Abstract
There is increased incidence of microsatellite instability (MSI) in patients who develop multiple primary colorectal cancers (CRC), although the association with hereditary nonpolyposis colon cancer (HNPCC) is unclear. This study aims to evaluate the underlying genetic cause of MSI in these patients. Microsatellite instability was investigated in 111 paraffin-embedded CRCs obtained from 78 patients with metachronous and synchronous cancers, and a control group consisting of 74 cancers from patients with a single CRC. Tumours were classified as high level (MSI-H), low level (MSI-L) or stable (MSS). MLH1, MSH2 and MSH6 gene expression was measured by immunohistochemistry. Methylation of the MLH1 promoter region was evaluated in MSI-H cancers that failed to express MLH1, and mutational analysis performed in MSI-H samples that expressed MLH1, MSH2 and MSH6 proteins. The frequency of MSI-H was significantly greater in the multiple, 58 out of 111 (52%), compared to the single cancers, 10 out of 74 (13.5%), P<0.01. Of the 32 patients from whom two or more cancers were analysed, eight (25%) demonstrated MSI-H in both cancers, 13 (41%) demonstrated MSI-H in one cancer and 11 (34%) failed to demonstrate any MSI-H. MSI-H single cancers failed to express MLH1 or MSH2 in seven out of nine (78%) cases and MSI-L/MSS cancers failed to express MLH1 or MSH2 in one out of 45 (2.2%) cases, all cancers expressed MSH6. MSI-H multiple cancers failed to express MLH1 or MSH2 in 21 out of 43 (48%) cases and MSI-L/MSS cancers failed to express MLH1 or MSH2 in four out of 32 (12.5%) cases. MSH6 expression was lost in five MSI-H multiple cancers, four of which also failed to express MLH1 or MSH2. Loss of expression of the same mismatch repair (MMR) gene was identified in both cancers from six out of 19 (31%) patients. Methylation was identified in 11 out of 17 (65%) multiple and three out of six (50%) single MSI-H cancers that failed to express MLH1. Mutational analysis of 10 MSI-H multiple cancers that expressed MLH1, MSH2 and MSH6 failed to demonstrate mutations in the MLH1 or MSH2 genes. We suggest that, although MSI-H is more commonly identified in those with multiple colorectal cancers, this does not commonly arise from a classical HNPCC pathway.
Keywords: microsatellite instability, synchronous colorectal cancer, metachronous colorectal cancer
Colorectal cancer (CRC) develops via two distinct genetic pathways. The ‘suppressor’ pathway involves loss of function of the tumour suppressor genes APC, DCC and p53, and the activation of the proto-oncogene k-ras. This accounts for 85% of sporadic CRC (Ilyas et al, 1999) and for cancers associated with familial adenomatous polyposis (FAP), that constitute 1% of all CRC (Vasen, 2000). In the ‘mutator’ pathway, mutations in mismatch repair (MMR) genes such as MLH1, MSH2, MSH6, PMS1 and PMS2 lead to microsatellite instability (MSI) in genes such as TGFβRII, ILGF, E2F-4 and BAX, whose coding regions are closely associated with microsatellite DNA (Markowitz et al, 1995; Souza et al, 1996, 1997; Rampino et al, 1997). Microsatellite instability is detected in 15% of sporadic CRC and 90% of cancers from patients with hereditary nonpolyposis colon cancer (HNPCC), which accounts for 1–2% of all CRC (Aaltonen et al, 1998; Percesepe et al, 2001; Samowitz et al, 2001).
Microsatellite instability is classified according to the level of instability detected as high (MSI-H), low (MSI-L) or stable (MSS) (Boland et al, 1998). In MSI-H sporadic cancers, there is predominantly loss of MLH1 function (Thibodeau et al, 1998), caused by transcriptional silencing of the gene brought about by methylation of the MLH1 promoter region (Herman et al, 1998). Hereditary nonpolyposis colon cancer associated cancers arise from MLH1 and MSH2 germline mutations and rarely involve mutations of other genes (Liu et al, 1996; Peltomaki and Vasen, 1997) or MLH1 promoter region methylation (Wheeler et al, 2000; Potocnik et al, 2001; Yamamoto et al, 2002).
At least half the patients with HNPCC will develop a metchronous CRC when treated by a standard colorectal resection (Lynch, 1996), and the Bethesda criteria, introduced to identify those with HNPCC advise MSI analysis in patients with multiple CRC (Rodriguez-Bigas et al, 1997). Microsatellite instability has been reported in 33–89% of cancers from patients with multiple cancers (Horii et al, 1994; Sengupta et al, 1997), including studies that excluded HNPCC clinically by the Amsterdam criteria (Brown et al, 1998; Masubuchi et al, 1999; Yamashita et al, 2000), although these results have been disputed (Pedroni et al, 1999). The Amsterdam criteria were not sufficiently selective (Beck et al, 1997), since the more rigorous Amsterdam criteria II (Vasen et al, 1999) and atypical HNPCC phenotypes associated with mutations in other MMR genes such as MSH6 (Wagner et al, 2001) were not considered. The underlying genetic cause in patients who develop metachronous or synchronous cancers is not clear, germline mutations in MLH1 and MSH2 account for 90% of cases of HNPCC, with mutations in MSH6 and PMS2 accounting for the remainder (Giardiello et al, 2001).
