T-DNA-type insertion events and that the virE2 gene product can enhance the transformation frequency.
MATERIALS AND METHODS
Plasmids.
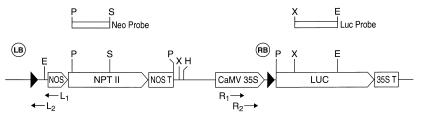
pNeoRBLuc (17) contains a left border sequence, the neomycin phosphotransferase gene (nptII) driven by the nos promoter (nopaline synthase), and the luciferase gene with the right border inserted between the promoter and the luciferase coding region (Fig. 1). The right border sequence (GATCCGGCAGGATATATACCGTTGTAATTCTGCA), corresponding to the right border sequence of LBA5269, was introduced as a synthetic duplex (18). pNeoLuc is the equivalent of pNeoRBLuc (17) with no right border sequence inserted between the cauliflower mosaic virus (CaMV) 35S promoter and the leader of the luciferase coding region. virD1, virD2, and virE2 genes from pTiA6 were subcloned into the expression vector pMF7 (19), which consists of the CaMV 35S promoter (0.5 kb), the Adh1 first intron (0.5 kb), and the nopaline synthase (nos) polyadenylation region (0.25 kb), yielding pInt-D1, pInt-D2, and pInt-E2. The construction of pInt-D1, pInt-D2, pInt-E2, pNeoRBLuc, and pNeoLuc was described previously (17). We have employed the “pInt” designation in this report to refer to the same plasmids previously designated “p35Sadh.”
Figure 1.
Schematic diagram of pNeoRBLuc. LB, left border; RB, right border. The top boxes above the diagram indicate the probe used for Southern blot analysis of transformants. Restriction sites are indicated as follows: E, EcoRI; H, HindIII; P, PstI; X, XbaI. (This figure is adapted from figure 2 of ref. 17.)
Maize Suspension Cells.
The suspension culture of Zea mays cv. Black Mexican Sweet (BMS) was maintained in N6 medium (20) supplemented with 30 g/liter sucrose and 2 mg/liter 2,4-dichlorophenoxyacetic acid (2,4-D) (2N63S). Maize cell suspensions used for experiments were taken from 3-day-old rapidly growing cultures.
Protoplasts were isolated from the cell suspension culture essentially according to Shillito et al. (21), using 2% cellulose RS and 1% Macerozyme R10 (Yakult Pharmaceutical, Japan). Protoplasts were transformed with 10 μg of pNeoRBLuc DNA or 10 μg of pNeoLuc DNA, with or without 20 μg of pInt-D1, 20 μg of pInt-D2, and/or 20 μg of pInt-E2. Paromomycin (200 μg/ml) was included in the medium 2 days after transformation. Independent microcalli that appeared about 4 weeks after transformation were transferred onto fresh plates supplemented with 200 μg/ml paromomycin. After two subcultures on the same medium, suspension cultures were initiated by inoculating about 100 mg of maize cells into 25 ml of liquid medium supplemented with 200 μg/ml paromomycin.
DNA Extraction and Southern Blot Hybridization.
Cell cultures were harvested by filtration 10 days after inoculation and frozen in liquid nitrogen. DNA was isolated as described (22). Approximately 10 μg of genomic DNA digested with EcoRI was applied to each lane of the gel. After separation on a 0.7% agarose gel, the DNA was transferred to Hybond+ membrane and hybridization was performed according to the conditions described by the manufacturer (Amersham). DNA probes were labeled with [α-32P]dCTP by using the Oligolabelling Kit of Pharmacia. The neo probe corresponded to a 0.3-kb PstI–SphI fragment of the nptII gene. The luc probe corresponded to a 0.7-kb XbaI–EcoRI fragment of the luciferase gene. For removal of probes prior to rehybridization, membranes were stripped with a solution of 0.5% SDS at 100°C for 5 min.
Sequence of Insert–Plant DNA Junction.
