Abstract
Thermus thermophilus possesses an aspartyl-tRNA synthetase (AspRS2) able to aspartylate efficiently tRNAAsp and tRNAAsn. Aspartate mischarged on tRNAAsn then is converted into asparagine by an ω amidase that differs structurally from all known asparagine synthetases. However, aspartate is not misincorporated into proteins because the binding capacity of aminoacylated tRNAAsn to elongation factor Tu is only conferred by conversion of aspartate into asparagine. T. thermophilus additionally contains a second aspartyl-tRNA synthetase (AspRS1) able to aspartylate tRNAAsp and an asparaginyl-tRNA synthetase able to charge tRNAAsn with free asparagine, although the organism does not contain a tRNA-independent asparagine synthetase. In contrast to the duplicated pathway of tRNA asparaginylation, tRNA glutaminylation occurs in the thermophile via the usual pathway by using glutaminyl-tRNA synthetase and free glutamine synthesized by glutamine synthetase that is unique. T. thermophilus is able to ensure tRNA aminoacylation by alternative routes involving either the direct pathway or by conversion of amino acid mischarged on tRNA. These findings shed light on the interrelation between the tRNA-dependent and tRNA-independent pathways of amino acid amidation and on the processes involved in fidelity of the aminoacylation systems.
Aminoacylation of tRNA is one of the biochemical processes exhibiting the highest accuracy. In most organisms, 20 aminoacyl-tRNA synthetases (aaRS), each with a particular specificity, provide the various aminoacyl-tRNA (aa-tRNA) involved in protein synthesis. With only a few exceptions, organisms encode a single aaRS for each amino acid, and each aaRS has strict charging specificity. Occasionally, two genes encode aaRS of the same specificity, and, in certain organisms, a given aaRS aminoacylates two tRNA of distinct specificity.
Duplication of synthetases first was exemplified with the discovery of two lysyl-tRNA synthetases (LysRS) encoded by distinct genes in Escherichia coli (1–3). Later, two threonyl- and two tyrosyl-tRNA synthetases (ThrRS, TyrRS), also of distinct genetic origins, were characterized in Bacillus subtilis (4–5), and two isoleucyl-tRNA synthetases (IleRS) were found in Staphylococcus aureus (6). The particular character of these duplications is manifested by their absence of phylogenic conservation, and the physiological implication of these duplications to date is not understood clearly. The two ThrRS and TyrRS from B. subtilis appear under different growth conditions. Each ThrRS is sufficient for normal cell growth and sporulation; however, only the thrS gene is expressed during vegetative growth whereas the thrZ gene is induced by a decreased level of charged tRNA promoted by Thr starvation (7). Similar interrelations were described for the two TyrRS from B. subtilis (8, 9). In contrast, one of the two LysRS from E. coli, the lysU product, is induced by heat shock, anaerobiosis, or low pH and is involved in adaptation of the organism to stress conditions whereas the lysS product is expressed constitutively (10–12). Finally, the IleRS from S. aureus encoded by a plasmidic gene differs from the chromosomal one by conferring resistance to the Ile analogue mupirocin (6).
The second exception to the rule of the unicity of the tRNA aminoacylation systems occurs in organisms deprived of a particular aaRS. In this case, the tRNA homologous to the missing aaRS first is mischarged by a heterologous aaRS, and the amino acid then is converted to match the specificity of tRNA. The biological significance of this process became evident when it was shown that it constitutes a pathway for synthesis of the amino acid. Various microorganisms, including the Gram+ bacteria, the Gram− bacterium Rhizobium meliloti, cyanobacteria, and archaebacteria, and also mitochondria and chloroplasts lack glutaminyl-tRNA synthetase (GlnRS) (13–15). tRNA glutaminylation occurs by ω amidation of glutamic acid mischarged on tRNAGln by glutamyl-tRNA synthetase (GluRS) (16, 17). A tRNA-dependent conversion of Asp into Asn resembling that of tRNA glutaminylation was reported recently in Haloferax volcanii (18); further, the absence of ORFs encoding GlnRS and asparaginyl-tRNA synthetase (AsnRS) in the genomes from archeon Methanococcus jannaschii (19) and from eubacterium Helicobacter pylori (20) suggests tRNA-dependent pathways of Gln and Asn formation in these bacteria. tRNA-dependent Gln synthesis was discovered in the late 1960s and is well documented whereas the partners involved in tRNA-dependent Asn synthesis are until now not characterized and the catalytic process remains unknown.
The existence of two genetically distinct aspartyl-tRNA synthetases (AspRS) in the thermophilic eubacterium Thermus thermophilus (21) that activate Asp and charge tRNAAsp but differ structurally has been reported recently (Mr of 68 and 51 for AspRS1 and AspRS2, respectively). To better understand the functional implication of this duplication, we investigated the involvement of these AspRS in tRNA charging. We demonstrate here that AspRS2 aminoacylates tRNAAsn and that T. thermophilus promotes tRNA asparaginylation via a pathway resembling the indirect route of tRNA glutaminylation reported earlier for various bacteria and organelles. The Asp-tRNAAsn amidotransferase (Asp-AdT) that promotes the conversion of Asp-tRNAAsn into Asn-tRNAAsn is described. Because T. thermophilus also contains an AsnRS capable of catalyzing direct tRNA asparaginylation, this eubacterium is able to aminoacylate a single tRNA by both direct and indirect pathways. In contrast, tRNA glutaminylation is promoted by a unique pathway involving GlnRS. Conservation of two pathways for tRNA asparaginylation in this thermophile is rationalized, and the results are discussed in the context of the phylogenetic interrelation between T. thermophilus, eubacteria, and archaebacteria.
EXPERIMENTAL PROCEDURES
General.
