Abstract
Epidermal growth factor receptor (EGFR) mutations are a potential predictor of the effectiveness of EGFR inhibitors for the treatment of lung cancer. Although EGFR mutations were reported to occur with high frequency in nonsmoking Japanese adenocarcinoma patients, the exact nature has not been fully elucidated. We examined EGFR gene mutations within exons 18–21 and their correlations to clinico-pathological factors and other genetic alterations in tumour specimens from 154 patients who underwent resection for lung cancer at Kyoto University Hospital. Epidermal growth factor receptor mutations were observed in 60 tumours (39.0%), all of which were adenocarcinoma. Among the patients with adenocarcinoma (n=108), EGFR mutations were more frequently observed in nonsmokers than former smokers or current smokers (83.0, 50.0, 15.2%, respectively), in women than men (76.3 vs 34.0%), in tumours with bronchio-alveolar component than those without bronchio-alveolar component (78.9 vs 42.9%), and in well or moderately differentiated tumours than poorly differentiated tumours (72.0, 64.4, 34.2%). No tumours with EGFR mutations had any K-ras codon 12 mutations, which were well-known smoking-related gene mutations. In conclusion, adenocarcinomas with EGFR mutation had a distinctive clinico-pathological feature unrelated to smoking. Epidermal growth factor receptor mutations may play a key role in the development of smoking-independent adenocarcinoma.
Keywords: epidermal growth factor receptor, lung adenocarcinoma, smoking, p53, K-ras, single-strand conformation polymorphism
Lung cancer is the leading cause of cancer death in many industrialised countries. Cigarette smoking is the most important cause of lung cancer, and a number of smoking-related gene alternations have been identified that are responsible for the development of lung cancer, such as mutations in K-ras (Vineis and Caporaso, 1995; Shields, 2002). However, lung cancer also develops in nonsmokers, and 30–40% of the lung cancer patients in Japan have never smoked history are female, and their major histological tumour type is adenocarcinoma (Sobue et al, 1994; Akazawa et al, 2003). While several reports have shown that the adenocarcinomas that occurred in nonsmokers were distinct from those that developed in smokers in terms of their histological subclassification, prognosis, gene expression pattern, and gene alterations (Tsuchiya et al, 1988; Hashimoto et al, 2000; Ahrendt et al, 2001; Bhattacharjee et al, 2001; Koga et al, 2001; Noda et al, 2001; Vahakangas et al, 2001; Yang et al, 2002), few significant genetic alterations have been reported in adenocarcinomas that developed in nonsmokers.
Recent laboratory studies have shown that the epidermal growth factor receptor (EGFR, also known as ErbB1 or HER1) plays a critical role in the development and progression of a variety of malignant tumours by promoting cell growth, and by preventing apoptosis through regulation of downstream effectors such as mitogen-activated protein kinase, protein kinase B, and signal transducer and activator of transcription 3 (Klapper et al, 2000). It has been shown in clinical studies that EGFR is overexpressed in 40–80% of non-small-cell lung carcinomas (NSCLCs) (Bunn and Franklin, 2002), and in preneoplastic lesions (Flanklin et al, 2002). These findings suggest that EGFR might have the potential to be an important molecular target for the diagnosis and treatment of NSCLC. However, it is important to note that the exact role of measuring EGFR status in clinical setting remains unclear: EGFR expression status may not be useful as a prognostic tool (Meert et al, 2002), and may not predict responsiveness to treatment with gefitinib, a small-molecule EGFR tyrosine kinase inhibitor (Kris et al, 2003).
Lynch et al (2004) and Paez et al (2004) recently identified specific mutations in the tyrosine kinase domain of the EGFR gene within exons 18, 19, and 21 in most NSCLC patients who responded to gefitinib. Furthermore, Pao et al (2004) reported the presence of a point mutation in exon 20 of the EGFR gene in an adenocarcinoma patient who responded to erlotinib, another EGFR tyrosine kinase inhibitor. Lynch et al (2004) and Pao et al (2004) also showed that cancer cells transfected with EGFR gene mutations showed enhanced tyrosine kinase activity in response to binding of epidermal growth factor and increased sensitivity to gefitinib and erlotinib, suggesting that specific EGFR mutations may predict responsiveness to this type of treatment.
On the other hand, Paez et al (2004) and Pao et al (2004) reported that EGFR mutations were more frequent in female than in male patients and in adenocarcinomas than in tumours of other histological types. Moreover, reports objecting peoples in East Asia (Huang et al, 2004; Kosaka et al, 2004; Shigematsu et al, 2005) reported a half of adenocarcinomas in East Asia patients had EGFR mutations and the absence of smoking history, mainly seen in female patients, were closely linked to EGFR mutations. These reports are not only important in determining which patients should receive EGFR-targeted treatment (Arteaga, 2004; Dancey, 2004) but also indicate that EGFR mutations may play a causal role in the development of lung adenocarcinoma in nonsmokers.
To confirm the correlation of EGFR mutations with smoking, we conducted a detailed study of EGFR gene mutations in NSCLC patients who underwent tumour resection at a particular Japanese hospital. In the study, mutations of p53 gene and K-ras gene codon 12, and promoter hypermethylation status of p16, RASSF1A, and APC1A gene, were also examined because these gene alterations had substantial role in pathogenesis of NSCLC and whether or not they correlated to EGFR mutations could help to further clarify the importance of EGFR mutations on pathogenesis of NSCLC. In addition, we report the detection procedure of EGFR mutation using polymerase chain reaction-single strand conformational polymorphism (PCR–SSCP) method (Orita et al, 1989). As PCR–SSCP method is very suitable for detecting mutations within a relatively limited region such as EGFR gene mutations and has been already used to detect p53 gene mutations commercially, the method can be more easily applied to detect EGFR mutations in clinical setting than direct sequence.