The junction regions of the introduced and plant genome DNAs were amplified by the thermal asymmetric interlaced (TAIL-) PCR method (23). Briefly, two PCRs were performed sequentially to amplify junction sequences, using nested T-DNA-specific primers on one side and an arbitrary degenerate (AD1) primer on the other side. Two sets of primers were used for two-step TAIL-PCR to determine the junction of right border-plant DNA: the primers AD1 and RB1 were used for the first reaction and AD1 and RB2 for the second reaction. For analysis of the left boundaries, primers AD2 and LB1 were used for the first reaction and primers AD2 and LB2 for the second reaction. TAIL-PCR reactions were performed as described (23). The amplified fragments were subcloned into pGEM-T (Promega) and sequenced.
Primer sequences: AD1, 5′-NGTCGA(G/C)(A/T)GANA(A/Y)GAA-3′, 128-fold degeneracy, average Tm = 46.6°C; RB1, 5′-GAGCATCGTGGAAAAAGAAGAC-3′; RB2, 5′-GACGCACAATCCCACTATCCTT-3′; LB1, 5′-TTCGGAACCACCATCAAACAG-3′; LB2, 5′-AATCAGCTGTTGCCCGTCTCAC-3′).
RESULTS
Experimental Design.
A target plasmid was designed to allow screening for VirD1-and-VirD2-mediated insertion events by a biochemical test. The target plasmid pNeoRBLuc contains, in sequence: a natural left border from a Ti plasmid; a chimeric neomycin phosphotransferase II gene with nopaline synthase promoter and terminator sequences (nos-NPTII-nos) that confers resistance to paromomycin; a CaMV 35S promoter; a synthetic right border sequence; and the luciferase coding region. To express virD1, virD2, and virE2 genes in plant cells, their respective open reading frames were placed under the control of the CaMV 35S promoter. This yielded plasmids pInt-D1, pInt-D2, and pInt-E2. As controls, maize protoplasts were transformed either with pNeoRBLuc alone or with a mixture of plasmids containing the virulence genes together with the borderless control plasmid, pNeoLuc. To identify candidate clones that contain vir-gene-mediated insertions, transformants were selected on paromomycin and screened for the absence of luciferase expression. Because a given cell could have undergone vir-gene-mediated events as well as one or more direct DNA insertions, both luciferase-positive and luciferase-negative transformants were analyzed by Southern blot hybridization using both luc and neo probes. This allows us to deduce the frequency of vir-gene-mediated transformation events among all types of transformants.
Analysis of Stable Transformants.
Table 1 summarizes results of multiple transformation experiments in maize calli. An average of 125 paromomycin-resistant clones appeared per plate when pNeoRBLuc was cotransformed with virD1 and virD2 genes. Similar numbers of calli were recovered when the borderless plasmid, pNeoLuc, was codelivered with virD1 and virD2 genes. An average of 75 calli were recovered per plate from the transformation with pNeoRBLuc alone. This slight change in efficiency can presumably be ascribed to a “carrier” effect because the total DNA concentration is severalfold higher when pInt-D1 and pInt-D2 are included.
The addition of the virE2 gene to the pool of virulence genes resulted in significant increase of paromomycin-resistant calli (average of 278 calli per plate). This increase depended on border sequences, because no increase was observed with the borderless plasmid pNeoLuc co-delivered with pInt-D1 and pInt-D2 (133 calli per plate versus 125 without pInt-E2).
From each set of experiments, 20 paromomycin-resistant clones were analyzed for luciferase activity (Table 1). Control experiments (1, 4, 5, and 6) yielded 0–5% luciferase-negative clones. In contrast, when virD1 and virD2 were codelivered with the target plasmid, 20–30% were found to be luciferase-negative, suggesting an ≈20–30% frequency of virD1–virD2-mediated integration events. There was no significant difference in the frequency of luciferase-negative calli recovered from experiments with or without the virE2 gene.
Southern Blot Analysis of Transformants.
