Abstract
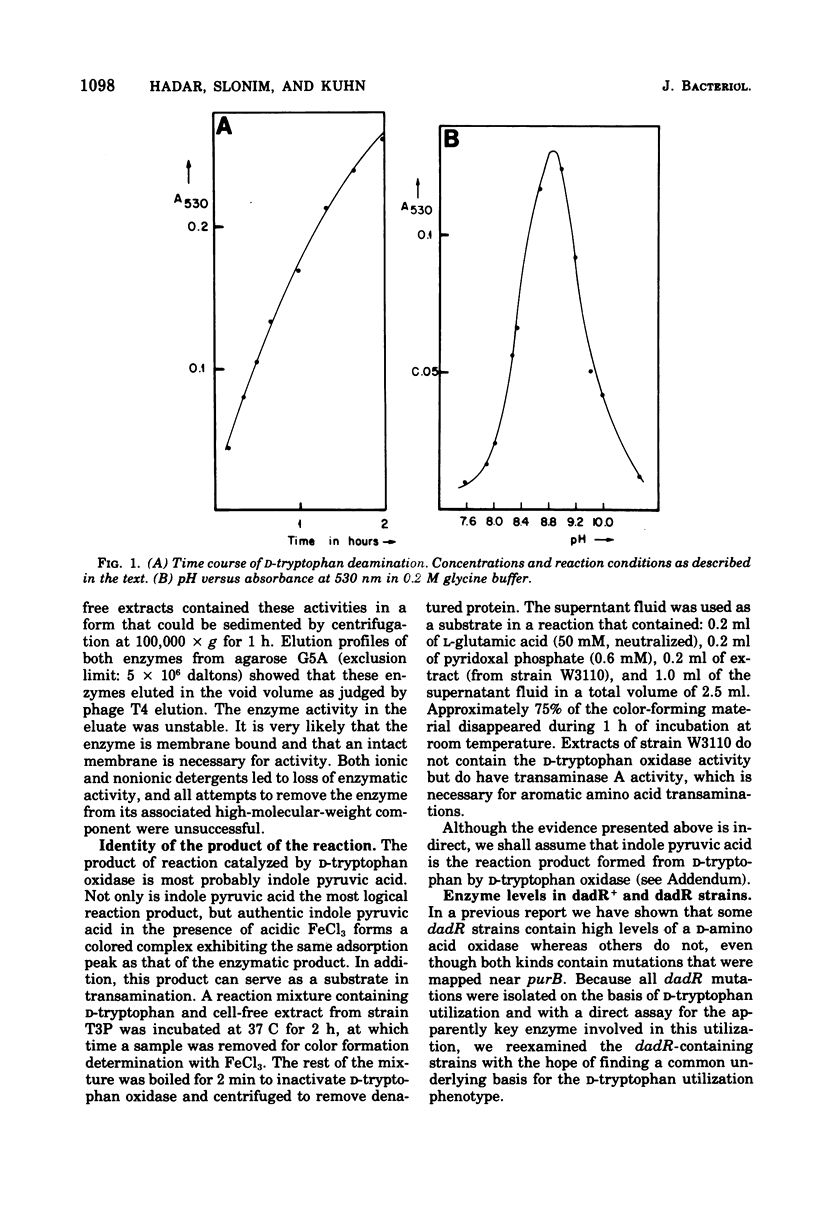
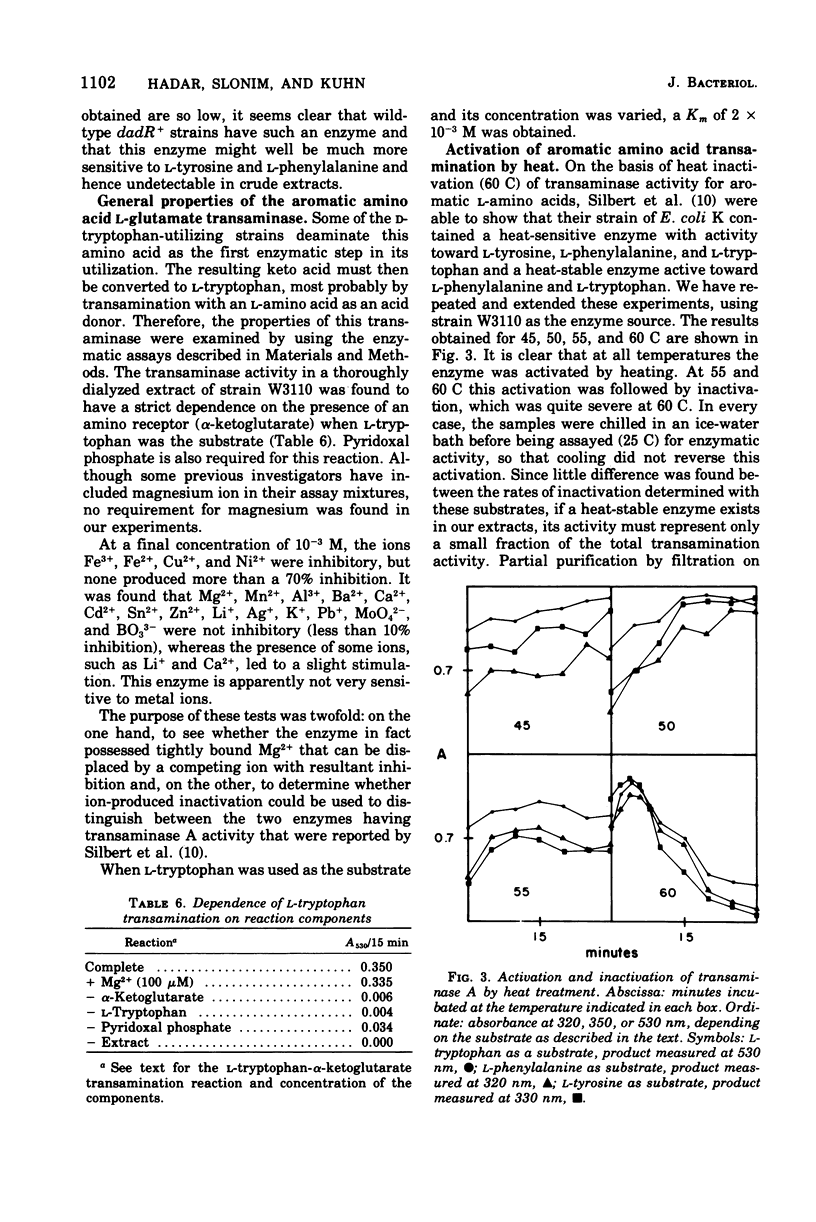
Mutants of Escherichia coli K-12 that require L-tryptophan (trp) are normally unable to utilize D-tryptophan to fulfill their requirement. However, secondary mutations (dadR) that confer this ability can be isolated. In such strains two distinct enzymes are found to be produced at high levels: D-amino acid oxidase (EC 1.4.3.3) and D-tryptophan oxidase. A convenient assay procedure for D-tryptophan oxidase is described. The two enzymes could be distinguished on the basis of their sensitivity to inhibition by L-phenylalanine and L-tyrosine. Strains that were trp dadR could not grow with D-tryptophan in the presence of L-phenylalanine, but further mutations, Fyo, could be isolated that allowed growth under these conditions. Some of them were characterized by further increases in the level of D-tryptophan oxidase activity and a sharp decrease in D-amino acid oxidase. These kinds of Fyo mutations lay in or near the dadR gene. The substrate specificity of the two enzymes toward a large number of compounds was examined. The transamination of aromatic keto acids was investigated. In the wild-type strain only a single enzyme, transaminase A (EC 2.6.1.5), was found, and it was irreversibly activated when subjected to elevated temperatures. The present state of our knowledge on D-amino acid utilization in E. coli is summarized.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames G. F., Lever J. Components of histidine transport: histidine-binding proteins and hisP protein. Proc Natl Acad Sci U S A. 1970 Aug;66(4):1096–1103. doi: 10.1073/pnas.66.