Abstract
All available comparative genomic hybridisation (CGH) analyses (n=31, until 12/2003) of human hepatocellular carcinomas (HCCs; n=785) and premalignant dysplastic nodules (DNs; n=30) were compiled and correlated with clinical and histological parameters. The most prominent amplifications of genomic material were present in 1q (57.1%), 8q (46.6%), 6p (22.3%), and 17q (22.2%), while losses were most prevalent in 8p (38%), 16q (35.9%), 4q (34.3%), 17p (32.1%), and 13q (26.2%). Deletions of 4q, 16q, 13q, and 8p positively correlated with hepatitis B virus aetiology, while losses of 8p were more frequently found in hepatitis C virus-negative cases. In poorly differentiated HCCs, 13q and 4q were significantly under-represented. Moreover, gains of 1q were positively correlated with the occurrence of all other high-frequency alterations in HCCs. In DNs, amplifications were most frequently present in 1q and 8q, while deletions occurred in 8p, 17p, 5p, 13q, 14q, and 16q. In conclusion, aetiology and dedifferentiation correlate with specific genomic alterations in human HCCs. Gains of 1q appear to be rather early events that may predispose to further chromosomal abnormalities. Thus, explorative CGH meta-analysis generates novel and testable hypotheses regarding the cause and functional significance of genomic alterations in human HCCs.
Keywords: hepatocellular carcinoma, hepatocarcinogenesis, hepatitis-B virus, hepatitis-C virus, comparative genomic hybridisation, meta-analysis
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers with steadily increasing incidence even in developed countries, such as Europe and the United States (El-Serag, 2002). It is believed that human hepatocarcinogenesis persists several decades normally starting from chronic liver disease over cirrhosis, premalignant precursor lesions (dysplastic nodules (DNs), ‘early’ HCC upto fully malignant HCC with angioinvasive and metastatic potential (reviewed in Kern et al, 2002a). In more than 80% of the cases, a well-defined aetiology (e.g. viral infection, aflatoxin B1 exposure, and chronic alcohol abuse) is associated with the development of HCC. Hepatitis B virus (HBV) infection is thought to contribute by two different mechanisms to hepatocarcinogenesis: chromosomal integrations of viral DNA with destabilising effects for the host genome (Luber et al, 1996) and expression of viral transactivating factors (HBxAg and preS/SAg; Kekule et al, 1990; Koike, 1995). The oncogenic potential of hepatitis C virus (HCV) infection has been linked to the viral transcriptional activator NS5A (Kato et al, 1997) and also to the core polypeptide (Moriya et al, 1998). Moreover, tumorigenic properties of aflatoxin B1 are linked to somatic G/T transversion in codon 249 of TP53 (Ozturk, 1999). Furthermore, several cellular factors have been implicated in the pathogenesis of HCC (e.g., TP53 (p53; Feitelson, 1998), CTNNB1 (β-catenin; Prange et al, 2003), RB1 (retinoblastoma; Zhang et al, 1994), COX2 (cyclooxygenase-2; Kern et al, 2002b), IGF2 (insulin-like growth factor-II; Breuhahn et al, 2004), and CDH1 (E-cadherin; Matsumura et al, 2001)).
Studies of the overall chromosomal alterations in carcinomas are based on loss of heterozygosity (LOH) analyses and comparative genomic hybridisation (CGH). Comparative genomic hybridization is a fluorescence-based technique that is used for the detection of chromosomal imbalances in tissues or cell populations. Areas representing loss of genomic material may contain tumour suppressor genes, while gains may harbour dominant protumorigenic factors (e.g. oncogenes and growth factors) relevant for respective tumour entity. The power of CGH lies in its potential for a more or less unbiased whole genome screening; the main limitation is its relatively low resolution. By CGH several chromosomal regions carrying tumour-relevant genes (e.g. oncogenes) have been identified in solid tumours and haematological malignancies (reviewed in Weiss et al, 2002).
However, many information regarding relevant chromosomal rearrangements that may play a central role in the development of HCC are still missing. Meanwhile, over 30 different in part small CGH studies have generated a wealth of over 700 analysed HCCs that await comprehensive and comprising interpretation. The aim of this study was to compile the information from all available CGH data of human DNs/HCCs and to correlate these alterations with aetiology and histological grading. The results demonstrate a significant association of specific genomic imbalances with viral aetiology and histological grade, and generate further testable hypotheses.