Paromomycin-resistant callus lines obtained after transformations listed in Table 1 were analyzed by Southern blot hybridization.
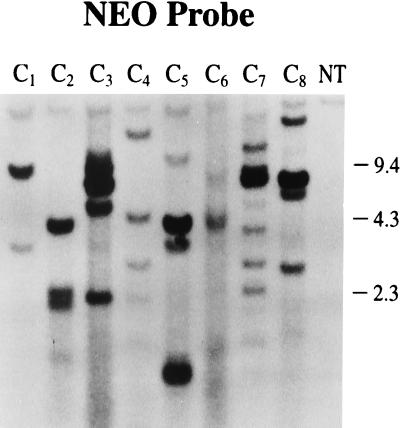
Control callus lines (Fig. 2) recovered from transformation of protoplasts with pNeoRBLuc alone (C1–C4) or with pNeoLuc codelivered with pInt-D1 and pInt-D2 plasmids exhibited high copy number. In addition, multiple copies of rearranged and/or fragmented DNA were seen.
Figure 2.
Southern blot analysis of callus lines transformed with control plasmids. DNA was digested with EcoRI and hybridized with the neo probe. Lane NT, nontransformed control callus. Lanes C1–C4, calli transformed with pNeoRBLuc. Lanes C5–C8, calli transformed with a mixture of pNeoLuc, pInt-D1, and pInt-D2.
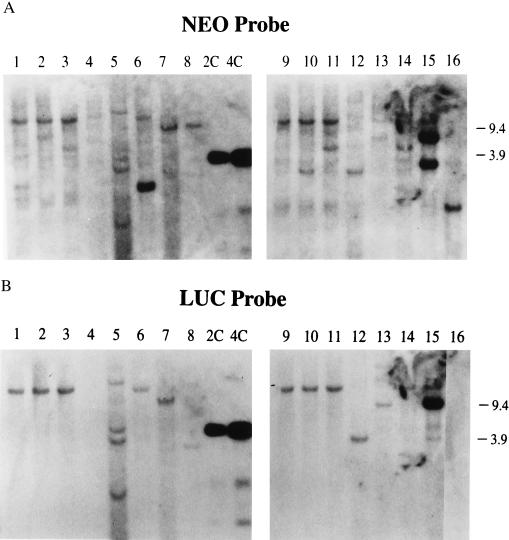
Southern blot analysis of DNA from paromomycin-resitant callus lines obtained after codelivery of target plasmid pNeoRBLuc with pInt-D1 and pInt-D2 with or without pInt-E2 plasmid DNAs is presented in Table 1 and Fig. 3. Of the four luciferase-negative calli from experiment 2A, three contain at least one insert hybridizing with the neo probe that does not hybridize to the luc probe (Fig. 3B). Therefore, these three callus lines appear to have undergone vir-mediated events.
Figure 3.
Southern blot analysis of callus lines transformed with target and helper plasmids. DNA was digested with EcoRI and hybridized with the neo probe (A) or the luc probe (B). Lanes 1, 2, 3, 4, 6, 9, 10, 11, 14, and 16 contain DNA from calli scoring negatively for luciferase activity. Lanes 1–8, calli transformed with a mixture of pNeoRBLuc, pInt-D1, and pInt-D2. Lanes 9–16, calli transformed with a mixture of pNeoRBLuc, pInt-D1, pInt-D2, and pInt-E2.
For comparison, we analyzed four events scoring positive for luciferase activity from experiment 2A. Three lines had undergone only non-vir-mediated events, but one (lane 6) had undergone both types of events. The estimated number of nptII gene copies was 1 or 2 per genome, as judged by a comparison with copy number standards included in the same hybridization filter. We note that there were fewer copies of transgenes in the callus lines expressing the luciferase activity (see lanes 5–8, 12, 13, and 15) recovered after codelivery of the target plasmid with the vir genes than in the callus lines recovered from transformation with pNeoRBLuc DNA alone (lanes C1–C4) or with the borderless plasmid codelivered with the virulence genes plasmid content (lanes C5–C8).