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F., Pittard J. Pathways of biosynthesis of aromatic amino acids and vitamins and their control in microorganisms. Bacteriol Rev. 1968 Dec;32(4 Pt 2):465–492. [PMC free article] [PubMed] [Google Scholar]
- Krajewska-Grynkiewicz K., Walczak W., Klopotowski T. Mutants of Salmonella typhimurium able to utilize D-histidine as a source of L-histidine. J Bacteriol. 1971 Jan;105(1):28–37. doi: 10.1128/jb.105.1.28-37.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J., Somerville R. L. Mutant strains of Escherichia coli K12 that use D-amino acids. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2484–2487. doi: 10.1073/pnas.68.10.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Rahmanian M., Claus D. R., Oxender D. L. Multiplicity of leucine transport systems in Escherichia coli K-12. J Bacteriol. 1973 Dec;116(3):1258–1266. doi: 10.1128/jb.116.3.1258-1266.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanian M., Oxender D. L. Derepressed leucine transport activity in Escherichia coli. J Supramol Struct. 1972;1(1):55–59. doi: 10.1002/jss.400010108. [DOI] [PubMed] [Google Scholar]
- SILBERT D. F., JORGENSEN S. E., LIN E. C. Repression of transaminase A by tyrosine in Escherichia coli. Biochim Biophys Acta. 1963 Jun 11;73:232–240. doi: 10.1016/0006-3002(63)90307-x. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- YANOFSKY C., LENNOX E. S. Transduction and recombination study of linkage relationships among the genes controlling tryptophan synthesis in Escherichia coli. Virology. 1959 Aug;8:425–447. doi: 10.1016/0042-6822(59)90046-7. [DOI] [PubMed] [Google Scholar]