MATERIALS AND METHODS
Comparative genomic hybridisation-studies
Overall, 785 different CGH analyses of HCCs are available from 31 studies (public NCBI database PubMed; published until 12/2003; see Supplementary online material). Inclusion criteria for the CGH studies were complete data profiles and explicit tumour classification (HCC, DN) as well as similar software and labelling systems and used thresholds (0.7–0.8 and 1.2–1.3). Information regarding the HBV status was present in 428 cases (244 HBV-positive and 184 HBV-negative cases). The HCV status was available in 338 cases (110 HCV-positive and 228 HCV-negative cases).
Tumour grading was given in 199 cases (126 cases with low grade (G1/2) and 73 cases with high-grade (G3/4) HCCs). Overall 30 CGH analyses of DNs were collected from four different studies (see Supplementary online material).
Data recording and statistical analyses
All CGH data were recorded in a standardised fashion: each chromosome arm was divided from 0 to 100 (centromere to telomere direction) and gains, losses, or normal ratio were evaluated in steps of five. From these data, the respective charts and ideograms were displayed. Moreover, the data were submitted to statistical analyses using SPSS. However, when only parts of a chromosomal arm were affected, it was scored as a complete loss of the arm in the summarising tables. When specific chromosomal aberrations occurred in 20% of all analysed cases, they were rated to be high frequency. These aberrations were specifically submitted to further examination. Statistical evaluation was performed using explorative χ2 test. P-values cited were not corrected for multiple testing. The list of known tumour relevant genes localised in the chromosomal regions with imbalances of highest frequency was determined using the OMIN database (Online Mendelian Inheritance of Man).
RESULTS
Predominant chromosomal alterations in human HCCs
The complete meta-analysis of all available CGH data (n=785 HCCs) revealed that gains of chromosomal material were most prevalent in 1q (57.1% of the cases), 8q (46.6%), 6p (22.3%), and 17q (22.2%), and losses were most frequently present in 8p (38%), 16q (35.9%), 4q (34.3%), 17p (32.1%), and 13q (26.2%) (Table 1). These data testify amplifications and deletions of chromosomal arms on which oncogenes (e.g. MYC on 8q24) and tumour suppressor genes (e.g. RB1 on 13q14) are located. Furthermore, several genes with known and potential protumorigenic functions (e.g. modulators of the WNT-signalling pathway like FZD3, WISP1, SIAH-1, and AXIN2) have been described to be grouped on respective chromosomal arms (Table 1).
Table 1. Frequencies of chromosomal alterations in human HCCs.
|
p-arm
|
q-arm
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Chromosome | Loss (%)a | Gain (%)a | High. Fre.b | Genesc | Loss (%)a | Gain (%)a | High. fre.b | Genesc |
| 1 | 15.4 | 5.2 | — | — | 0.6 | 57.1 | q21.1–q44 | WNT14, FASL |
| 2 | 1.4 | 7.1 | — | — | 2.9 | 8 | — | — |
| 3 | 3.9 | 5 | — | — | 1.9 | 8.8 | — | — |
| 4 | 10.6 | 6 | — | — | 34.3 | 1.7 | q21.1–q35 | LEF1, CCNA |
| 5 | 1.7 | 13.6 | — | — | 7.8 | 11.1 | — | — |
| 6 | 1 | 22.3 | — | PIM1, CDKN1A | 15 | 7.9 | — | — |
| 7 | 0.9 | 15 | — | — | 3.1 | 16.8 | — | — |
| 8 | 38 | 4.6 | p21.1–p22 | FZD3, PLK3 | 1.9 | 46.6 | q22.1–q24.3 | MYC, WISP1 |
| 9 | 14 | 3.3 | — | — | 11.1 | 2.9 | — | — |
| 10 | 2.7 | 8.3 | — | — | 11.1 | 4.1 | — | — |
| 11 | 5.4 | 4.3 | — | — | 10.2 | 9.4 | — | — |
| 12 | 6.5 | 2.4 | — | — | 2.9 | 6.9 | — | — |
| 13 | 0 | 0 | — | — | 26.2 | 7.4 | q14.1–q22 | RB1, BRCA3 |
| 14 | 0 | 0 | — | — | 11.3 | 4.1 | — | — |
| 15 | 0 | 0 | — | — | 5.4 | 4.6 | — | — |
| 16 | 16.8 | 3.4 | — | — | 35.9 | 1.8 | q12.1–q24 | SIAH1, CDH1 |
| 17 | 32.1 | 2.9 | p13 | p53, HIC1 | 3.7 | 22.2 | q23–q25 | AXIN2, TIMP2 |
| 18 | 4.1 | 5.5 | — | — | 10.8 | 5 | — | — |
| 19 | 6.9 | 5 | — | — | 3.8 | 10.4 | — | — |
| 20 | 2 | 14.9 | — | — | 0.9 | 18.6 | — | — |
| 21 | 0 | 0 | — | — | 8.8 | 2.2 | — | — |
| 22 | 0 | 0 | — | — | 6.4 | 2.8 | — | — |
| X | 5 | 11.2 | — | — | 4.5 | 15 | — | — |
| Y | 5.1 | 2.3 | — | — | 5.6 | 2.3 | — | — |
Frequencies ⩾20% are highlighted in bold.