MATERIALS AND METHODS
Patients and data collection
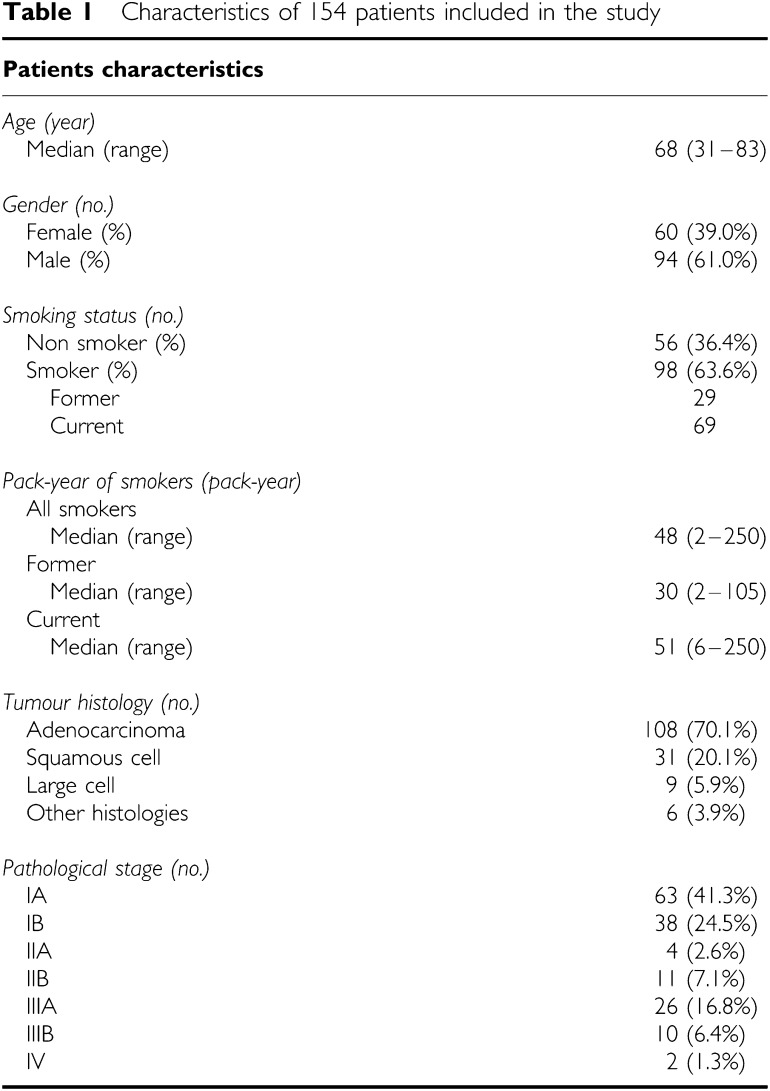
A total of 154 consecutive patients with NSCLC who underwent resection at the Department of Thoracic Surgery, Kyoto University Hospital, from January 2003 to November 2004 were included in the present study (Table 1). Clinical data of patients involved were obtained from the inpatient and outpatient medical records, chest X-ray films, whole-body computed tomography films, bone scanning data, and operation records. On smoking status, we classified patients into nonsmokers and smokers, and subclassified smokers into former smoker (who stop smoking at least 6 months before the time diagnosis of NSCLC) and current smoker. In total, 142 patients underwent complete lobe or segment in which the tumour existed and also received hilar and mediastinal lymphnode dissections. A total of 13 patients received partial lung resection and lymphnode sampling. No patient was exposed to gefitinib before his or her tumour was resected. Pathological staging was determined using the current tumour-node-metastasis classification system (UICC, 1997). The histological type and differentiation grade of the tumours in these patients were determined using the. Pathological diagnosis were performed by two pathologists, unaware of the genetic information, of Kyoto University Hospital Laboratory of Anatomic Pathology, and finally confirmed by one pathologist (TM) according to the WHO classification system (Travis et al, 1999). Many of adenocarcinomas were classified into mixed subtype according to WHO classification system. Thus, to clarify the impact of EGFR gene mutations on subtypes of adenocarcinoma, classification according to the presence or absence of each component (BAC, papillary, acinar, and solid carcinoma with mucin (solid)) in a tumour was also employed in this study. A written informed consent to perform genetic analyses was obtained from all patients before surgery, and the study itself was approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine.
Table 1. Characteristics of 154 patients included in the study.
| Patients characteristics | |
|---|---|
| Age (year) | |
| Median (range) | 68 (31–83) |
| Gender (no.) | |
| Female (%) | 60 (39.0%) |
| Male (%) | 94 (61.0%) |
| Smoking status (no.) | |
| Non smoker (%) | 56 (36.4%) |
| Smoker (%) | 98 (63.6%) |
| Former | 29 |
| Current | 69 |
| Pack-year of smokers (pack-year) | |
| All smokers | |
| Median (range) | 48 (2–250) |
| Former | |
| Median (range) | 30 (2–105) |
| Current | |
| Median (range) | 51 (6–250) |
| Tumour histology (no.) | |
| Adenocarcinoma | 108 (70.1%) |
| Squamous cell | 31 (20.1%) |
| Large cell | 9 (5.9%) |
| Other histologies | 6 (3.9%) |
| Pathological stage (no.) | |
| IA | 63 (41.3%) |
| IB | 38 (24.5%) |
| IIA | 4 (2.6%) |
| IIB | 11 (7.1%) |
| IIIA | 26 (16.8%) |
| IIIB | 10 (6.4%) |
| IV | 2 (1.3%) |
Tumour sample collection
Tumour tissues were frozen immediately after resection, and were stored at −80°C until DNA extraction. A part of each tumour tissue was used for formalin-fixed, paraffin-embedded tissue block to confirm that tumour cells were sufficiently included within the sample. Genomic DNA was extracted using the FastDNA® Kit (Qbiogene Inc., USA) as recommended by the manufacturer. As EGFR mutations were reported to be somatic, corresponding nonmalignant lung tissues were analysed in only six cases harbouring EGFR mutations in their tumour tissues, and all had no EGFR mutations in their nonmalignant lung tissues.