This study investigates the incidence of MSI in patients from the general population with multiple primary colorectal cancers, and the relevance of MLH1, MSH2 and MSH6 gene expression in identifying those patients who may have HNPCC.
MATERIALS AND METHODS
Patients
From the Thames cancer registry, 754 patients who developed either synchronous (393) or metachronous (361) colorectal cancers between 1972 and 1997 were identified, 387 females and 367 males, with a median age at diagnosis of 70 (range 32–94) years. There were 986 cancers in the left colon and 694 cancers in the right colon. The pathology departments of all hospitals identified by the registry were contacted, 12 of these agreed to supply tissue for this study and provided us with 111 samples of paraffin-embedded cancer tissue and corresponding normal mucosa from 78 patients (44 females, 34 males, median age 72 (range 35–91) years). A cancer was synchronous when detected within 2 years of the first (index) cancer, and metachronous when it occurred later. There were 52 synchronous cancers from 29 patients and 59 metachronous cancers from 49 patients, with two or more cancers from 32 patients available for analysis. There were 56 cancers in the left colon and 51 in the right side of the colon; the site of origin of four cancers was undetermined. A control group of 74 patients (31 females, 43 males, median age 74, range 38–100 years) with single CRC resected at least 7 years earlier was identified. There were 42 cancers in the left colon and 29 in the right colon; the site of origin of three cancers was undetermined. Paraffin-embedded cancer tissue and the corresponding normal mucosa were obtained from the resected specimens. Regional ethics committee approval for this study was obtained on the condition that all data were anonymous and that contact with patients to obtain family history was strictly prohibited.
Analysis of MSI
DNA was extracted from the paraffin-embedded tissue using a standard technique (Sengupta et al, 1997) and microsatellite DNA was amplified by polymerase chain reaction (PCR) at the mononucleotide loci Bat 26, Bat 25 and Bat 40, and the dinucleotide loci D2S123, D5S346 and D17S250. Each PCR reaction mixture consisted of: 2′-deoxynucleoside 5′-triphosphates 0.2 mmol l−1 (Pharmacia, St Albans, UK); 1 × PCR buffer containing Tris-HCl 50 mmol l−1 (pH 9); KCL 50 mmol l−1; MgCl2 7 mmol l−1; (NH4)2SO4 16 mmol l−1 (HT Biotechnologies, Cambridge, UK): 0.5 U SuperTaq poymerase (HT Biotechnologies); 25 pmol of each primer and 100 ng of genomic DNA and sterile water to make a uniform volume of 50 μl. The reaction was performed using an Omnigene thermal cycler (Hybaid, Teddington, UK). PCR products were analysed by single-stranded conformational polymorphism (SSCP) on a multiphor II system (Amersham Pharmacia Biotech) using an ExcelGel® DNA Analysis Kit (Amersham Pharmacia Biotech) and a Plus One ™ DNA silver staining kit (Amersham Pharmacia Biotech) on a Hoefer automated gel stainer. MSI was identified by the presence of a characteristic ladder pattern in tumour DNA but not in the corresponding normal mucosa. Cancers which demonstrated MSI in >30% of the loci investigated were designated as MSI-H, those with MSI in <30% of the loci were considered MSI-L and those failing to demonstrate instability at any locus were classified as MSS, in accordance with the guidelines laid down by the National Cancer Institute workshop on MSI (Boland et al, 1998). Cancers were categorised as MSI-H, while MSI-L and MSS were grouped together.
Immunohistochemistry (IHC)
Formalin-fixed, paraffin-embedded cancer tissue sections 5 μm thick were mounted on superfrost slides (BDH Laboratory Supplies) and held at 37°C overnight. Paraffin was removed and the tissue rehydrated using xylene and ethanol. Slides were subjected to microwave antigen retrieval in 10 mM citrate buffer (pH 6) at 85°C for 35 min and cooled in phosphate-buffered saline (PBS), pH 7.4 (Sigma). Endogenous peroxidase activity was blocked with 2% hydrogen peroxide in methanol and slides were washed with PBS prior to overnight incubation with the appropriate antibody at a dilution of 1 : 100. Commercially available monoclonal antibodies against the nuclear proteins MLH1 (PharMingen International, Clone G168-15), MSH2 (Calbiochem-Novabiochem International, Clone FE11) and MSH6 (Serotec, Clone GTBP.P1/2.D4) were applied, followed by staining with Strept ABC complex/HRP Duet kit (DAKO Ltd, Denmark) in conjunction with diamino benzedene (DAB) 180 mg in 300 ml PBS with 300 μl H2O2. Tissues were counterstained with Mayer's haematoxylin prior to mounting with DePex (BDH, Poole, UK). Slide scoring was performed without prior knowledge of MSI status. Loss of expression was determined by failure of the cancer cell nuclei, but not the normal mucosa or stromal cells to stain. Sections that did not stain or showed equivocal staining were excluded from further analysis.