Regions with highest frequency of imbalances on the respective chromosomal arm are highlighted in bold.
Examples of known tumour-relevant genes located on the respective chromosomal high-frequency region.
Hepatocellular carcinoma aetiology correlates with specific genomic imbalances
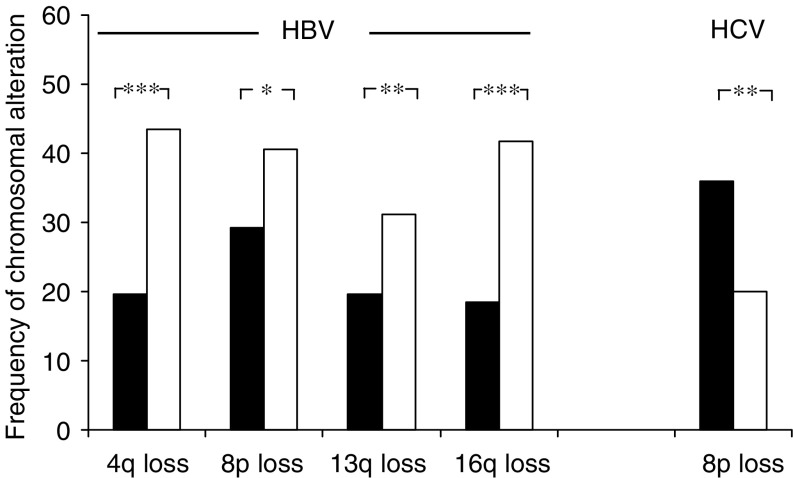
In 428 CGH analyses of HCCs, the HBV status was given (244 HBV-positive and 184 HBV-negative cases). When HBV-positive and -negative cases were compared, losses at 4q (43.4 vs 19.6%), 16q (41.8 vs 18.5%), 13q (31.1 vs 19.6%), and 8p (40.6 vs 29.3%) were positively correlated with HBV aetiology (Figures 1 and 2; Table 2). The 338 HCCs with known HCV status consisted of 110 HCV-positive and 228 HCV-negative cases. Among these cases, only losses of 8p were more frequent in HCV-negative cases (36 vs 20%) (Figure 2; Table 2). No other significant association of genomic imbalances in HCCs with viral infections was found (data not shown).