Mutation detection and nucleotide sequence analysis of the EGFR and p53 genes
Polymerase chain reaction-single strand conformational polymorphism (Orita et al, 1989) was used to screen for mutations in the EGFR gene within exons 18–21 and for mutations in the p53 gene within exons 5–8. Polymerase chain reaction amplification was performed using the HotStarTaq Master Mix (Qiagen, Germany); the primers used and PCR conditions are listed in Table 2. Single-strand conformational polymorphism analyses were performed using the GenePhor System and GeneGel Excel 12.5/24 (Amersham Biosciences, Sweden) following the manufacturer's protocol; the gel temperature was maintained at 10°C for SSCP analysis of exon 21 of the EGFR gene and exon 6 of the p53 gene, 15°C for exons 18, 19, and 20 of the EGFR gene, and 18°C for exons 5, 7, and 8 of the p53 gene. After the gels were stained with silver carbonate, altered bands were cut from the gels and DNA fragments were eluted for direct sequencing. Each mutation within exon 19 of the EGFR gene was expediently named according to the start point of the amino-acid change and its order of detection, as shown in Table 3.
Table 2. PCR primers and parameters.
| Gene | Forward (5′–3′) | Reverse (5′–3′) | Product size (bp) | Number of cycle | Annealing condition |
|---|---|---|---|---|---|
| EGFR | |||||
| Exon 18 | TACACCCAGTGGAGAAGCTCC | CCCCACCAGACCATGAGAG | 169 | 30 | 58°C, 30 s |
| Exon 19 | CAATTGCCAGTTAACGTCTTCC | GGAGATGAGCAGGGTCTAGAG | 239 | 30 | 58°C, 30 s |
| Exon 20 | CACACTGACGTGCCTCTC | CTTATCTCCCCTCCCCGTA | 252 | 30 | 56°C, 30 s |
| Exon 21 | AGGGCATGAACTACTTG | CCTCCTTACTTTGCCTCCTTC | 167 | 35 | 55°C, 30 s |
| p53 | |||||
| Exon 5 | TTCAACTCTGTCTCCTTCCT | CAGCCCTGTCGTCTCTCCAG | 248 | 30 | 55°C, 30 s |
| Exon 6 | GCCTCTGATTCCTCACTGAT | TTAACCCCTCCTCCCAGAGA | 181 | 30 | 55°C, 30 s |
| Exon 7 | CTTGCCACAGGTCTCCCCAA | TGTGCAGGGTGGCAAGTGGC | 196 | 30 | 59°C, 30 s |
| Exon 8 | TTCCTTACTGCCTCTTGCTT | CGCTTCTTGTCCTGCTTGCT | 201 | 30 | 55°C, 30 s |
| K-ras | ACTGAATATAAACTTGTGGTAGTTGGACCT | CTGTATCAAAGAATGGTCCTGCACCAGTA | 162 | 30 | 58°C, 30 s |
| p16 | |||||
| Unmethylated | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 151 | 35 | 60°C, 30 s |
| Methylated | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | 150 | 35 | 65°C, 15 s |
| RASSF1A | |||||
| Methylated | GGGTTTTGCGAGAGCGCG | GCTAACAAACGCGAACCG | 169 | 35 | 64°C, 50 s |
| APC1A | |||||
| Methylated | TATTGCGGAGTGCGGGTC | TCGACGAACTCCCGACGA | 98 | 35 | 62°C, 10 s |
Table 3. Types of EGFR gene mutations found in this study.
| Exon | Type of sequence | Alteration | Nucleotide alteration | Amino-acid alteration | No. of cases |
|---|---|---|---|---|---|
| 18 | Substitution | 2156G>C | G719A | 1 | |
| Substitution | 2159C>T | S720F | 1 | ||
| 19 | Type 1 | ||||
| 1a | Deletion | 2235–2249delGGAATTAAGAGAAGC | E746-A750del | 14 | |
| 1b | Deletion | 2236–2250delGAATTAAGAGAAGCA | E746-A750del | 8 | |
| 1c | Deletion+ | 2235–2249delGGAATTAAGAGAAGC | E746-A750del | 1 | |
| Substitution | 2251A>G | T751A | |||
| 1d | Deletion+ | 2235–2236delGC | E746-A750del | 1 | |
| Substitution | 2242–2248delAGAGAAG | ins I and P | |||
| 2241A>C | |||||
| 1e | Deletion | 2235–2236delGC | E746-T751del | 2 | |
| 2239–2252delTAAGAGAAGCAAC | ins I | ||||
| 1f | Deletion+ | 2235–2236delGC | E746-T751del | 1 | |
| Substitution | 2242–2251delAGAGAAGCAA | ins I and P | |||
| 2241A>C | |||||
| 1g | Deletion+ | 2237–2254delAATTAAGAGAAGCAACAT | E746-S752del | 1 | |
| Substitution | 2255C>T | ins V | |||
| Type 2 | |||||
| 2a | Deletion+ | 2240–2248delTAAGAGAAG | L747-A750 | 1 | |
| Substitution | 2239T>C | insP | |||
| 2b | Deletion+ | 2240–2251delTAAGAGAAGCAA | L747-T751del | 2 | |
| Substitution | 2239T>C | ins S | |||
| 2c | Deletion | 2239–2253delTTAAGAGAAGCAACA | L747-T751del | 1 | |
| 2d | Deletion | 2239–2256delTTAAGAGAAGCAACATCT | L747-S752del | 1 | |
| 2e | Deletion | 2240–2257delTAAGAGAAGCAACATCTC | L747-P753del | 1 | |
| ins S | |||||
| 20 | Insertion+ | 2308ins/dup(CCAGCGTGG) | ins779(ASV) | 1 | |
| Substitution | 2310C>T, 2315C>G | P782R | |||
| Insertion+ | 2311ins/dup(GCGTGGACA) | ins780(SVD) | 1 | ||
| Substitution | 2315C>G | P782R | |||
| 21 | Substitution | 2573T>G | L858R | 20 | |
| Substitution | 2572–2573CT>AG | L858R | 1 | ||
| Substitution | 2573–2574TG>GT | L858R | 1 | ||
Detection of mutations in codon 12 of the K-ras gene
We used a modified mutagenic PCR–RFLP method (Hatzaki et al, 2001) for screening mutations in codon 12 of the K-ras gene. PCR primers and the amplification conditions are shown in Table 2. The PCR products of mutated K-ras genes were sequenced for confirmation of mutation.