Methylation analysis
Methylation-specific PCR (MSP) following bisulfite modification was performed on samples that failed to express the MLH1 gene using published primer sequences (Herman et al, 1998) designed to amplify either the methylated or unmethylated MLH1 promoter region. For bisulphite modification, 5 μl genomic DNA (100–350 ng of genomic DNA) was incubated for 10 min at 37°C with 1 μl of salmon sperm DNA and 1 μl of 10 N NaOH, and made up to 50 μl with deionised H2O. Freshly prepared 10 mM hydroquinone (30 μl) (Sigma) and 3 M NaHSO3− (520 μl) (Sigma) were added, covered with a layer of mineral oil and incubated at 50°C for 18 h. DNA was cleaned using the QIAquick Gel Extraction Kit (Qiagen) in accordance with the manufacturer's instructions. The clean DNA was incubated with 1.5 μl 10 M NaOH at room temperature for 10 min (final concentration 0.3 M) prior to incubation at −30°C overnight with 1 μl of glycogen (Ambion), 5 μl of 3 M sodium acetate and 125 μl of 100% ethanol. DNA was subsequently washed with 70% ethanol, air dried and re-suspended in sterile, deionised H2O. Following MSP, the PCR products were analysed using 2% agarose in Tris-borate-EDTA gel containing ethidium bromide at a concentration of 0.05 mg per 100 ml (BDH-Merck) and visualised under ultraviolet light.
Mutational analysis
Mutational analysis was performed on multiple cancer samples with MSI-H that expressed the MLH1 and MSH2 proteins, as demonstrated by immunohistochemistry. All 19 MLH1 and 16 MSH2 exons underwent PCR amplification using published primer sequences (Kolodner et al, 1994; Wijnen et al, 1996) and were analysed by SSCP and silver staining.
Statistical analysis
The age at diagnosis of the cancers in the groups was compared by Mann–Whitney U-test, while categorical data were compared by Fisher's exact test. All statistical analysis was performed using GraphPad InStat version 3.00 for Windows 95, GraphPad Software, San Diego, CA USA, www.graphpad.com.
RESULTS
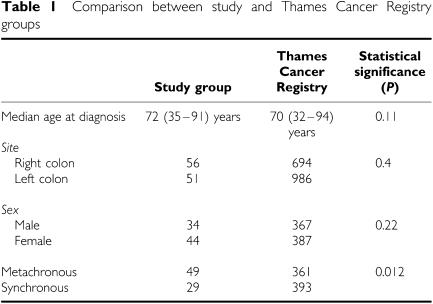
The study group was representative of the Thames cancer registry data. The age at diagnosis, (P=0.11), gender (P=0.4) and site distribution (P=0.22) of the study group were comparable to those of the whole group in the Registry. There were proportionally more patients with metachronous CRCs in the study group, compared to the Registry (P=0.012) (Table 1).
Table 1. Comparison between study and Thames Cancer Registry groups.
| Study group | Thames Cancer Registry | Statistical significance (P) | |
|---|---|---|---|
| Median age at diagnosis | 72 (35–91) years | 70 (32–94) years | 0.11 |
| Site | |||
| Right colon | 56 | 694 | 0.4 |
| Left colon | 51 | 986 | |
| Sex | |||
| Male | 34 | 367 | 0.22 |
| Female | 44 | 387 | |
| Metachronous | 49 | 361 | 0.012 |
| Synchronous | 29 | 393 |
Microsatellite status
MSI-H was detected in 10 (13.5%). single cancers, MSI-L in 19 (25.5%) and MSS in 45 (61%) cancers. There was no difference between the ages of patients with MSI-H (median 80, range 68–89 years) and with MSI-L/MSS (73.5, range 31–100 years) (P=0.2). There were more female patients with MSI-H (8 vs 2) than with MSI-L/MSS (23 vs 41) (P=0.01). MSI-H was detected in 58 (52%) multiple cancers, a significantly higher incidence than in the single cancers (P<0.001), MSI-L in 29 (26%) and MSS in 24 (22%) multiple cancers. The ages of patients with MSI-H multiple cancers (72.5, range 40–87 years) were similar to those in the MSI-L/MSS group (median 73, range 35–91 years), P=0.78. There was no difference in the sex distribution in MSI-H (30 females vs 28 males) and in MSI-L/MSS (30 females vs 23 males) cancers (P=0.7).