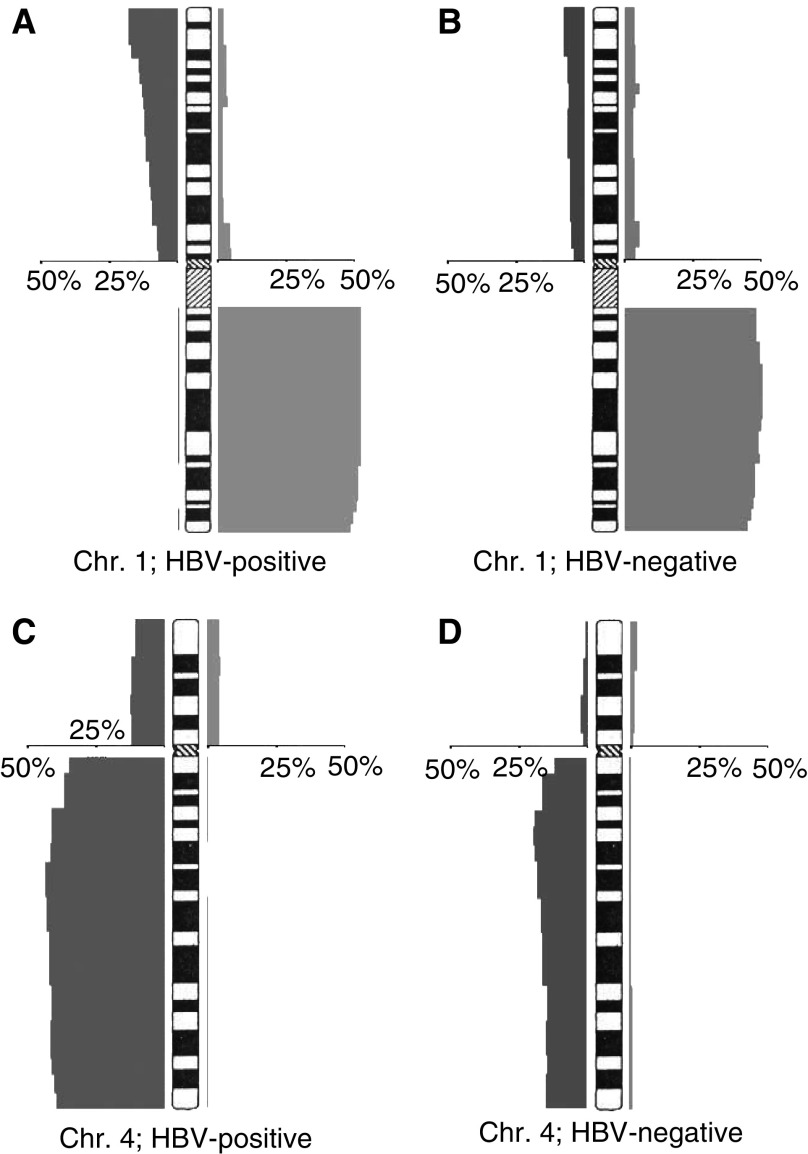
Figure 1.
Distribution of genomic imbalances (n=428) with regard to HBV aetiology (for details, see Table 2). Data are exemplarily demonstrated for chromosomes 1 (no differences between (A) HBV-positive and (B) HBV-negative HCCs) and four (significant difference between (C) HBV-positive and (D) HBV-negative HCCs).
Figure 2.
Graphical comparison of significant high-frequency genomic alterations in HBV- or HCV-negative (black bars) and HBV- or HCV-positive human HCCs (white bars). *P<0.05; **P<0.01; ***P<0.001.
Table 2. Comparison of significant high-frequency genomic imbalances (⩾20%; see Table 1) in human HCCs with HBV- (n=428) and HCV-aetiology (n=338).
|
HBV
*
|
HCV*
|
|||||
|---|---|---|---|---|---|---|
| Chromosome | Negative (n=184) | Positive (n=244) | Negative (n=228) | Positive (n=110) | ||
| 1q (gain; %) | 50.5 | P=0.625 | 53.3 | 53.5 | P=0.201 | 45.5 |
| 4q (loss; %) | 19.6 | P<0.0005 | 43.4 | 26.3 | P=0.896 | 27.3 |
| 6p (gain; %) | 21.2 | P=0.488 | 24.2 | 24.1 | P=0.121 | 16.4 |
| 8p (loss; %) | 29.3 | P=0.019 | 40.6 | 36.0 | P=0.004 | 20.0 |
| 8q (gain; %) | 37.0 | P=0.05 | 46.7 | 42.1 | P=0.194 | 34.5 |
| 13q (loss; %) | 19.6 | P=0.008 | 31.1 | 28.5 | P=0.363 | 23.6 |
| 16q (loss; %) | 18.5 | P<0.0005 | 41.8 | 28.1 | P=0.898 | 27.3 |
| 17p (loss; %) | 25.0 | P=0.108 | 32.4 | 24.1 | P=0.19 | 30.9 |
| 17q (gain; %) | 17.4 | P=0.389 | 20.9 | 19.3 | P=0.766 | 17.3 |
Significant differences are highlighted in bold (P<0.05).
Histological grade of HCCs correlates with genomic imbalances
Next we analysed whether the histological grade (tumour differentiation) of the HCCs correlated with the observed pattern of chromosomal aberrations (Table 3). In 199 cases the histological grade was specified (126 low-grade (G1/G2) and 73 high-grade (G3/G4) HCCs). Losses of 13q (43.8 vs 11.9%) and 4q (45.2 vs 23%) were significantly more frequent in high-grade HCCs while the other high-frequency genomic imbalances did not correlate with the tumour grade (data not shown).