E. coli and T. thermophilus DNA were from K12 and HB8 strains, respectively. Oligonucleotides were synthesized by Nucleic Acids Products Supply (Göttingen, Germany). Rabbit antibodies directed against T. thermophilus AspRS1 and AspRS2 were obtained as described (21).
Enzyme Purifications and Analysis.
All steps were conducted at 4°C, and the buffers contained 5 mM 2-mercaptoethanol and 0.1 mM each of EDTA and diisopropylfluorophosphate. Asp-AdT was isolated from 250 g of cells disrupted as described (21). The 105,000 × g supernatant was adsorbed on DEAE-cellulose, and the proteins were eluted with a potassium phosphate gradient from 20 mM (pH 7.2) to 250 mM (pH 6.5). Because the enzyme was not retained on phosphocellulose, the dialyzed fractions were adsorbed on hydroxyapatite, and the proteins were eluted with a potassium phosphate gradient from 10 to 200 mM (pH 6.8). The proteins from active fractions then were subjected to three successive chromatographies in 50 mM Tris⋅HCl buffer (pH 7.5): (i) on Heparine-Ultrogel and elution with a gradient from 0 to 500 mM KCl; (ii) on Cibacron Blue-Sepharose and elution with a gradient from 0 to 1 M KCl; and (iii) on mono Q-Sepharose and elution with a gradient from 0 to 500 mM KCl. Pure enzyme (0.4 mg) was obtained by a final 12% PAGE.
GlnRS was purified from 115 g of cells. The 105,000 × g supernatant was submitted to DEAE-cellulose chromatography as above. The proteins from active fractions then were fractionated on phosphocellulose by elution with a gradient from 0 to 500 mM KCl and on hydroxyapatite by elution with a gradient from 20 to 200 mM potassium phosphate (pH 6.8). Pure enzyme (6 mg) was obtained by a final chromatography on Heparine-Ultrogel after elution with a gradient from 0 to 500 mM KCl.
Glutamine synthetase (GlnS) was purified from the same extract as Asp-AdT. The enzyme was retained on DEAE- and phosphocellulose and was eluted respectively at 230 mM potassium phosphate and 830 mM KCl. Pure enzyme (9 mg) was obtained by a final chromatography on Heparine-Ultrogel after elution with a gradient from 0 to 500 mM KCl. S100 protein extracts from T. thermophilus, E. coli, and yeast were obtained by chromatography of the 105,000 × g supernatant on DEAE-cellulose (21). Molecular masses and subunit structures were determined by gel-filtration and by native and SDS/PAGE as described (21).
Enzymatic Characterizations.
tRNA aminoacylation. The standard reaction mixture contained 100 mM Na-Hepes (pH 7.2), 30 mM KCl, 2 mM ATP, 10 mM MgCl2, 0.05 mM l-[14C]Asp, Gln, or Glu or [3H]Asn, 1–4 mg/ml unfractionated or enriched T. thermophilus tRNA, and 5–300 μg/ml of protein. For Km determinations, 8 or 5 μM l-[3H]Asp or [3H]Asn were present, as were pure tRNAAsp or enriched tRNAAsn from T. thermophilus at concentrations in the Km range and adequate amounts of enzymes. The kcat values were determined with saturating substrates concentrations. The kinetics of aa-tRNA formation were determined at 70°C as described (21).
tRNA-dependent amino acid amidation.
The Asp amidation mixture of 50 μl contained 100 mM Na-Hepes (pH 7.2), 30 mM KCl, 10 mM ATP, 12 mM MgCl2, 2 mM NH4Cl, l-Asn or Gln, 0.05 nmol [14C]Asp-tRNAAsn obtained by aspartylation of enriched tRNAAsn with AspRS2 followed by phenol extraction in 0.3 M sodium acetate (pH 4.5), and either 20 μg of a dialyzed S100 protein extract or 0.2 μg pure Asp-AdT. The Glu amidation mixture deprived of aa-tRNA contained 25 μM l-[14C]Glu, 0.1 nmol enriched tRNAGln, and 0.2 μg T. thermophilus GluRS. After 2–4 h of incubation at 70°C, the reaction was stopped by phenol-chloroform extraction; the aa-tRNA was deacylated by 30 min incubation at 70°C in the presence of 50 mM KOH before neutralization with HCl. The released amino acids were concentrated in a Speed Vac, were fractionated by TLC on cellulose plates (20 × 20 cm) extended by a 3 MM Whatman paper sheet of equal size with isopropanol/formic acid/water (80/20/4, vol/vol/vol) solvent, and were identified by scanning with a Fuji Bioimager.
Contributions of AspRS2 and AsnRS to tRNA asparaginylation were determined at 70°C in standard aminoacylation and amidation mixtures containing either 50 μM l-[14C]Asp without or with 50 μM unlabeled Asn and 2 mM Gln or 50 μM l-[3H]Asn without or with 50 μM unlabeled Asp; the reactions were conducted with 32 μg of proteins from S100 extract and 100 μg of unfractionated T. thermophilus tRNA. The kinetics of formation of [3H]Asn-tRNAAsn by AsnRS and of [14C]Asn-tRNAAsn by AspRS2 and Asp-AdT were determined as described above.
tRNA-independent amino acid amidation.
The reactions were conducted in the amidation mixture containing 25 μM l-[14C]Asp or [14C]Glu and 10 μg of crude extract or S100 protein extract or 0.5 μg of pure T. thermophilus GlnS for 2 h at 70°C. The amino acids were analyzed as described above.
Isolation and Sequencing of tRNAAsn from T. thermophilus.
Pure tRNAAsn (40 nmol/mg) was obtained by two-dimensional PAGE under denaturing conditions starting from a tRNA fraction enriched by BD-cellulose chromatography and was sequenced by using conventional methods (22).