In 46 patients with multiple cancers, only a single cancer was available for analysis, 29 (63%) demonstrated MSI-H (median 71, range 53–87 years) and 17 (37%) MSI-L/MSS (median 73 years, range 53–87); the ages were not different in these groups (P=0.9). There was no difference in MSI-H in metachronous (35/62) or synchronous (23/49) cancers (P=0.3). In 32 patients, the MSI status of two or more cancers obtained from a single patient was available; in eight, MSI-H was detected in both cancers (one of whom demonstrated MSI-H in three synchronous cancers); in 13 MSI-H was identified in a single cancer from a pair and MSI-L/MSS in the corresponding cancer, and in 11 patients both cancers demonstrated MSI-L/MSS. There was no difference between the groups in the ages at diagnosis (Table 2).
Table 2. MSI status of paired cancers and statistical comparison of age at cancer presentation.
Immunohistochemistry
Of the single cancers, 19 paraffin-embedded tissue samples were inadequate for immunohistochemistry. Of nine MSI-H cancers, six failed to express MLH1, one failed to express MSH2, and two expressed MLH1 and MSH2. One of the MSI-L/MSS cancers failed to express MSH2 and the remaining 45 expressed both MLH1 and MSH2. All 55 cancers expressed MSH6 (Table 3). Of the multiple cancers, 36 paraffin-embedded samples were inadequate for immunohistochemistry. Of 43 MSI-H cancers, 14 failed to express the MLH1, seven failed to express MSH2, while 22 expressed MLH1 and MSH2. Of 32 MSI-L/MSS cancers, 28 expressed MLH1 and MSH2, three failed to express MLH1 and one failed to express MSH2. Five MSI-H cancers failed to express the MSH6, of which one failed to express MLH1, three failed to express MSH2 and only one cancer failed to express MSH6 alone (Table 3).
Table 3. Immunohistochemistry results for multiple and single cancers.
|
MSI-H
|
MSI-L/MSS
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
Negative staining
|
Negative staining
|
|||||||
| Positive staining | MLH1 | MSH2 | MSH6 | Positive staining | MLH1 | MSH2 | MSH6 | |
| Multiple cancers | ||||||||
| Number | 22 | 14 | 7 | 5 | 28 | 3 | 1 | 0 |
| Median age (range) (years) | 72.5 (40–84) years | 73 (35–91) years | ||||||
| Single cancers | ||||||||
| Number | 2 | 6 | 1 | 0 | 45 | 0 | 1 | 0 |
| Median age (range) (years) | 80 (68–89) years | 73.5 (38–100) years | ||||||
There were 19 paired multiple cancers available for immunohistochemistry. In seven patients, both cancers demonstrated MSI-H; three failed to express MSH2 and two failed to express MLH1 in either cancer. One patient with three MSI-H synchronous cancers showed normal MLH1, MSH2 and MSH6 expression in their cancers, the last patient demonstrated loss of MLH1 in one tumour, but MLH1, MSH2 and MSH6 expression in the other. MSH6 expression was lost in two cancers that also failed to express MSH2, the corresponding cancer pair also failed to express MSH2, but expressed MSH6 (Table 4). In five patients, a single cancer demonstrated MSI-H and the other showed MSI-L/MSS; one lost MLH1 expression in both cancers, one showed loss of MLH1 expression in the MSI-H cancers but not in the MSI-L/MSS cancer, while three patients expressed MLH1, MSH2 and MSH6 in both cancers. In seven patients, both cancers demonstrated MSI-L/MSS, a single cancer did not express MSH2, the remaining 13 cancers expressed MLH1, MSH2 and MSH6.
Table 4. Immunohistochemistry and methylation data for ‘paired’ cancers.