Table 3. Comparison of significant high-frequency genomic imbalances (⩾20%; see Table 1) in human HCCs with regard to tumour grade (n=199; low grade: G1 and G2 tumours; high grade: G3 and G4 tumours).
|
Tumour grade
*
|
|||
|---|---|---|---|
| Chromosome | Low (G1+G2) | High (G3+G4) | |
| 1q (gain; %) | 54.0 | P=0.459 | 60.3 |
| 4q (loss; %) | 23.0 | P=0.001 | 45.2 |
| 6p (gain; %) | 19.8 | P=1.0 | 19.2 |
| 8p (loss; %) | 32.5 | P=0.876 | 34.2 |
| 8q (gain; %) | 39.7 | P=0.299 | 47.9 |
| 13q (loss; %) | 11.9 | P<0.0001 | 43.8 |
| 16q (loss; %) | 23.8 | P=0.053 | 37.0 |
| 17p (loss; %) | 33.3 | P=0.443 | 39.7 |
| 17q (gain; %) | 19.8 | P=0.224 | 27.4 |
Significant differences are highlighted in bold (P<0.05).
Correlations between different genomic imbalances
We next tested whether any of the high-frequency imbalances (⩾20%) were statistically connected. Altogether, 498 CGH analyses of HCCs given as individual profiles were accessible to this analysis. Deletions of genomic material on 4q, 13q, and 16q frequently coincided. Furthermore, gains of 1q were positively associated with all other high-frequency alterations, except gains on chromosome 17q (Table 4).
Table 4. Correlation analysis of genomic aberrations in human HCCs (in ⩾20%; n=498).
| Chromosome | 1q (gain) | 4q (loss) | 6p (gain) | 8p (loss) | 8q (gain) | 13q (loss) | 16q (loss) | 17p (loss) | 17q (gain) |
|---|---|---|---|---|---|---|---|---|---|
| 1q | |||||||||
| gain | — | 43 | 30.6 | 45.7 | 52.6 | 30.2 | 38.1 | 36.8 | — |
| Ø | — | 19.8 | 11.1 | 25.6 | 30.4 | 16.4 | 23.7 | 21.3 | — |
| 4q | |||||||||
| Loss | 73.4 | — | — | 51.6 | — | 42 | 53.2 | 41 | 29.8 |
| Ø | 41.9 | — | — | 28.7 | — | 13.9 | 19.4 | 23.9 | 15.5 |
| 6p | |||||||||
| Gain | 76.9 | — | — | 52.1 | — | — | — | — | — |
| Ø | 45.9 | — | — | 32.6 | — | — | — | — | — |
| 8p | |||||||||
| Loss | 66.3 | 46.6 | 33.7 | — | — | — | 48.3 | 41 | — |
| Ø | 46.6 | 26.3 | 17.2 | — | — | — | 23.1 | 24.4 | — |
| 8q | |||||||||
| Gain | 65.5 | — | — | — | — | — | — | — | — |
| Ø | 43.5 | — | — | — | — | — | — | — | — |
| 13q | |||||||||
| Loss | 68.7 | 57.3 | — | — | — | — | 52.7 | — | — |
| Ø | 48.2 | 25.3 | — | — | — | — | 24.8 | — | — |
| 16q | |||||||||
| Loss | 65 | 56.4 | — | 55.2 | — | 41.1 | — | 42.9 | — |
| Ø | 47.8 | 22.4 | — | 28.7 | — | 16.4 | — | 24.2 | — |
| 17p | |||||||||
| Loss | 66.9 | 44.8 | — | 51.3 | — | — | 46.8 | — | — |
| Ø | 48.8 | 28.5 | — | 31.1 | — | — | 25.6 | — | — |
| 17q | |||||||||
| Gain | — | 55 | — | — | — | — | — | — | — |
| Ø | — | 28.2 | — | — | — | — | — | — | — |
Only significant differences between distinct chromosomal imbalances (first line) and the possibles status of all other high-frequency alterations (gain/loss vs no changes) are shown (P<0.05).