DNA Sequence Analysis of Border Regions of vir-Mediated Integration Inserts.
The clones of four putative vir-mediated integration events, isolated by TAIL PCR (see Materials and Methods), were sequenced outward from a primer just inside T-DNA (Fig. 4). Each sequence matched that of pNeoRBLuc up to the third nucleotide of the T-DNA border, followed by novel, presumably maize, DNA sequence. Sequence analysis of the left junction revealed that the junctions were located in or near the 25-bp repeats. This result demonstrates that insertion of DNA into the maize genome had occurred through T-strand excision and integration. Analysis of the structure of the integrated T-DNA also revealed that, in the presence of virE2, the left end was intact and contained part of the left border sequence. In the absence of VirE2, the left (3′) end was subjected to short deletions.
Figure 4.
Sequence analysis of vir-mediated insert/plant DNA junctions. Sequence 1, clone 2; sequence 2, clone 6; sequence 3, clone 10; sequence 4, clone 12 as indicated on the Southern blots; LB, donor DNA sequence around the left border; RB, donor DNA sequence around the right border. Sequences of the junctions found in transformants are shown above the sequences of the borders of pNeoRBLuc. Sequences presumably originating from the maize genome are underlined. Donor plasmid sequence is italicized except for border repeat sequences, shown in roman type and overlined. Arrows in LB sequence indicate break points for left border clones 1–4.
DISCUSSION
We show evidence of T-DNA formation in planta and its integration into the maize genome after cotransformation of maize protoplasts with plant expression cassettes of Agrobacterium vir genes together with a plasmid harboring the border sequences flanking the selectable marker. About 60% of the transformed maize calli exhibited vir-mediated inserts. Sequence analysis of the junctions between the transgene and plant DNA confirmed that these events occurred through T-strand excision and integration. In some experiments 50% of transformed maize calli had undergone only vir-mediated events (unpublished data).
Taken together with our earlier agrolistic transformation results, the findings reported here demonstrate that, independent of DNA delivery mechanism, once the target and helper DNAs arrive in the plant cell, vir gene products can act upon the target DNA and will result in more simple (and therefore presumably more stable) transgene arrangements in the plant genome. If any helper (vir gene) DNA is integrated during the process, it should be easy to eliminate it through breeding, because it will be integrated at a different locus.
The vir-mediated events have some of the typical features of T-DNA insertions found so far in dicotyledonous plants (6, 8): (i) the right border of the insert ends just after the first 3 nucleotides of the 25-bp repeat; (ii) the right transgene/plant junction is more precisely conserved than the left one; and (iii) the site of insertion in the plant genome does not seem to be specific. The precise integration at the right end of the T-strand has been attributed to the fact that VirD2 remains attached to the 5′ end of the T-strand after cleavage at the right border sequence in Agrobacterium. Such precise insertion also favors the interpretation that VirD2 may be functionally involved in T-strand integration into the plant genome. The finding that the right end of the integrated DNA was conserved in our transformed lines indicates that whatever role VirD2 may play in the Agrobacterium transformation system, it plays a similar role in maize cells. One feature of a typical T-DNA insertion in dicotyledonous plants (6) was, however, not found for vir-mediated inserts in maize: the plant DNA sequences adjacent to the insert were not A+T-rich. This may merely reflect the fact that maize genome has a higher G+C content than the tobacco genome, in which the insertion sites have been studied previously.
In callus lines recovered from a transformation with the target plasmid and the vir plasmids and having undergone non-vir-mediated events, we observed a surprisingly low copy number of the nptII and luc genes compared with that in control lines with or without vir plasmids. This could be explained by a cleavage of the transforming DNA at the border sequences by the Vir proteins before integration into the plant genome. Indeed, the phenomenon must be related to the presence of the border sequences, because no decrease in copy number was observed in callus lines recovered from transformation with the borderless plasmid and the vir genes.