| Patient number | MSI status | MSH2 expression | MLH1 expression | MSH6 expression | MLH1 methylation | Age at presentation |
|---|---|---|---|---|---|---|
| 1 | H | × | ✓ | × | — | 55 |
| H | × | ✓ | ✓ | — | 54 | |
| 2 | H | × | ✓ | × | — | 40 |
| H | × | ✓ | ✓ | — | 43 | |
| 3 | H | × | ✓ | ✓ | — | 53 |
| H | × | ✓ | ✓ | — | 42 | |
| 4 | H | ✓ | × | ✓ | × | 50 |
| H | ✓ | × | ✓ | × | 65 | |
| 5 | H | ✓ | × | ✓ | ✓ | 67 |
| H | ✓ | × | ✓ | ✓ | 73 | |
| 6 | H | ✓ | ✓ | ✓ | — | 76 |
| H | ✓ | ✓ | ✓ | — | 76 | |
| H | ✓ | ✓ | ✓ | — | 76 | |
| 7 | H | ✓ | × | ✓ | × | 62 |
| L | ✓ | × | ✓ | × | 62 | |
| 8 | H | ✓ | × | ✓ | ✓ | 79 |
| H | ✓ | ✓ | ✓ | × | 78 | |
| 9 | H | ✓ | × | ✓ | ✓ | 82 |
| S | ✓ | ✓ | ✓ | × | 73 | |
| 10 | H | ✓ | ✓ | ✓ | — | 75 |
| L | ✓ | ✓ | ✓ | — | 76 | |
| 11 | S | ✓ | ✓ | ✓ | — | 84 |
| H | ✓ | ✓ | ✓ | — | 84 | |
| 12 | H | ✓ | ✓ | ✓ | — | 60 |
| L | ✓ | ✓ | ✓ | — | 62 | |
| 13 | S | × | ✓ | ✓ | — | 64 |
| L | ✓ | ✓ | ✓ | — | 67 | |
| 14 | L | ✓ | ✓ | ✓ | — | 73 |
| S | ✓ | ✓ | ✓ | — | 73 | |
| 15 | L | ✓ | ✓ | ✓ | — | 70 |
| L | ✓ | ✓ | ✓ | — | 72 | |
| 16 | S | ✓ | ✓ | ✓ | — | 77 |
| S | ✓ | ✓ | ✓ | — | 78 | |
| 17 | L | ✓ | ✓ | ✓ | — | 69 |
| L | ✓ | ✓ | ✓ | — | 73 | |
| 18 | S | ✓ | ✓ | ✓ | — | 57 |
| L | ✓ | ✓ | ✓ | — | 59 | |
| 19 | L | ✓ | ✓ | ✓ | — | 70 |
| L | ✓ | ✓ | ✓ | — | 73 |
✓=positive, × =negative, —=not tested.
In six patients, there was loss of expression of the same MMR gene in paired cancers. Their ages at diagnosis (median 54.5, range 40–73 years) were lower than in those with a single MSI-H cancer (median 75.5, range 60–84 years) (P=0.003).
Methylation
Methylation analysis was carried out on all cancers that failed to express the MLH1 gene protein. Of the single cancer group, three of the six (50%) MSI-H cancers and 11 of 17 (65%) MSI-H multiple cancers demonstrated promoter region methylation. Of three patients with paired cancers that failed to express MLH1 in either cancer, two failed to demonstrate methylation (patients no. 4 and 7) and the other (patient no. 5) showed promoter region methylation in both cancers (Table 4).
Mutational analysis
In 10 MSI-H multiple cancers that expressed the MLH1 and MSH2 protein, no mutations were identified in either gene.
DISCUSSION
The incidence of MSI-H in multiple cancerss (52%) is significantly higher than in single cancers (13.5%), consistent with other reports (Sengupta et al, 1997; Brown et al, 1998; Masubuchi et al, 1999; Pedroni et al, 1999; Yamashita et al, 2000). The presence of patients with HNPCC, known to be associated with MSI-H in >90% of cancers, might account for this. Patients with multiple CRCs due to HNPCC are expected to demonstrate concordant MSI-H and loss of expression of the same mismatch repair gene in all cancers. We identified concordant MSI-H in only 25% of paired cancers, the remaining patients having either one or both MSI-L/MSS cancers. Since the incidence of HNPCC in patients with MSI-L/MSS cancers is very low (Terdiman et al, 2001), it is unlikely that HNPCC is responsible for the development of multiple cancers in 75% of patients. Of the 19 paired cancers from which tissue was available for immunohistochemical analysis, five patients demonstrated MSI-H with loss of a concordant MMR genes and one demonstrated concordant loss of MLH1 expression in MSI-H and MSI-L cancers, suggesting that 31% of patients might have HNPCC. In addition, these patients were younger (median age 54.5 years) at the time of diagnosis than those with paired cancers in which only one demonstrated MSI-H (75.5 years). Since the median age of diagnosis of an HNPCC-related CRC is 42 years (Aarnio et al, 1995), this supports our hypothesis. However, when MLH1 promoter region methylation analysis was performed on those MSI-H cancers that failed to express MLH1, one patient with a paired MSI-H cancer demonstrated concordant methylation. Since methylation accounts for loss of MLH1 function in up to 90% of sporadic CRC and is uncommon in HNPCC (Wheeler et al, 2000), it is more likely that this patient has developed two sporadic cancers rather than having an underlying germline mutation, suggesting that, on combined immunohistochemistry and MSI analysis, only 26% of patients might have HNPCC. In addition, the relatively high incidence of methylation in MLH1-negative cancers (65%) throughout the group suggests that the majority of these patients have a sporadic MSI-H cancer and that an underlying germline mutation is not responsible.