Measurements of the Binding Capacity of Asp-tRNAAsn and Asn-tRNAAsn to Elongation Factor Tu (EF-Tu).
All buffers contained 10 mM MgCl2 and 5 mM 2-mercaptoethanol. EF-Tu (8.3 mg) tagged to six His residues at the Ct (23) was incubated for 10 min at room temperature with 2 ml of agarose beads substituted by Ni++ nitriloacetic acid in 50 mM Tris⋅HCl buffer (pH 7.5) containing 50 mM each of NH4Cl and KCl and 0.1 mM GTP. EF-Tu was activated by 30 min incubation at 37°C with 3 ml of the buffer containing 5 mM GTP, 5 mM phosphoenolpyruvate, and 0.3 mg pyruvate kinase. Enriched tRNA (200 nmol) containing ≈4 nmol of [14C]aa-tRNA was adsorbed on the matrix in a 2-ml column before incubation for 30 min at 37°C. tRNA and aa-tRNA not retained by EF-Tu were eluted with the incubation buffer containing 0.1 M NaCl and 0.1 mM GTP. Bound aa-tRNA was eluted with 0.1 M Na borate buffer (pH 7.5) containing 1 M NaCl and 0.1 mM GTP. The amino acid acylating the tRNA was identified by TLC after alkaline hydrolysis of the aa-tRNA isolated by phenol-chloroform extraction and ethanol precipitation. Protection against alkaline hydrolysis of aa-tRNA by EF-Tu was analyzed at 37°C by incubating 26 pmol of [14C]Asp-tRNAAsn or Asn-tRNAAsn with 765 pmol of activated EF-Tu in 50 mM Tris⋅HCl buffer (pH 8.0) and 50 mM KCl. Deacylations were followed as described (21).
Analysis of the Presence of Asparagine synthetase (AsnS) Genes in Genomic DNA from T. thermophilus and E. coli.
The analysis was performed by PCR amplification (21) of genomic sequences encoding strictly conserved protein sequences in the known AsnS A and B of various phylae including archaea (24–27): 110HSVYVDQWDWERV122 and 236AFELSSMGIRVD247 in AsnS A and 230GVLLSGGLDSS240 and 314HIETYDVTTIRA325 in AsnS B (the numbering corresponds to the sequences in E. coli enzymes). Amplification was conducted with 0.1 μg of genomic DNA and 400 pmol of sense and antisense primers corresponding to the genomic sequences from E. coli (sense and antisense primers for AsnS A: 5′-CACTCGGTCTATGTTGACCAGTGGGACTGGGAACGCGTA-3′ and 5′-ATCTACACGGATCCCCATGGAGGAAAGCTCAAACGC-3′; and for AsnS B: 5′-GGTGTG CTGCTTTCTGGTGGTCTGGATTCCTCA-3′ and 5′-AGCGCGAATAGTGGTCACATCATAAGTTTCGATGTG-3′) or was synthesized by taking into account the codon usages in T. thermophilus and E. coli (degenerated sense and antisense primers for AsnS A: 5′-CACTCG/ CGTG/CTACGTG/CGACCAGTGGGACTGGGAGCG-3′ and 5′GTCG/CACG/CCGGATG/CCCCATG/CGAG/CGAG/CAGCTCA/GAAG/CGC-3′; and for AsnS B: 5′-GGG/ T/CGTG/CCTG/CCTG/T/CTCG/T/CGGG/T/CGGG/ T/CCTG/CGAT/CTCG/CTC-3′ and 5′-A/C/GGCG/CCGA/ GATA/C/GGTG/CGTG/CACA/GTCA/GTAA/C/GGTT/ CTCGATGTG-3′). The primers frame DNA fragments comprising, respectively, 408 and 285 nucleotides in AsnS A and B genes from E. coli.
RESULTS
Evidence for a Dual Specificity for tRNA Charging of AspRS2 from T. thermophilus.
Analysis of the extents of aminoacylation of unfractionated tRNA from T. thermophilus by AspRS1 and AspRS2 indicates higher plateaus for AspRS2 than for AspRS1 (0.66 and 0.42 nmol/mg, respectively). Because T. thermophilus contains only one tRNAAsp species well charged by AspRS1, this result suggests that AspRS2 acylates another tRNA in addition to tRNAAsp. This tRNA, purified by two-dimensional PAGE, was charged by Asp and Asn when incubated in aminoacylation mixtures containing AspRS2 and AsnRS, respectively. Sequencing revealed the presence of Asn anticodon (GUU) and N6-threonyl-carbamoyladenosine at position 37, found in all tRNAAsn until now sequenced. T. thermophilus tRNAAsn presents 76% identity with E. coli tRNAAsn. This establishes aspartylation of tRNAAsn by AspRS2. Table 1 shows that AspRS2 aspartylates pure tRNAAsn as efficiently as tRNAAsp whereas AspRS1 aspartylates it three orders of magnitude less efficiently. Thus, AspRS1 possesses a restricted specificity, and AspRS2 possesses a dual specificity in tRNA charging.
Table 1.
Kinetic constants of aminoacylation of tRNAAsp and tRNAAsn by AspRS1, AspRS2, and AsnRS from T. thermophilus at 70°C. L = (kcat/Km) of cognate tRNA/(kcat/Km) of noncognate tRNA
| Parameters | AspRS1
|
AspRS2
|
AsnRS
|
|||
|---|---|---|---|---|---|---|
| tRNA
|
tRNA
|
tRNA
|
||||
| Asp | Asn | Asp | Asn | Asp | Asn | |
| Km (μM) | 0.03 | 3.4 | 0.073 | 0.063 | 23 | 0.048 |
| kcat (s−1) | 2.7 | 0.12 | 0.24 | 0.092 | 0.024 | 0.4 |
| kcat/Km (s × μM)−1 | 90 | 0.04 | 3.3 | 1.5 | 0.001 | 8.3 |
| L | 1 | 2250 | 1 | 2 | 8300 | 1 |
Evidence for Conversion of Asp Mischarged on tRNAAsn into Asn in T. thermophilus.