We found that the transformation efficiency increased when virE2 was added to the pool of virulence genes. VirE2 protein binds in a cooperative manner to single-stranded DNA and in doing so is believed to protect the T-strand physically from nucleases. The interaction of VirE2 protein with single-stranded DNA has been shown to protect the DNA from either exonuclease or endonuclease in vitro (24). Furthermore, VirE2 contains a nuclear localization signal that probably assists in nuclear transport of the T-strand into the plant nucleus. There are fewer T-DNA molecules in tobacco protoplasts cocultivated with an Agrobacterium carrying a mutated virE2 gene as compared with the wild-type Agrobacterium (25).
The proportion of vir-mediated events was found to be independent of the presence of the virE2 gene. These results are consistent with the finding that in Agrobacterium-mediated transformation of tobacco, the efficiency of T-DNA integration is independent of VirE2 (14). Interestingly, deletions were found at the 3′ end of the integrated insert in transformation experiments lacking the virE2 gene. When VirE2 was present, the left border region suffered very little or no deletion. Since the calli in our studies were selected for the presence of the nptII gene, which is very close to the left border, it was not possible to select callus containing T-DNA-like inserts with larger deletions.
Our experiments confirm that VirE2 is not essential to the integration process, but it does seem to protect the T-DNA strand from degradation, particularly at the 3′ end.
Our results clearly show that vir-mediated transformation can simplify the pattern of integration of a foreign gene delivered to protoplasts. Furthermore, the addition of virE2 improves the transformation efficiency of vir-mediated integration but only if the target DNA carries a border repeat. These findings will aid in the production of transformed plants with a simple pattern of integration of the gene(s) of interest and with no extraneous DNA.
Table 1.
Recovery of vir-mediated and non-vir-mediated events
| Exp. | Target ± vir genes | Repeat | No. of callus lines* (average) | Luc-negative,† % | Southern‡ | Lanes§ | Type of event¶ |
|---|---|---|---|---|---|---|---|
| 1 | T | A | 80 | 0 | 4 Luc+ | C1–C4 | 4 NV |
| B | 75 (75) | 5 | |||||
| C | 69 | 5 | |||||
| 2 | T + D1 + D2 | A | 120 | 20 | 4 Luc− | 1–4 | 3 V + NV, 1 NT |
| 4 Luc+ | 5–8 | 3 NV, 1V + NV | |||||
| B | 135 (125) | 30 | |||||
| C | 119 | 20 | |||||
| 3 | T + D1 + D2 + E2 | A | 270 | 25 | 5 Luc− | 9, 10, 11, 14, 16 | 5 V |
| 3 Luc+ | 12, 13, 15 | 3 V + NV | |||||
| B | 256 (278) | 30 | |||||
| C | 310 | 20 | |||||
| 4 | C | A | 76 | 0 | 4 Luc+ | Data not shown | 4 NV |
| B | 82 (77) | 0 | |||||
| C | 72 | 5 | |||||
| 5 | C + D1 + D2 | A | 121 | 0 | 4 Luc+ | C5–C8 | 4 NV |
| B | 134 (125) | 0 | |||||
| C | 119 | 5 | |||||
| 6 | C + D1 + D2 + E2 | A | 131 | 5 | |||
| B | 114 (133) | 0 | |||||
| C | 155 | 0 |
Maize protoplasts were transformed with pNeoRBLuc (target plasmid = T) or with pNeoLuc (control plasmid = C) with or without virulence genes virD1; D2, virD2; and E2, virE2.
Callus lines: number of individual callus lines recovered and scored 4 weeks after transformation.
Luc-negative, callus lines not expressing luciferase in 20 lines examined.
Luc+, line contains luciferase gene detected by southern blot analysis; Luc−, no luciferase gene detected by Southern blot analysis.
NV, non-vir-mediated event; V, vir-mediated event; NT, not transformed (escape).