Although the majority of the patients in our group do not appear to have classical HNPCC, the role of alternative MMR genes such as MSH6, PMS1, PMS2, MLH3 and EXO1 should be considered (Akiyama et al, 1997; Miyaki et al, 1997; Thompson et al, 2004). Since PMS1, MLH3 and EXO1 have only been implicated in the development of colorectal cancers in case reports (Liu et al, 1996;Hienonen et al, 2003;Thompson et al, 2004), they are unlikely to be implicated in the development of cancer in our group. However, MSH6 mutations may be involved in 5% of all HNPCC families and give rise to atypical HNPCC with a delayed onset of cancer. Although extra-colonic cancers, particularly endometrial, appear to be more prevalent in this group (Plaschke et al, 2004), MSH6 mutations have been implicated in the development of 22% of Amsterdam criteria-positive criteria patients with MSI-L cancers (Wu et al, 1999). Our finding that MSH6 was expressed in all MSI-L and MSS cancers suggests that it is unlikely to play a significant role in this group. Of the MSI-H cancers, only five multiple cancers demonstrated loss of MSH6 expression, four of these also failed to express MSH2 or MLH1. In two cases, the paired cancer continued to express MSH6 normally, while also failing to express MSH2, suggesting a secondary mutation possibly in the intragenic C8 mononucleotide repeat region of MSH6 as a result of loss of MLH1 or MSH2 function. In only one case was MSH6 expression lost in isolation, rendering a single patient a candidate for an underlying MSH6 germline mutation. Mutational analysis in these cases may help clarify the role of MSH6. Mutations of PMS2 have been shown to be important in a small group of HNPCC patients (Thompson et al, 2004). Although primary mutations of PMS2 are uncommon, the inclusion of PMS2 immunohistochemistry may aid the identification of MLH1 germline mutations in MSI-H cancers that continue to express MLH1 (Thompson et al, 2004). The addition of PMS2 immunohistochemistry may have improved the low concordance between MSI-H and loss of MMR gene expression in multiple cancers (49%). The sensitivity of immunohistochemical analysis in our single cancers is in keeping with other series (Terdiman et al, 2001;Lindor et al, 2002), suggesting that this discrepancy is not due to experimental error. Missense mutations or in-frame deletions have been shown to account for the continued expression of a nonfunctioning MMR protein (Peltomaki and Vasen, 1997; Debniak et al, 2000; de Jong et al, 2004); therefore SSCP analysis of all MLH1 and MSH2 exons in 10 patients was undertaken to verify the immunohistochemistry result. Although intronic DNA was not analysed and SSCP is only sensitive for 70% of mutations, being particularly prone to miss large deletions (Grompe, 1993), we feel that it is unlikely that mutations, if present, would not have been identified in at least some of the 10 cancers studied. PMS2 analysis may elucidate the underlying genetic cause in this group.
In conclusion, although there is an increased incidence of MSI-H in those who develop multiple colorectal cancers, the majority are unlikely to be caused by HNPCC. In such cases, a combination of MSI analysis and immunohistochemistry analysis should be performed on both cancers to identify those who should undergo more extensive genetic evaluation.
References
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la CA (1998) Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease (see comments). N Engl J Med 338: 1481–1487 [DOI] [PubMed] [Google Scholar]
- Aarnio M, Mecklin JP, Aaltonen LA, Nystrom-Lahti M, Jarvinen HJ (1995) Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer 64: 430–433 [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y (1997) Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res 57:3920–3923 [PubMed] [Google Scholar]
- Beck NE, Tomlinson IP, Homfray T, Hodgson SV, Harocopos CJ, Bodmer WF (1997) Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal cancer even if the Amsterdam criteria are not fulfilled (see comments). Br J Surg 84: 233–237 [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–5257 [PubMed] [Google Scholar]
- Brown SR, Finan PJ, Hall NR, Bishop DT (1998) Incidence of DNA replication errors in patients with multiple primary cancers. Dis Colon Rectum 41: 765–769 [DOI] [PubMed] [Google Scholar]
- de Jong AE, van Puijenbroek M, Hendriks Y, Tops C, Wijnen J, Ausems MG, Meijers-Heijboer H, Wagner A, van Os TA, Brocker-Vriends AH, Vasen HF, Morreau H (2004) Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res 10: 972–980 [DOI] [PubMed] [Google Scholar]