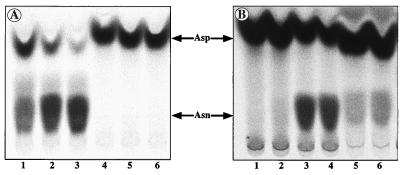
To understand the functional significance of the tRNA mischarging promoted by AspRS2, we investigated the possible conversion of Asp bound to tRNAAsn into Asn. tRNAAsn enriched by two successive BD-cellulose chromatographies, and depleted of all traces of contaminating tRNAAsp, was charged with Asp by AspRS2. Asp-tRNAAsn isolated by phenol extraction then was incubated with a S100 protein extract from T. thermophilus, ATP, and either ammonia, Gln, or Asn as amide group donors. Analysis by TLC of the amino acid released by deacylation of the aa-tRNA revealed formation of Asn (Fig. 1A). Thus, mischarging of tRNAAsn by AspRS2 is corrected by conversion of Asp into Asn. Incubation of Asp under similar conditions but in the absence of tRNA does not give rise to formation of Asn whereas incubation with protein extracts from E. coli or yeast promote conversion (Fig. 1B). This reveals that, in T. thermophilus, Asn is formed by tRNA-dependent ω amidation of Asp and, in E. coli and yeast, is formed by ω amidation of free Asp. Implication of AspRS2 but not of AspRS1 in the tRNA-dependent conversion was established firmly by analysis of the effects of anti-AspRS1 and anti-AspRS2 antibodies. When the reaction was performed in a mixture containing a crude protein extract, unfractionated tRNA, ATP, and ammonia or Gln to allow tRNA aspartylation, either by AspRS2 or by AspRS1 before Asp amidation, anti-AspRS2 antibodies totally abolished the conversion whereas anti-AspRS1 antibodies were without effect (data not shown).
Figure 1.
Conversion of Asp into Asn in T. thermophilus, E. coli, and yeast. (A) tRNA-dependent and -independent conversions in T. thermophilus. The reactions were conducted with a S100 protein extract in the presence (lanes 1–3) or absence (lanes 4–6) of tRNAAsn and 2 mM NH4Cl (lanes 1 and 4), or Asn (lanes 2 and 5), or Gln (lanes 3 and 6). (B) tRNA-independent conversion in T. thermophilus, E. coli, and yeast. The reactions were conducted with a S100 protein extract from T. thermophilus (lanes 1 and 2), E. coli (lanes 3 and 4), or yeast (lanes 5 and 6), at 70°C (lanes 1 and 2) or 37°C (lanes 3 to 6), in the presence of 2 mM NH4Cl (lanes 1, 3, and 5) or Gln (lanes 2, 4, and 6).
Isolation and Structural Properties of Asp-AdT from T. thermophilus.
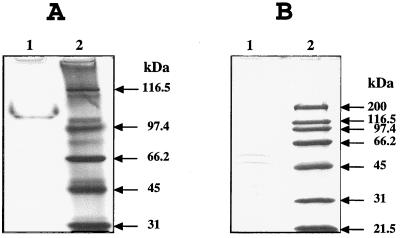
Conversion of tRNA-bound Asp into Asn by the crude extract from T. thermophilus prompted us to search for the Asp-AdT. Pure enzyme was obtained by a seven-step purification procedure. Gel-filtration and native PAGE revealed a Mr of 110 (Fig. 2A) whereas SDS/PAGE showed two polypeptide chains, α and β, with Mr of 51 and 57, respectively, in a 1/1 stoichiometry (Fig. 2B). The heterodimeric αβ structure of this enzyme differs from the homodimeric α2 one found for AsnS of various origins. Analysis by Edman’s degradation of the Nt ends revealed a starting Met in both chains excluding a proteolytic origin of the heterodimeric structure. No significant homologies were found between the 40 first residues of Asp-AdT polypeptide chains and AsnS. However, the Nt sequence of the α chain shows 70% identity with the Nt end of the B subunit of the Glu-tRNAGln amidotransferase (Glu-AdT) from B. subtilis (28) and 47–60% identity with the Nt sequences of proteins HP0658 from H. pylori, MJ0160 from M. jannaschii, MG100 from Mycoplasma genitalium and its homologue from Mycoplasma pneumoniae, and PET112 from Saccharomyces cerevisiae. These proteins are also unique homologues of Glu-AdT (28), suggesting the presence of the same subunit in the two enzymes. In contrast, no significant identity was found between the Nt sequence of the β chain of Asp-AdT and the A and C chains of Glu-AdT and other known proteins, indicating high divergences in these subunits of Asp- and Glu-AdT. It cannot be excluded that, at least in archaebacteria, a same transamidase shares both Asp- and Glu-AdT activities.
Figure 2.
Analysis of the Asp-AdT from T. thermophilus. (A) Native PAGE in a 10–20% gel. (B) Denaturing PAGE in a 10% gel containing 0.1% SDS. Lanes: 1, 0.8 μg (A) and 0.6 μg (B) protein were analyzed; 2, Mr markers.
The Two Pathways of tRNA Asparaginylation in T. thermophilus.