Promoter hypermethylation analysis
To detect promoter hypermethylation of the p16, RASSF1A, and APC1A genes, methylation-specific PCR method was used (Herman et al, 1996). PCR primers and the amplification conditions were listed in Table 2 (Herman et al, 1996; Burbee et al, 2001; Yu et al, 2002).
Statistical analyses
The significance of differences in categorical data was tested using the χ2 or Fisher's exact test. Differences between continuous variables were examined using the Mann–Whitney U-test. To determine which of gender or smoking history, or which component of subtype of adenocarcinoma affected EGFR mutations, logistic regression analyses were performed. StatView software (version 5, SAS Institute, USA) was used to carry out all statistical calculations. All statistical tests were two-sided, and differences were considered to be statistically significant if the P-value was less than 0.05.
RESULTS
Epidermal growth factor receptor mutations in non-small-cell lung carcinomas
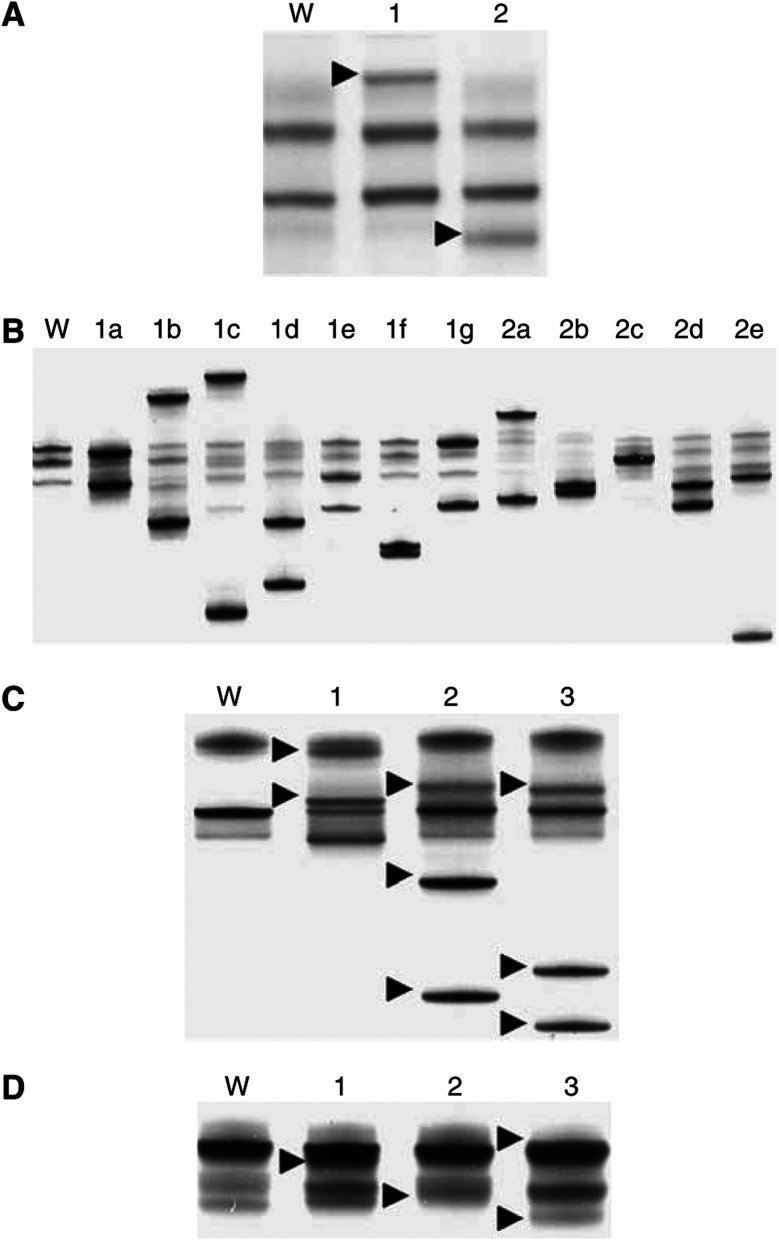
A total of 61 mutations in the EGFR gene were found in 60 of our patients (39.0%). Mutations occurred within exon 18 in two patients (1.3%), exon 19 in 34 patients (21.9%), exon 20 in three patients (1.9%), and exon 21 in 22 patients (14.2%), respectively. One patient had mutations within exon 19 and 20. Polymerase chain reaction-single strand conformational polymorphism analysis revealed two types of altered bands in exon 18 (Figure 1A), 12 types in exon 19 (Figure 1B), three types in exon 20 (Figure 1C), and three types in exon 21 (Figure 1D), and nucleotide sequencing confirmed the presence of the corresponding mutations shown in Table 3. The mutations identified within exon 18 were point mutation of 2156G>C (n=1), which substituted alanine for glycine at codon 719, and point mutation of 2159C>T (n=1), which substituted phenylalanine for serine at codon 720 (Table 3). A total of 12 types of mutations were discovered around codon 747–750 within exon 19, and 2235–2249del (n=14) and 2236–2250del (n=8) were major types (Table 3). The mutations identified within exon 20 were point mutation of 2361G>A and 2407C>A (silent mutation) observed in one patient who had deletion mutation within exon 19, and two types of duplication/insertion with point mutation (Table 3). The mutations identified within exon 21 were point mutation of 2573T>G (n=20), 2572–2573CT>AG (n=1), and 2573–2574TG>GT (n=1). All these mutations provided amino-acid substitution of arginine in the place of leucine at codon 858 (Table 3). Epidermal growth factor receptor mutations were exclusively observed in adenocarcinoma patients; the incidence of EGFR mutations in adenocarcinoma patients was 55.6% (60/108).