- Debniak T, Kurzawski G, Gorski B, Kladny J, Domagala W, Lubinski J (2000) Value of pedigree/clinical data, immunohistochemistry and microsatellite instability analyses in reducing the cost of determining hMLH1 and hMSH2 gene mutations in patients with colorectal cancer. Eur J Cancer 36: 49–54 [DOI] [PubMed] [Google Scholar]
- Giardiello FM, Brensinger JD, Petersen GM (2001) AGA technical review on hereditary colorectal cancer and genetic testing. Gastroenterology 121: 198–213 [DOI] [PubMed] [Google Scholar]
- Grompe M (1993) The rapid detection of unknown mutations in nucleic acids. Nat Genet 5: 111–117 [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB (1998) Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95: 6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hienonen T, Laiho P, Salovaara R, Mecklin JP, Jarvinen H, Sistonen P, Peltomaki P, Lehtonen R, Nupponen NN, Launonen V, Karhu A, Aaltonen LA (2003) Little evidence for involvement of MLH3 in colorectal cancer predisposition. Int J Cancer 106: 292–296 [DOI] [PubMed] [Google Scholar]
- Horii A, Han HJ, Shimada M, Yanagisawa A, Kato Y, Ohta H, Yasui W, Tahara E, Nakamura Y (1994) Frequent replication errors at microsatellite loci in tumors of patients with multiple primary cancers. Cancer Res 54: 3373–3375 [PubMed] [Google Scholar]
- Ilyas M, Straub J, Tomlinson IP, Bodmer WF (1999) Genetic pathways in colorectal and other cancers. Eur J Cancer 35: 335–351 [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Hall NR, Lipford J, Kane MF, Rao MR, Morrison P, Wirth L, Finan PJ, Burn J, Chapman P (1994) Structure of the human MSH2 locus and analysis of two Muir-Torre kindreds for msh2 mutations (published erratum appears in Genomics 1995;28(3):613). Genomics 24: 516–526 [DOI] [PubMed] [Google Scholar]
- Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, Jass JR, Hopper J, Gallinger S, Bapat B, Redston M, Thibodeau SN (2002) Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 20: 1043–1048 [DOI] [PubMed] [Google Scholar]
- Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, Jass JR, Dunlop M, Wyllie A, Peltomaki P, de la CA, Hamilton SR, Vogelstein B, Kinzler KW (1996) Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients (see comments). Nat Med 2: 169–174 [DOI] [PubMed] [Google Scholar]
- Lynch HT (1996) Is there a role for prophylactic subtotal colectomy among hereditary nonpolyposis colorectal cancer germline mutation carriers? Dis Colon Rectum 39: 109–110 [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B (1995) Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability (see comments). Science 268: 1336–1338 [DOI] [PubMed] [Google Scholar]
- Masubuchi S, Konishi F, Togashi K, Okamoto T, Senba S, Shitoh K, Kashiwagi H, Kanazawa K, Tsukamoto T (1999) The significance of microsatellite instability in predicting the development of metachronous multiple colorectal carcinomas in patients with nonfamilial colorectal carcinoma. Cancer 85: 1917–1924 [PubMed] [Google Scholar]
- Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T (1997) Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 17: 271–272 [DOI] [PubMed] [Google Scholar]
- Pedroni M, Tamassia MG, Percesepe A, Roncucci L, Benatti P, Lanza Jr G, Gafa R, Di Gregorio C, Fante R, Losi L, Gallinari L, Scorcioni F, Vaccina F, Rossi G, Cesinaro AM, Ponz dL (1999) Microsatellite instability in multiple colorectal tumors. Int J Cancer 81: 1–5 [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen HF (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113: 1146–1158 [DOI] [PubMed] [Google Scholar]
- Percesepe A, Borghi F, Menigatti M, Losi L, Foroni M, Di Gregorio C, Rossi G, Pedroni M, Sala E, Vaccina F, Roncucci L, Benatti P, Viel A, Genuardi M, Marra G, Kristo P, Peltomaki P, Ponz DL (2001) Molecular screening for hereditary nonpolyposis colorectal cancer: a prospective, population-based study. J Clin Oncol 19: 3944–3950 [DOI] [PubMed] [Google Scholar]
- Plaschke J, Engel C, Kruger S, Holinski-Feder E, Pagenstecher C, Mangold E, Moeslein G, Schulmann K, Gebert J, von Knebel DM, Ruschoff J, Loeffler M, Schackert HK (2004) Lower incidence of colorectal cancer and later age of disease onset in 27 families with pathogenic MSH6 germline mutations compared with families with MLH1 or MSH2 mutations: the German Hereditary Nonpolyposis Colorectal Cancer Consortium. J Clin Oncol 22: 4486–4494 [DOI] [PubMed] [Google Scholar]
- Potocnik U, Glavac D, Golouh R, Ravnik-Glavac M (2001) Causes of microsatellite instability in colorectal tumors: implications for hereditary non-polyposis colorectal cancer screening. Cancer Genet Cytogenet 126: 85–96 [DOI] [PubMed] [Google Scholar]
- Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M (1997) Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science 275: 967–969 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S (1997) A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines (see comments). J Natl Cancer Inst 89: 1758–1762 [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Curtin K, Lin HH, Robertson MA, Schaffer D, Nichols M, Gruenthal K, Leppert MF, Slattery ML (2001) The colon cancer burden of genetically defined hereditary nonpolyposis colon cancer. Gastroenterology 121: 830–838 [DOI] [PubMed] [Google Scholar]
- Sengupta SB, Yiu CY, Boulos PB, De Silva M, Sams VR, Delhanty JD (1997) Genetic instability in patients with metachronous colorectal cancers. Br J Surg 84: 996–1000 [DOI] [PubMed] [Google Scholar]
- Souza RF, Appel R, Yin J, Wang S, Smolinski KN, Abraham JM, Zou TT, Shi YQ, Lei J, Cottrell J, Cymes K, Biden K, Simms L, Leggett B, Lynch PM, Frazier M, Powell SM, Harpaz N, Sugimura H, Young J, Meltzer SJ (1996) Microsatellite instability in the insulin-like growth factor II receptor gene in gastrointestinal tumours. Nat Genet 14: 255–257 [DOI] [PubMed] [Google Scholar]
- Souza RF, Yin J, Smolinski KN, Zou TT, Wang S, Shi YQ, Rhyu MG, Cottrell J, Abraham JM, Biden K, Simms L, Leggett B, Bova GS, Frank T, Powell SM, Sugimura H, Young J, Harpaz N, Shimizu K, Matsubara N, Meltzer SJ (1997) Frequent mutation of the E2F-4 cell cycle gene in primary human gastrointestinal tumors. Cancer Res 57: 2350–2353 [PubMed] [Google Scholar]
- Terdiman JP, Gum Jr JR, Conrad PG, Miller GA, Weinberg V, Crawley SC, Levin TR, Reeves C, Schmitt A, Hepburn M, Sleisenger MH, Kim YS (2001) Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology 120: 21–30 [DOI] [PubMed] [Google Scholar]
- Thibodeau SN, French AJ, Cunningham JM, Tester D, Burgart LJ, Roche PC, McDonnell SK, Schaid DJ, Vockley CW, Michels VV, Farr Jr GH, O'Connell MJ (1998) Microsatellite instability in colorectal cancer: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res 58: 1713–1718 [PubMed] [Google Scholar]
- Thompson E, Meldrum CJ, Crooks R, McPhillips M, Thomas L, Spigelman AD, Scott RJ (2004) Hereditary non-polyposis colorectal cancer and the role of hPMS2 and hEXO1 mutations. Clin Genet 65: 215–225 [DOI] [PubMed] [Google Scholar]
- Vasen HF (2000) Clinical diagnosis and management of hereditary colorectal cancer syndromes (in process citation). J Clin Oncol 18: 81S–92S [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin JP, Lynch HT (1999) New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116: 1453–1456 [DOI] [PubMed] [Google Scholar]
- Wagner A, Hendriks Y, Meijers-Heijboer EJ, de Leeuw WJ, Morreau H, Hofstra R, Tops C, Bik E, Brocker-Vriends AH, van Der MC, Lindhout D, Vasen HF, Breuning MH, Cornelisse CJ, van Krimpen C, Niermeijer MF, Zwinderman AH, Wijnen J, Fodde R (2001) Atypical HNPCC owing to MSH6 germline mutations: analysis of a large Dutch pedigree. J Med Genet 38: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JM, Loukola A, Aaltonen LA, Mortensen NJ, Bodmer WF (2000) The role of hypermethylation of the hMLH1 promoter region in HNPCC versus MSI+ sporadic colorectal cancers. J Med Genet 37: 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen J, Khan PM, Vasen H, Menko F, van der KH, van den BM, Leeuwen-Cornelisse I, Nagengast F, Meijers-Heijboer EJ, Lindhout D, Griffioen G, Cats A, Kleibeuker J, Varesco L, Bertario L, Bisgaard ML, Mohr J, Kolodner R, Fodde R (1996) Majority of hMLH1 mutations responsible for hereditary nonpolyposis colorectal cancer cluster at the exonic region 15–16. Am J Hum Genet 58: 300–307 [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Berends MJ, Mensink RG, Kempinga C, Sijmons RH, Der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM (1999) Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet 65: 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Min Y, Itoh F, Imsumran A, Horiuchi S, Yoshida M, Iku S, Fukushima H, Imai K (2002) Differential involvement of the hypermethylator phenotype in hereditary and sporadic colorectal cancers with high-frequency microsatellite instability. Genes Chromosomes Cancer 33: 322–325 [DOI] [PubMed] [Google Scholar]
- Yamashita K, Arimura Y, Kurokawa S, Itoh F, Endo T, Hirata K, Imamura A, Kondo M, Sato T, Imai K (2000) Microsatellite instability in patients with multiple primary cancers of the gastrointestinal tract. Gut 46: 790–794 [DOI] [PMC free article] [PubMed] [Google Scholar]