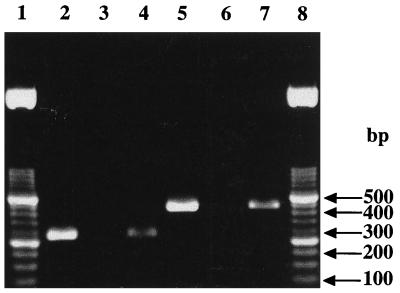
In addition to the indirect route, tRNAAsn also can be asparaginylated directly in T. thermophilus with Asn by AsnRS (29). AsnRS acylates the homologous tRNAAsn 6-fold more efficiently than AspRS2 and the heterologous tRNAAsp four orders of magnitude less efficiently (Table 1). Like AspRS1, but in contrast to AspRS2, AsnRS has restricted tRNA charging specificity. However, the functionality of the direct route of asparaginylation depends on the ability of the organism to synthesize free Asn. To our surprise, all biochemical and genetic attempts to characterize AsnS were unsuccessful. First, as reported above, the crude protein extract from T. thermophilus was unable to convert Asp into Asn when tRNA was absent (Fig. 1B). Second, the genes encoding AsnS A or B, which are well characterized in various organisms, could not be detected in the genome of T. thermophilus by PCR amplification of sequences encoding highly conserved peptides (Fig. 3). T. thermophilus is unable to convert free Asp into Asn by ω amidation. As a consequence, the sources of Asn used by AsnRS for tRNA asparaginylation are the cell-import and/or the protein degradation.
Figure 3.
Detection of AsnS A and B genes in genomic DNA from E. coli and T. thermophilus. Lanes: 2–4, amplification of sequences of AsnS A genes in DNA from E. coli (lanes 2 and 4) and T. thermophilus (lane 3); 2, amplification with nondegenerated primers; 3 and 4, amplification with degenerated primers; 5–7, amplification of sequences of AsnS B genes in DNA from E. coli (lanes 5 and 7) and T. thermophilus (lane 6); 5, amplification with nondegenerated primers; 6 and 7, with degenerated primers; 1 and 8, ladder.
Binding Properties of Asp-tRNAAsn and Asn-tRNAAsn on EF-Tu.
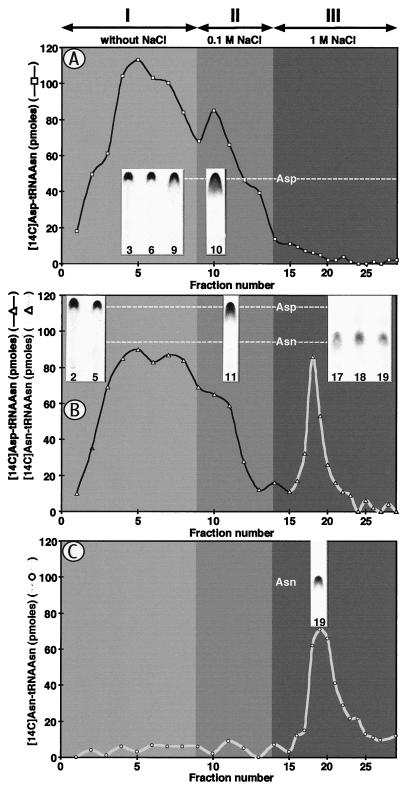
The aa-tRNA released from aaRS are directed by EF-Tu to the ribosomes, where codon-anticodon interactions specify incorporation of amino acids into polypeptide chains, and it generally is believed that EF-Tu binds aa-tRNA without specificity (30). Thus, recognition of Asp-tRNAAsn by EF-Tu should result in misincorporation of Asp into polypeptide chains. We analyzed the binding capacity of Asp-tRNAAsn and Asn-tRNAAsn on EF-Tu. Enriched tRNAAsn aspartylated by AspRS2 was not retained on EF-Tu-substituted Sepharose (Fig. 4A). However, when Asp-AdT, ATP, and amide group donors were added to the aspartylation mixture, two species of aa-tRNA appeared differing in their binding capacity to EF-Tu. The first one displayed the same behavior as Asp-tRNAAsn and had no affinity for EF-Tu, and the second one was retained by EF-Tu and was eluted at high salt concentration like Asn-tRNAAsn (Fig. 4 B and C). Analysis by TLC of the amino acid released by alkaline hydrolysis of the aa-tRNA showed Asp in the species not retained by EF-Tu and Asn in the species retained by EF-Tu (Fig. 4 A–C). Finally, acquisition of the binding capacity of Asp-tRNAAsn to EF-Tu by conversion of Asp into Asn also was revealed by comparing the protection induced by EF-Tu against alkaline hydrolysis of the ester bond before and after amidation of Asp. Asp-tRNAAsn was not protected by EF-Tu against hydrolysis (half-life 20 min at pH 8.0 and 37°C, in the absence and presence of EF-Tu) whereas conversion of Asp into Asn induced protection by increasing the half-life of aa-tRNA by one order of magnitude. After conversion, EF-Tu stabilized the aa-tRNA as Asn-tRNAAsn (data not shown).
Figure 4.
Interaction of Asp-tRNAAsn and Asn-tRNAAsn with EF-Tu. Aminoacylated were 1.5 nmol tRNAAsn with [14C]Asp and AspRS2 (A and B) or with [14C]Asn and AsnRS (C); the [14C]aa-tRNA then were isolated and adsorbed on activated EF-Tu substituted Sepharose (A and C) or were incubated for 2 h in the amidation mixture containing Gln before adsorption on the matrix (B). Adsorption, washing, and elution with buffers I, II, and III were conducted as described in Experimental Procedures. The insets show the position after TLC of the amino acid released by alkaline hydrolysis of the [14C]aa-tRNA.