Figure 1.
Single-strand conformation polymorphism of the EGFR gene. Each band alteration corresponds to a specific gene mutation. (A) Exon 18. W: wild type, 1: 2156G>C, 2: 2159C>T. Allow heads: altered bands. (B) Exon 19. W: wild type. The designated types of mutation (1a–1g, 2a–2e) in exon 19 were those described in Table 3. (C) Exon 20. W: wild type, 1: 2361G>A and 2407C>A, 2: 2308ins/dup(CCAGCGTGG) with 2310C>T and 2315C>G, 3: 2311ins/dup(GCGTGGACA) with 2315C>G. Allow heads: altered bands. (D) Exon 21. W: wild type, 1: 2573T>G, 2: 2573–2573CT>AG, 3: 2573–2574TG>GT. Allow heads: altered bands.
Other genetic alterations
In total, 52 mutations within exons 5–8 of the p53 gene were observed in 51 patients (33.1%). The frequency distribution of these mutations was as follows: 12 missense point mutations and one duplication/insertion within exon 5; seven missense point mutations, two deletions, and two duplication/insertion within exon 6; four missense point mutations, three duplication/insertion, and two deletions within exon 7; and 15 missense point mutations, and four deletions within exon 8.
Mutations within codon 12 of the K-ras gene were observed in 10 patients (6.5%) with the following frequency: substitution of cystein (TGT) in the place of glycine (n=4), aspartic acid (GAT) (n=3), serine (AGT) (n=1), valine (GTT) (n=1), and phenylalanine (TTT) (n=1). All 10 patients with K-Ras mutated tumours were smokers.
Promoter hypermethylation of p16, RASSF1A, or APC1A genes was observed in 64 (41.6%), 78 (50.6%), or 82 (53.2%) of 154 patients, respectively.
Correlation of clinico-pathological characteristics and other genetic alterations with EGFR mutations in lung adenocarcinoma
As EGFR mutations were observed exclusively in adenocarcinoma patients, we investigated the relationship between the clinical features and other genetic alterations in these adenocarcinoma patients (n=108) and their mutations (Table 4). The incidence of EGFR mutations was higher in female patients than in male patients (76.3 vs 34.0%, P<0.001, odds ratio: 6.3, 95% confidence intervals: 2.7–14.7). The incidence of EGFR mutations was higher in nonsmokers than in former smokers (83.0 vs 50.0%, P=0.008, odds ratio: 4.9, 95% confidence intervals: 1.5–15.7), and higher in former smokers than in current smokers (50.0 vs 15.2%, P=0.007, odds ratio: 5.6, 95% confidence intervals: 1.4–21.7). There was a significant correlation between gender and smoking status in these patients; 48 of 53 male patients (90.6%) were smokers, whereas only seven of 55 female patients (12.7%) were smokers (P<0.001, odds ratio: 65.8, 95% confidence intervals: 23.3–186.1). Logistic regression analysis revealed absence of smoking history, not female, affected EGFR gene mutation (P<0.001, odds ratio: 11.4, 95% confidence intervals: 2.7–47.3).
Table 4. Relationship of EGFR gene mutations to clinicopathological characteristics and other genetic/epigenetic alterations in 108 patients with adenocarcinoma.
Epidermal growth factor receptor mutations were more frequently found in patients with lower pathologic stage disease although there was no statistical significance (P=0.059) (Table 4).
The histological subtype of adenocarcinomas according to WHO classification did not correlate statistically with their EGFR incidence of mutations. Epidermal growth factor receptor mutations were, however, more frequently observed in tumours with BAC component than those without BAC component (78.9 vs 42.9%, P<0.001, odds ratio: 5.0, 95% confidence intervals: 2.0–12.6). By contraries, EGFR mutations were less frequently observed in tumours with solid component than those without solid component (34.2 vs 67.1%, P=0.001, odds ratio: 0.25, 95% confidence intervals: 0.11–0.61). Papillary component and acinar component did not correlate to the incidence of EGFR gene mutations (Table 4). Logistic regression analysis revealed that BAC component positively related to EGFR gene mutations (P=0.006, odds ratio: 3.9, 95% confidence intervals: 1.5–10.1) and solid component inversely related to EGFR gene mutations (P=0.035, odds ratio: 0.36, 95% confidence intervals: 0.14–0.93). The differentiation grade of the tumours correlated significantly with their incidence of EGFR mutations. Thus, incidence of EGFR mutations was lower in poorly differentiated tumours than in well-differentiated tumours (34.2 vs 72.0%, P=0.005, odds ratio: 4.9, 95% confidence intervals: 1.5–15.9) or than in moderately differentiated tumours (34.2 vs 64.4%, P=0.008, odds ratio: 3.5, 95% confidence intervals: 1.3–9.2) (Table 4).
K-ras gene mutations were not detected in any of the EGFR mutated tumours and this negative correlation was statistically significant (P=0.001), while EGFR gene mutation status did not correlate with p53 gene mutation status or with promoter hypermethylation status of p16, RASSF1A, or APC1A gene (Table 4).
The detailed type of EGFR mutation did not correlate with gender, smoking status, p-stage, histological subtype, grade of tumour differentiation (data not shown).
DISCUSSION
In the present study, we detected EGFR gene mutations in 60 of 154 Japanese patients (39.0%) who underwent resection for NSCLC. The demonstration of a high prevalence of these mutations in our Japanese patients was consistent with previous data that NSCLC occurred in people in the East Asia including Japan showed higher prevalence of EGFR mutation (19–40%) than those in other patient groups (4–10%) (Table 5). Our study clearly showed that EGFR mutations were not only observed in advanced NSCLC that may be considered for gefitinib treatment but also in early NSCLC. This suggests that such mutations is involved in the early stage of oncogenesis of NSCLC, so that EGFR mutations should be investigated further in regard to oncogenesis of NSCLC, as well as considered for design of clinical trial of or selecting candidates of EGFR-targeting drugs.