ABBREVIATIONS
- CaMV
cauliflower mosaic virus
- TAIL
thermal asymmetric interlaced
References
- 1.Ahl Goy P, Duesing J H. Bio/Technology. 1995;13:454–458. [Google Scholar]
- 2.Raskin I. Proc Natl Acad Sci USA. 1996;93:3164–3166. doi: 10.1073/pnas.93.8.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrish F M, Fromm M E. Curr Opin Biotechnol. 1992;3:141–146. [Google Scholar]
- 4.Vasil V, Castillo A M, Fromm M E, Vasil I K. Bio/Technology. 1992;10:667–674. [Google Scholar]
- 5.Register J C, III, Peterson D J, Bell P J, Bullock W P, Evans I J, Frame B, Greenland A J, Higgs N S, Jepson I, Jiao S, Lewnau J L, Sillick J M, Wilson H M. Plant Mol Biol. 1994;25:951–961. doi: 10.1007/BF00014669. [DOI] [PubMed] [Google Scholar]
- 6.Koncz C, Németh K, Rédei G P, Schell J. In: Homologous Recombination and Gene Silencing in Plants. Paszkowski J, editor; Paszkowski J, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 167–189. [Google Scholar]
- 7.Zupan J R, Zambryski P. Plant Physiol. 1995;107:1041–1047. doi: 10.1104/pp.107.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tinland B. Trends Plant Sci. 1996;1:178–184. [Google Scholar]
- 9.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasper F, Koncz C, Schell J, Steinbiss H-H. Proc Natl Acad Sci USA. 1994;91:694–698. doi: 10.1073/pnas.91.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheiffele P, Pansegrau W, Lanka E. J Biol Chem. 1995;270:1269–1276. doi: 10.1074/jbc.270.3.1269. [DOI] [PubMed] [Google Scholar]
- 12.Tinland B, Hohn B. In: Genetic Engineering, Principles and Methods. Setlow J K, editor; Setlow J K, editor. New York: Plenum; 1995. pp. 209–229. [Google Scholar]
- 13.Tinland B, Schoumacher F, Gloeckler V, Bravo Angel A M B, Hohn B. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi L, Hohn B, Tinland B. Proc Natl Acad Sci USA. 1996;93:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Citovsky V, Zambryski P. Annu Rev Microbiol. 1993;47:167–197. doi: 10.1146/annurev.mi.47.100193.001123. [DOI] [PubMed] [Google Scholar]
- 16.Chilton M-D. Proc Natl Acad Sci USA. 1993;90:3119–3120. doi: 10.1073/pnas.90.8.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen G, Chilton M-D. Proc Natl Acad Sci USA. 1996;93:14978–14983. doi: 10.1073/pnas.93.25.14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Haaren M J J, Sedee N J A, De Boer H A, Schilperoort R A, Hooykaas P J J. Plant Mol Biol. 1989;13:523–531. doi: 10.1007/BF00027312. [DOI] [PubMed] [Google Scholar]
- 19.Callis J, Fromm M E, Walbot V. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 20.Chu C C, Wang C C, Sun C S, Hsu C, Yin K G, Chu C Y, Bi F Y. Sci Sin. 1975;18:659–668. [Google Scholar]
- 21.Shillito R D, Carswell G K, Johnson C M, DiMaio J J, Harms C T. Bio/Technology. 1989;7:581–587. [Google Scholar]
- 22.Hall G, Allen G C, Loer D S, Thompson W F, Spiker S. Proc Natl Acad Sci USA. 1991;88:9320–9324. doi: 10.1073/pnas.88.20.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y-G, Mitsukawa N, Oosumi T, Whittier R F. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 24.Citovsky V, Wong M L, Zambryski P. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yusibov V M, Steck T R, Gupta V, Gelvin S B. Proc Natl Acad Sci USA. 1994;91:2994–2998. doi: 10.1073/pnas.91.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]