The Pathway of tRNA Glutaminylation in T. thermophilus.
tRNA glutaminylation is achieved in various organisms by either the direct or the indirect pathway, resembling those of asparaginylation in T. thermophilus. We examined the pathway of tRNA glutaminylation in this organism and found that Gln-tRNAGln is formed by the direct pathway involving GlnRS and free Gln. The GlnRS we purified consists of a monomer of Mr 66 whose Nt sequence of 41 residues, including the HIGH signature sequence of class 1 aaRS, shows 68% identity with GlnRS from E. coli and Haemophilus influenzae. In contrast, alignment with GlnRS from Lupinus luteus, Homo sapiens, S. cerevisiae, and Dictyostelium discoideum upstream of the Nt extension, which characterizes eukaryotic aaRS, shows only 36–46% identity. Further, GlnS, which catalyzes conversion of free Glu into Gln in the presence of ATP and either Gln or ammonia as amide group donors, was purified to homogeneity. Like the E. coli enzyme, it consists of a dodecamer with identical polypeptide chains of Mr 45 whose Nt sequence of 55 residues presents 41 and 49% identity with GlnS chains from E. coli and Thermotoga maritima, respectively. Finally, GluRS from T. thermophilus is deprived of charging capacity of tRNAGln, and no Glu-AdT activity could be detected. Thus, in contrast to tRNA asparaginylation, T. thermophilus does not use the indirect pathway for tRNA glutaminylation. However, this conclusion does not prove lack of Glu-AdT because discrimination by GluRS of tRNAGlu and tRNAGln prevents formation of the Glu-tRNAGln substrate.
Relative Contributions of the Two AspRS and of the Two Pathways of Asparaginylation in tRNA Aminoacylation in T. thermophilus.
The presence in T. thermophilus of two AspRS and of two pathways for tRNA asparaginylation prompted us to examine their contribution in the overall tRNA charging. When unfractionatad tRNA is aspartylated with a crude protein extract, anti-AspRS2 antibodies decrease the rate of aspartylation by 15%, and anti-AspRS1 antibodies decrease the rate of aspartylation by 85% (21). Thus, under conditions approaching physiological conditions, AspRS1 does 85% of tRNA aspartylation whereas the contribution of AspRS2 is minor. We estimated the contribution of AsnRS to the overall tRNA asparaginylation to a similar extent. Indeed, (i) the rate of tRNA asparaginylation of unfractionated tRNA with [3H]Asn by a crude protein extract decreases by ≈20% when tRNAAsn is charged in parallel with unlabeled Asp by AspRS2, and (ii) the rate of formation of [14C]Asn-tRNAAsn promoted by AspRS2 and Asp-AdT in a crude extract decreases drastically when tRNAAsn is charged in parallel with unlabeled Asn by AsnRS. Thus, under standard growth conditions, contribution of AspRS2 to tRNA asparaginylation is minor. However, under Asn starvation, its contribution may become essential because the organism is incapable of synthesizing free Asn.
DISCUSSION
Distribution of the Two-Step Pathway of tRNA Asparaginylation and Glutaminylation in the Phylae.
In T. thermophilus, synthetase duplication clearly relates to a peculiar function in tRNA aminoacylation. AspRS1, the canonical AspRS, ensures tRNA aspartylation whereas the main role of AspRS2 is charging of tRNAAsn by Asp, which is then converted into Asn by Asp-AdT:
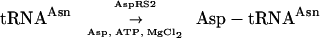 |
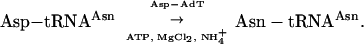 |
Comparison of AspRS polypeptide chains show that T. thermophilus AspRS2 structurally is related more to archaebacterial AspRS than to eubacterial and eukaryotic AspRS (21). The polypeptide chains of AspRS2 from T. thermophilus and archaebacterial AspRS are significantly shorter than those from eubacterial and eukaryotic AspRS (422–438 and 500–600 residues, respectively). Of interest, archaebacteria also use the indirect pathway of tRNA asparaginylation, as revealed by the tRNA-dependent conversion of Asp into Asn in H. volcanii (18) and the lack of gene encoding AsnRS in M. jannaschii (19). Thus, the small archaebacterial AspRS exhibit dual specificity for tRNA charging whereas the large eubacterial and eukaryotic AspRS possess restricted specificity. Indirect asparaginylation probably also occurs in the mesophilic eubacterium H. pylori, which has no AsnRS (20), and in Deinococcus radiodurans [accompanying paper (ref. 31)]. The indirect pathway of asparaginylation resembles that of glutaminylation in which Glu mischarged on tRNAGln by GluRS is converted into Gln by Glu-AdT (13–17, 28). However, the two pathways are not distributed uniformly in the various phylae. Archaebacteria deprived of GlnRS and AsnRS use the indirect pathways for Gln-tRNAGln and Asn-tRNAAsn formation. In contrast, many eubacteria are deprived of GlnRS and use the indirect pathway for glutaminylation but catalyze asparaginylation with AsnRS. Finally, eukaryotes use the direct pathways for glutaminylation and asparaginylation except in the organelles devoid of GlnRS, where glutaminylation occurs via the two-step pathway. Surprisingly, the distribution of GlnS and AsnS does not coincide with that of GlnRS and AsnRS in the phylae. Except for T. thermophilus deprived of AsnS, all organisms we analyzed contain GlnS and AsnS, even when amino acids can be formed by the tRNA-dependent pathway and regardless the presence or absence of GlnRS and AsnRS. Thus, no interrelation emerges between the nature of the pathway, either direct or indirect, used by the organisms for Gln-tRNAGln and Asn-tRNAsn formation and their ability to synthesize free Gln and Asn. The properties of the asparaginylation system of D. radiodurans (31) and T. thermophilus show that coexistence of direct and indirect pathways of asparaginylation is related to the absence of AsnS.
Discrimination of Asn-tRNAAsn Against Asp-tRNAAsn by EF-Tu.