Table 5. Summary of previous reports on EGFR gene mutations in NSCLC.
| Reference | Patient group | No. of patients |
Frequency in
|
Predilection in non/former smokers | Histological features in Ada | Respose to TKIbof patients with EGFR mutated tumour | Correlation to p53 | Correlation to K-ras | |
|---|---|---|---|---|---|---|---|---|---|
| Overall | Ada | ||||||||
| Lynch et al (2004) | USA | 25 +16 TKIb receivers | 2/25 | 2/22 | Yes | More frequent in BACc | 8 patients in 9 responders not in 7 nonresponders | — | — |
| Paez et al (2004) | USA Japan | 119 +9 TKIb receivers | 16/119 (13%) 1/61(2%) in USA 15/58 (26%) in Japan | 15/70 (21%) 1/29(3%) in USA 14/41(34%) in Japan | Yes | — | 5 patients in 5 responders not in nonresponders | — | — |
| Pao et al (2004) | USA | 96 +17 TKI b responders | 11/96 (11%) | — | Yes | — | 12 patients in 17 responders | — | — |
| Kosaka et al (2004) | Japan | 277 | 111/277 (40%) | 110/224 (49%) | Yes | More frequent in well-to-mederately differentiated Ada | — | Independent | Mutually exclusive |
| Huang et al (2004) | Taiwan | 101 | 39/101 (39%) | 38/69 (55%) | Yes | — | 7 responders in 9 patients | — | — |
| Marchetti et al (2005) | Italy | 860 | 37/860 (4%) | 37/375 (10%) | Yes | More frequent in Ad with BACc features | — | — | Mutually exclusive |
| Han et al (2005) | Korea | 90 | 17/90 (19%) | 14/65 (22%) | No | — | 11 responders in 17 patients | — | — |
| Shigematsu et al (2005) | Japan Taiwan USA Australia | 519 | 120/519 (23%) 107/361 (30%) in East Asia 13/158(8%) in others | 114/289 (39%) 102/214 (48%) in East Asia 12/75(16%) in others | Yes | No correlation to Ad with BACc fearures | — | — | Mutually exclusive |
| Qin et al (2005) | China | 41 | 10/41 (24%) | 7/17 (41%) | No | — | — | — | — |
| Yang et al (2002) | USA | 219 | 26/219 (12%) | 25/164 (15%) | Yes | — | — | — | — |
Ad=adenocarcinoma.
TKI=tyrosine kinase inhibitor.
BAC=bronchio-alveolar carcinoma.
A total of 15 base pair deletions within exon 19 and a point mutation within exon 21 were two of the major types of mutations that were found in our patients. Among 11 types of EGFR mutations within exon 19, mutations of the type 1 series, in which the start point for amino acid deletion was E746, were most frequent. This finding was somewhat different from the findings in US patients, in whom mutations that had their start point for amino acid deletion at L747 were frequent (Lynch et al, 2004; Paez et al, 2004), but was similar to those in other reports (Huang et al, 2004; Kosaka et al, 2004; Pao et al, 2004; Han et al, 2005; Marchetti et al, 2005; Qin et al, 2005; Shigematsu et al, 2005; Yang et al, 2005). These differences may reflect ethnic and/or social differences among patient groups although the net effect of a deletion in the type 1 or 2 series on sensitivity to EGFR tyrosine kinase inhibitors seems similar (Huang et al, 2004; Lynch et al, 2004; Paez et al, 2004; Pao et al, 2004; Han et al, 2005).
It is interesting that both mutations in exons 19 and 21 were found with higher frequency (34/154 and 22/154, respectively) in our patients than in patients outside of the East Asia (Table 5), despite the type of each mutation were different (a deletion on exon 19 and a point mutation on exon 21). This finding suggests that one or just a few substances with DNA editing capacity may mediate both deletions and point mutations within the tyrosine kinase domain of the EGFR. Clarification of the mechanism by which EGFR mutations occur can lead to advances in our understanding of oncogenesis and its prevention.
Smoking history strongly affected the incidence of EGFR mutations in our study. In addition to higher incidence of EGFR mutations in adenocarcinomas developed in nonsmokers, we showed that the incidence of EGFR mutations was higher in former smokers than in current smokers. Former smokers had lower pack-year (Table 1) and may be less affected by smoking. Probably smoking is not involved in mechanisms of EGFR gene mutations.
Although the incidences of EGFR mutations were higher in female patients and in nonsmokers, a history of smoking strongly correlated with gender in our study. We showed that the absence of smoking history, but not female, independently affected EGFR gene mutations. This result is similar to the data of Kosaka et al (2004), but is different from the data of Marchetti et al, which indicated the absence of smoking history and female sex independently influenced to EGFR mutations. This difference may be derived from social difference on smoking between Japan and Italy. At least, in NSCLC occurred in Japanese people, the absence of smoking history, not female gender, seems a critical factor that links to the prevalence of EGFR mutations.
We found that BAC component positively related to EGFR mutations, solid component inversely related to EGFR gene mutations, and that well-to-moderately differentiated adenocarcinomas had higher incidence of EGFR gene mutations than poorly differentiated adenocarcinomas. These findings are consistent with Lynch et al (2004), Kosaka et al (2004), and Marchetti et al (2005). Adenocarcinomas that develop in nonsmokers frequently display features of BAC and papillary type tumours, whereas those that develop in smokers frequently include poorly differentiated and solid subtype tumours (Hashimoto et al, 2000; Yang et al, 2002; Nordquist et al, 2004). High incidence of harbouring EGFR mutations in nonsmokers adenocarcinoma well-explains this predilection on pathological findings. Thus, including BAC component and well-to-moderate differentiation grade seem histological features of adenocarcinomas with EGFR mutations.
K-ras mutations were not observed in any of the EGFR-mutated tumours in our patients. This feature is similar to reports referring EGFR mutations and K-ras mutations (Table 5). Both K-ras and EGFR are important molecules that are responsible to the regulation of the mitogen-activated protein kinase pathway. But K-ras mutations are linked to the development of adenocarcinomas in smokers (Ahrendt et al, 2001), and are rarely observed in adenocarcinomas in nonsmokers (Noda et al, 2001). This is in marked contrast to EGFR mutations, which occur more frequently in nonsmokers. Probably, EGFR mutations have a similar significance to K-ras mutations in oncogenesis of lung adenocarcinomas. Unlike K-ras mutations involved in adenocarcinomas developed in smokers, however, EGFR gene mutations may play a key role in the development of adenocarcinomas in nonsmokers.
Similar to Kosaka et al (2004), EGFR mutations did not correlate to p53 gene mutations in our study. Moreover, we showed that EGFR mutations did not correlate to promoter hypermethylation status of p16, RASSF1A, or APC1A genes. In the multistep progression of sporadic colorectal carcinomas, K-ras mutations are thought to occur independently at a different step from that of p53 mutations (Klump et al, 2004). Similar situation is seen in K-ras mutations, p53 mutations, and promoter hypermethylation of p16 gene in pancreatic cancer (Moore et al, 2003). In NSCLC, EGFR, p53, p16, RASSF1A, or APC1A can be involved in oncogenesis at a different level from one another (Rom et al, 2000). Therefore, alterations of these genes can independently occur in lung adenocarcinomas, unlike mutations of EGFR and K-ras.
We used the PCR–SSCP method (Orita et al, 1989) to screen for EGFR mutations. Advantages of the PCR–SSCP method are: it is fast and easily employed for screening numerous samples simultaneously, it indicates type-specific mutations without nucleotide sequence because one altered SSCP-band pattern can correspond to a specific mutation, and it has higher sensitivity than direct sequence sufficient for clinical use (Marchetti et al, 2005). The PCR–SSCP method can be theoretically applied not only to resected tumour samples but also to sputum, pleural effusion, and biopsy specimens; as such, this technique can be used to preselect appropriate patients for EGFR tyrosine kinase inhibitor treatment.
This study has several limitations. As only surgically resected tumours were involved in the study, the incidence of EGFR mutations in the study could not indicate the incidence in whole NSCLCs. Mutations outside exons 18–21 were not examined, and PCR–SSCP method has possibility to overlook mutations that is not reflected on band pattern alterations, so that the incidence of EGFR may be underestimated. The incidence of EGFR mutation and types of mutations detected in our study are, however, quite similar to previous reports; therefore, we think our study has sufficient validity.
In conclusion, mutations in the EGFR gene were found in approximately half of our Japanese adenocarcinoma patients, and were more common in tumours developed in nonsmokers. Adenocarcinomas with EGFR mutations displayed inclusion of bronchio-alveolar component, or well-to-moderately differentiated features, which are also one of the histological features of adenocarcinomas in nonsmokers. Adenocarcinomas with EGFR mutations negatively correlated with K-ras mutations that are known to be associated with smoking. Thus, EGFR mutations may play a role in the aetiology of adenocarcinoma in nonsmokers.
Acknowledgments
We thank Drs Kazumasa Takenaka, Seiji Matsumoto, Shinya Ito, and Masatsugu Nakagawa (Department of Thoracic Surgery, Kyoto University Hospital) for their help in collecting clinical samples and for their technical assistance, and Drs Hiromichi Katakura and Masashi Adachi (Department of Thoracic Surgery, Kyoto University Hospital) for their help in writing this report. We thank Drs Daisuke Harada, Hirokazu Kotani, and Yoshiaki Ueda (Laboratory of Anatomic Pathology, Kyoto University Hospital) for pathological diagnosis of tumours included in this study. No funding for this project was received from any sponsors.
References
- Ahrendt SA, Decker PA, Alawi EA, Zhu Yr YR, Sanchez-Cespedes M, Yang SC, Haasler GB, Kajdacsy-Balla A, Demeure MJ, Sidransky D (2001) Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer 92: 1525–1530 [DOI] [PubMed] [Google Scholar]
- Akazawa Y, Satoh H, Takeda YY, Takiguchi K, Ishikawa H, Ohtsuka M, Sekizawa K (2003) Significantly lower rate of smoking in female compared to male patients with lung adenocarcinoma. Eur J Cancer Care 12: 283–285 [DOI] [PubMed] [Google Scholar]
- Arteaga CL (2004) Selecting the right patient for tumor therapy. Nat Med 10: 577–578 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 98: 13790–13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn Jr PA, Franklin W (2002) Epidermal growth factor receptor expression, signal pathway, and inhibitors in non-small cell lung cancer. Semin Oncol 29(Suppl 14): 38–44 [DOI] [PubMed] [Google Scholar]
- Burbee DG, Forbacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White E, Minna JD (2001) Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 93: 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancey JE (2004) Predictive factors for epidermal growth factor receptor inhibitors – the bull's-eye hits the arrow. Cancer Cell 5: 411–415 [DOI] [PubMed] [Google Scholar]
- Flanklin WA, Veve R, Hirsch FR, Helfrich BA, Bunn Jr PA (2002) Epidermal growth factor receptor family in lung cancer and premalignancy. Semin Oncol 29(Suppl 4): 3–14 [DOI] [PubMed] [Google Scholar]