The fact that tRNAAsn is charged efficiently with Asp is in keeping with the rule that tRNA mischarged by the amino acid homologous to the synthetase is not corrected (32); corrections occur only when tRNA is charged with noncognate amino acids. The lack of binding of Asp-tRNAAsn to EF-Tu when Asp is not amidated prevents misincorporation of Asp into polypeptide chains. A similar behavior has been reported for Glu-tRNAGln formed by GluRS in Euglena gracilis chloroplasts before amidation of Glu (33). It is believed currently that EF-Tu binds aa-tRNA without specificity and regardless of the nature of the amino acid acylating tRNA, except Sec-tRNASec, which binds to the SEL B factor but not to EF-Tu (34). However, we show here that the EF-Tu is able to discriminate the aspartylated tRNAAsn from that which is asparaginylated. Thus, EF-Tu has refined its specificity to the cellular context, in particular when mischarging is of physiological importance, to improve the accuracy in protein synthesis. Recognition of aa-tRNA by EF-Tu involves the amino acid moiety and the acceptor stem (35). The lack of binding when tRNAAsn is mischarged with Asp may result from an unfavorable presentation of the amino acid by the noncognate tRNA because of the negatively charged β carboxyl group of Asp and structural peculiarities of the acceptor arm. The antideterminants of Asp-tRNAAsn and probably also of Glu-tRNAGln for binding on EF-Tu differ from those of Sec-tRNASec, which involve solely the length of the acceptor arm (34).
Evolution of the Pathways of tRNA Asparaginylation and Glutaminylation.
AspRS1 and AsnRS charge the noncognate tRNAAsn and tRNAAsp only three to four orders of magnitude less efficiently than the cognate ones (Table 1). This poor discrimination contrasts with the significantly higher selection reported for homologous systems (36) and supports arguments for the recent introduction of AspRS1 and AsnRS in T. thermophilus. Thus, the archaebacterial type AspRS2 probably constitutes a remnant from ancestral AspRS.
It has been proposed that Asn was one of the latest amino acids added to the cell repertoire and that AAU and AAC codons first specified Asp (37). Thus, the ancestor of tRNAAsp and tRNAAsn was charged with Asp by the primitive AsxRS before Asn appeared. tRNA asparaginylation was acquired with the capacity of conversion of Asp charged on tRNAGUU with Asp-AdT. If AsnRS appeared according to the scheme proposed for GlnRS (38, 39), it emerged in eukaryotes by duplication of the AsxRS gene and acquired specificity for Asn and tRNAGUU. Eubacterial acquisition of AsnRS by horizontal gene transfer then was correlated with refinement of specificity of AsxRS for aspartylation of tRNAGUC. This would not be possible in T. thermophilus because tRNA asparaginylation depended on the primitive pathway because of the lack of AsnS. Further, the imported AsnRS was not sufficiently accurate in discrimination of tRNAAsp and tRNAAsn to ensure survival of the organism. Accuracy then was improved by acquisition of a modern AspRS, which lowered the pool of free tRNAAsp. Thus, AspRS1 and AsnRS were conserved because of their efficiency and the increased accuracy they promoted when present together. In contrast, glutamylation and glutaminylation specificities split because GlnS, which was probably more easily acquired than AsnS because of the involvement of Gln in essential cellular functions (40), promoted emergence of the direct route of glutaminylation after acquisition of GlnRS and allowed ancestral GlxRS to restrict its specificity to tRNAGlu.
Position of T. thermophilus in the Phylogenetic Tree.
Because archaebacteria do not contain AsnRS and GlnRS whereas most eubacteria contain AsnRS and only Gram− eubacteria contain GlnRS, acquisitions of the modern pathways of asparaginylation and glutaminylation by prokaryotes were delayed in time. Eubacteria acquired AsnRS and GlnRS after the split of the archaebacteria, which conserved the AspRS and GluRS of dual specificity and refined the tRNA-dependent pathways of Asn and Gln formation. The idea that GlnRS emerged in eukaryotes from ancestral GlxRS before its transfer into eubacteria is suggested by the stronger structural similarities of eubacterial GlnRS with eukaryotic GluRS than with eubacterial GluRS (38); because we could not find such structural interconnection between eubacterial AsnRS and eukaryotic and eubacterial AspRS, AsnRS appeared in eukaryotes and was transferred to eubacteria probably before GlnRS. As a consequence, most eubacteria have lost the primitive pathway of asparaginylation. GlnRS appeared later, in the ancestor of the Gram− bacteria after splitting of the Gram+ phylum. By exhibiting archeal and eubacterial Gram− metabolic peculiarities, T. thermophilus constitutes a link in evolution of the prokaryotes. Similar properties of D. radiodurans reported elsewhere (31) support arguments for a peculiar position of the Thermus–Deinococcus group (41) in the Gram− eubacteria.
Acknowledgments
The authors thank J. Reinbolt (Institut de Biologie Moléculaire et Cellulaire, Strasbourg, France) for protein sequencing, G. Keith (Institut de Biologie Moléculaire et Cellulaire) for help in tRNA sequencing, R. Leberman (European Molecular Biology Laboratory, Grenoble, France) for generous gifts of AsnRS and [14C]Asn, S. Ribeiro and M. Sprinzl (University of Bayreuth, Germany) for generous gift of EF-Tu, F. Fasiolo and S. Lodmell for critical reading and reviewing of the manuscript, and R. Giegé for support. This work was supported by grants from Centre National de la Recherche Scientifique et Technique and from Association de la Recherche contre le Cancer. H.D.B. is a recipient of a grant from Ministère de l’Enseignement Supérieur et de la Recherche.