- Han SW, Kim TY, Hwang PG, Jeong S, Kim J, Choi IS, Oh DY, Kim HJ, Kim DW, Chung DH, Im SA, Kim YT, Lee JS, Heo DS, Bang YJ, Kim NK (2005) Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol 23: 2493–2501 [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Tokuchi Y, Hayashi M, Kobayashi Y, Nishida K, Hayashi S, Ishikawa Y, Nakagawa K, Hayashi J, Tsuchiya E (2000) Different subtypes of human lung adenocarcinoma caused by different etiological factors. Evidence from p53 mutational spectra. Am J Pathol 157: 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzaki A, Razi E, Anagnostopoulou K, Iliadis K, Kodaxis A, Papaioannou D, Labropoulos S, Vasilaki M, Kosmidis P, Saetta A, Mihalatos M, Nasioulas G (2001) A modified mutagenic PCR-RFLP method for K-ras codon 12 and 13 mutations detection in NSCLC patients. Mol Cell Probes 15: 243–247 [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, Wu YC, Chen YR, Tsai SF (2004) High frequency of epidermal growth factor receptor mutations with complez patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 10: 8195–8203 [DOI] [PubMed] [Google Scholar]
- Klapper LN, Kirschbaum MH, Sela M, Yarden Y (2000) Biochemical and clinical implications of the ErbB/HER signaling network of growth factor receptors. Adv Cancer Res 77: 25–79 [PubMed] [Google Scholar]
- Klump B, Nehls O, Okech T, Hsieh C-J, Gaco V, Gittinger DS, Sarbia M, Borchard F, Greschniok A, Gruenagel HH, Porshen R, Gregor M (2004) Molecular lesions in colorectal cancer: impact on prognosis? Original data and review of the literature. Int J Colorectal Dis 19: 23–42 [DOI] [PubMed] [Google Scholar]
- Koga T, Hashimoto S, Sugio K, Yoshino I, Mojtahedzadeh S, Matsuo Y, Yonemitsu Y, Sugimachi K, Sueishi K (2001) Clinicopathological and molecular evidence indicating the independence of bronchioloalveolar components from other subtypes of human peripheral lung adenocarcinoma. Clin Cancer Res 7: 1730–1738 [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T (2004) Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 64: 8919–8923 [DOI] [PubMed] [Google Scholar]
- Kris MG, Natale RB, Herbst RS, Lynch Jr TJ, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC (2003) Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA 290: 2149–2158 [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Marchetti A, Martella C, Felicioni L, Barassi F, Salvatire S, Chella A, Camolese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F (2005) EGFR mutations in non-small cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol 23: 857–865 [DOI] [PubMed] [Google Scholar]
- Meert AP, Martin B, Delmotte P, Berghmans T, Lafitte JJ, Mascaux C, Paesmans M, Steels E, Verdebout JM, Sculier JP (2002) The role of EGF-R expression on patient survival in lung cancer: a systematic review with meta-analysis. Eur Respir J 20: 975–981 [DOI] [PubMed] [Google Scholar]
- Moore PS, Beghelli S, Zamboni G, Scarpa A (2003) Genetic abnormalities in pancreatic cancer. Mol Cancer 2: 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N, Matsuzoe D, Konno T, Kawahara K, Yamashita Y, Shirakusa T (2001) K-ras gene mutations in non-small cell lung cancer in Japanese. Oncol Rep 8: 889–892 [DOI] [PubMed] [Google Scholar]
- Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G (2004) Improved survival in never-smokers vs current smokers with primary adenocarcinoma of the lung. Chest 126: 347–351 [DOI] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86: 2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck ML, Sellers WR, Johnson BE, Meyerson M (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H (2004) EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 101: 13306–13311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin BM, Chen X, Zhu JD, Pei DQ (2005) Identification of EGFR kinase domain mutations among lung cancer patients in China: implication for targeted cancer therapy. Cell Res 15: 212–217 [DOI] [PubMed] [Google Scholar]
- Rom WN, Hay JG, Lee TC, Jiang Y, Tchou-Wong KM (2000) Molecular and genetic aspects of lung cancer. Am J Respir Crit Care Med 161: 1355–1367 [DOI] [PubMed] [Google Scholar]
- Shields PG (2002) Molecular epidemiology of smoking and lung cancer. Oncogene 21: 6870–6876 [DOI] [PubMed] [Google Scholar]
- Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, FUjisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF (2005) Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 97: 339–346 [DOI] [PubMed] [Google Scholar]
- Sobue T, Suzuki T, Fujimoto I, Matsuda M, Doi O, Mori T, Furuse K, Fukuoka M, Yasumitsu T, Kuwahara O, Kono K, Taki T, Kuwabara M, Nakahara K, Endo S, Sawamura K, Kurata M, Ichitani M, Hattori S (1994) Case–control study for lung cancer and cigarette smoking in Osaka, Japan: comarison with the results from Western Europe. Jpn J Cancer Res 85: 464–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis WD, Colby TV, Corrin B, Shimosato Y, Brambilla E (eds) (1999) World Health Organization: Histological Typing of Lung and Pleural Tumors; International Histological Classification of Tumors, 3rd edn New York: Springer [Google Scholar]
- Tsuchiya E, Chan JK, Chan SH, Saw D, Ho JH, Tominaga S (1988) Comparative histopathology of resected bronchial cancers of women in Hong Kong and Japan. Int J Cancer 41: 661–665 [DOI] [PubMed] [Google Scholar]
- UICC (1997) Lung and pleural tumours. In UICC: TNM Classification of Malignant Tumours Sobin LH, Wittekind Ch (eds). 5th edn, pp 91–100. New York: Wiley [Google Scholar]
- Vahakangas KH, Bennett WP, Castren K, Welsh JA, Khan MA, Blomeke B, Alavanja MC, Harris CC (2001) p53 and K-ras mutations in lung cancers from former and never-smoking women. Cancer Res 61: 4350–4356 [PubMed] [Google Scholar]
- Vineis P, Caporaso N (1995) Tobacco and cancer: epidemiology and the laboratory. Environ Health Perspect 103: 156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Cerhan JR, Vierkant RA, Olson JE, Vachon CM, Limburg PJ, Parker AS, Anderson KE, Sellers TA (2002) Adenocarcinoma of the lung is strongly associated with cigarette smoking: further evidence from a prospective study of women. Am J Epidemiol 156: 1114–1122 [DOI] [PubMed] [Google Scholar]
- Yang SH, Mechanic LE, Yang P, Landi MT, Bowman ED, Wampfler J, Merzaman D, Hong KM, Mann F, Dracheva T, Fukuoka J, Travis W, Caporaso NE, Harris CC, Jen J (2005) Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res 11: 2106–2110 [DOI] [PubMed] [Google Scholar]
- Yu J, Ni M, Xu J, Zhang H, Gao B, Gu J, Chen J, Zhang L, Wu M, Zhen S, Zhu J (2002) Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer 2: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]