ABBREVIATIONS
- aaRS
aminoacyl-tRNA synthetase
- aa-tRNA
aminoacyl-tRNA
- AsnS
asparagine synthetase
- Asp-AdT
Asp-tRNAAsn amidotransferase
- EF-Tu
elongation factor Tu
- GlnS
glutamine synthetase
- Glu-AdT
Glu-tRNAGln amidotransferase
References
- 1. Hirshfield I N, Bloch P L, VanBogelen R A, Neidhardt F C. J Bacteriol. 1981;146:345–351. doi: 10.1128/jb.146.1.345-351.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark R L, Neidhardt F C. J Bacteriol. 1990;172:3237–3243. doi: 10.1128/jb.172.6.3237-3243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lévêque F, Plateau P, Dessen P, Blanquet S. Nucleic Acids Res. 1990;18:305–312. doi: 10.1093/nar/18.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Putzer H, Brakhage A A, Grunberg-Manago M. J Bacteriol. 1990;172:4593–4602. doi: 10.1128/jb.172.8.4593-4602.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henkin T M, Glass B L, Grundy F J. J Bacteriol. 1992;174:1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbart J, Perry C R, Slocombe B. Antimicrob Agents Chemother. 1993;37:32–38. doi: 10.1128/aac.37.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putzer H, Gendron N, Grunberg-Manago M. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy F Y, Hodil S E, Rollins S M, Henkin T M. J Bacteriol. 1997;179:2587–2594. doi: 10.1128/jb.179.8.2587-2594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser P, Danchin A, Kunst F, Débarbouillé M, Vertès A, Dedonder R. DNA Sequence: Sequencing Mapping. 1990;1:251–261. doi: 10.3109/10425179109020780. [DOI] [PubMed] [Google Scholar]
- 10.Lévêque F, Gazeau M, Fromant M, Blanquet S, Plateau P. J Bacteriol. 1991;173:7903–7910. doi: 10.1128/jb.173.24.7903-7910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neidhardt F C, VanBogelen R A. Biochem Biophys Res Commun. 1981;100:894–900. doi: 10.1016/s0006-291x(81)80257-4. [DOI] [PubMed] [Google Scholar]
- 12.Ito K, Kowakami K, Nakamura Y. Proc Natl Acad Sci USA. 1993;90:302–306. doi: 10.1073/pnas.90.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapointe J, Duplain J, Proulx M. J Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schön A, Kannangara C G, Gough S, Söll D. Nature (London) 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 15.Gagnon Y, Lacoste L, Champagne L, Lapointe J. J Biol Chem. 1996;271:14856–14863. doi: 10.1074/jbc.271.25.14856. [DOI] [PubMed] [Google Scholar]
- 16.Wilcox M. Eur J Biochem. 1969;11:405–412. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 17.Jahn D, Kim Y-C, Ishino Y, Söll D. J Biol Chem. 1990;256:8059–8064. [PubMed] [Google Scholar]
- 18.Curnow A W, Ibba M, Söll D. Nature (London) 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 19.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 20.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 21.Becker H D, Reinbolt J, Kreutzer R, Giegé R, Kern D. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 22.Keith G. In: Journal of Chromatography Library. Gehrke C W, Kuo K C, editors. New York: Elsevier; 1990. pp. A103–A141. [Google Scholar]
- 23.Ribeiro S, Nock S, Sprinzl M. Anal Biochem. 1995;228:330–335. doi: 10.1006/abio.1995.1359. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura M, Yamada M, Hirota Y, Sugimoto K, Oka A, Takanami M. Nucleic Acids Res. 1981;9:4669–4676. doi: 10.1093/nar/9.18.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colombo G, Villafranca J J. J Biol Chem. 1986;261:10587–10591. [PubMed] [Google Scholar]
- 26.Andrulis I L, Chen J, Ray P N. Mol Cell Biol. 1987;7:2435–2443. doi: 10.1128/mcb.7.7.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scofield M A, Lewis W S, Schuster S M. J Biol Chem. 1990;265:12895–12902. [PubMed] [Google Scholar]
- 28.Curnow A, W, Hong K W, Yuan R, Kim S I, Martins O, Winkler W, Henkin T, M, Söll D. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seignovert L, Härtlein M, Leberman R. Eur J Biochem. 1996;239:501–508. doi: 10.1111/j.1432-1033.1996.0501u.x. [DOI] [PubMed] [Google Scholar]
- 30.Wagner T, Sprinzl M. Eur J Biochem. 1980;108:213–221. doi: 10.1111/j.1432-1033.1980.tb04714.x. [DOI] [PubMed] [Google Scholar]
- 31.Curnow A L, Tumbula D L, Pelaschia J T, Min B, Söll D. Proc Natl Acad Sci USA. 1998;21:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietrich A, Kern D, Bonnet J, Giegé R, Ebel J-P. Eur J Biochem. 1976;70:147–158. doi: 10.1111/j.1432-1033.1976.tb10965.x. [DOI] [PubMed] [Google Scholar]
- 33.Stanzel M, Schön A, Sprinzl M. Eur J Biochem. 1994;219:435–439. doi: 10.1111/j.1432-1033.1994.tb19956.x. [DOI] [PubMed] [Google Scholar]
- 34.Baron C, Böck A. In: tRNA: Structure, Biosynthesis and Function. Söll D, RajBandhary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 529–544. [Google Scholar]
- 35.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F C, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 36.Rogers K C, Söll D. Biochemistry. 1993;32:14210–14219. doi: 10.1021/bi00214a021. [DOI] [PubMed] [Google Scholar]
- 37.Wong J T. Proc Natl Acad Sci USA. 1975;72:1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamour V, Quevillon S, Diriong S, N′Guyen V C, Lipinsky M, Mirande M. Proc Natl Acad Sci USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogers K, Söll D. J Mol Evol. 1995;40:476–481. doi: 10.1007/BF00166615. [DOI] [PubMed] [Google Scholar]
- 40.Taylor B. Annu Rev Biochem. 1978;47:1127–1162. doi: 10.1146/annurev.bi.47.070178.005403. [DOI] [PubMed] [Google Scholar]
- 41.Hensel R, Demharter W, Kandler O, Kroppenstedt R, M, Stakebrandt E. Int J Syst Bacteriol. 1986;36:444–453. [Google Scholar]