Hepatocellular carcinomas with gains on 1q (n=293; 58.8%) were compared to cases without genomic alterations of 1q (n=205; 41.2%) regarding the occurrence of additional chromosomal imbalances per case. A significantly higher number of chromosomal imbalances were found in HCCs with 1q gains (6.74 imbalances per case) than in HCCs without 1q gains (3.40 imbalances per case; P<0.05).
Genomic macroimbalances in human DNs
Four different studies have analysed a limited number of DNs (n=30) by CGH. Altogether, DNs showed genomic imbalances, although at a lower frequency as compared to HCCs. Gains were detectable in 1q (30%) and 8q (10%), while losses were most prevalent at 8p (16.7%), 17p (16.7%), 5p, 13q, 14q, and 16q (all 10%); (Figure 3). In DNs only gains on 1q were frequently detectable in more than 20% of all analysed cases.
Figure 3.
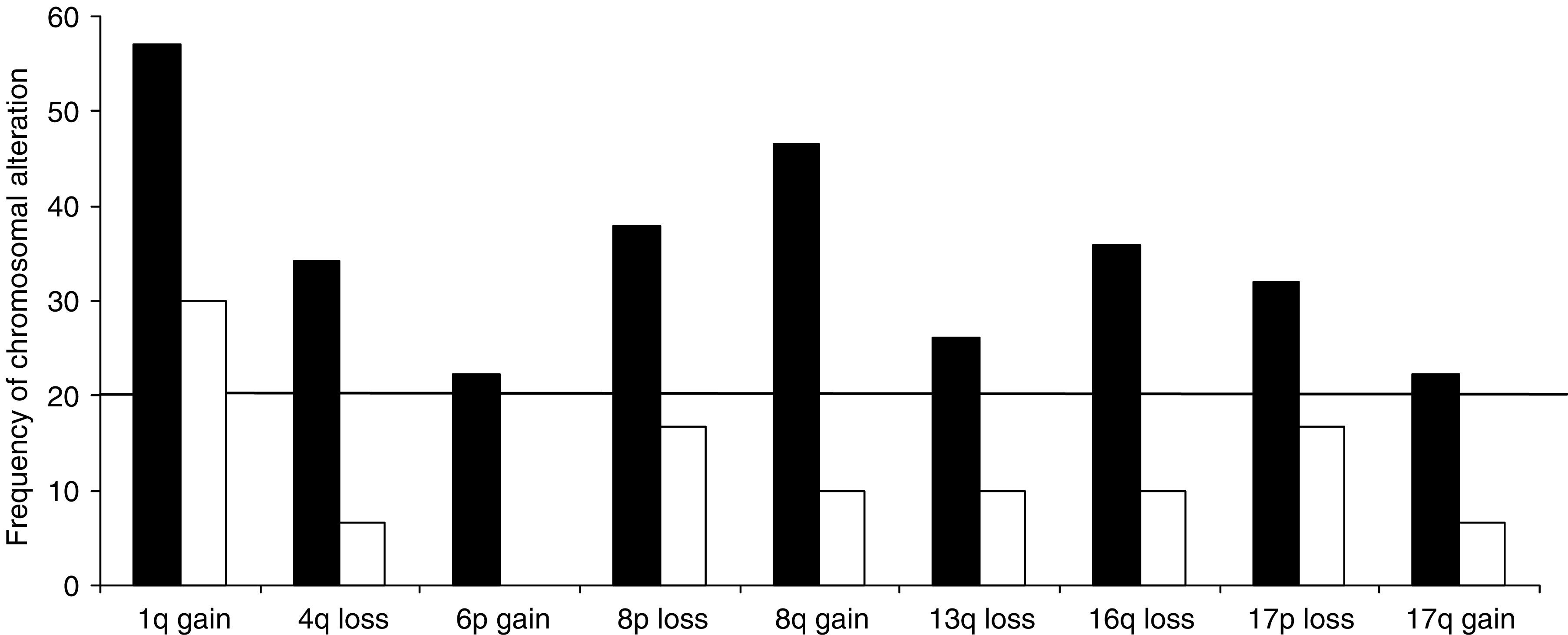
Direct comparison of chromosomal high-frequency alterations (⩾20%) in HCCs (black bars) and related DNs (white bars).
DISCUSSION
This meta-analysis of CGH data of human HCCs has unravelled several correlations between aetiology as well as histological grade and genomic imbalances. However, this analysis is afflicted with several restrictions as a result of not identical criteria in different studies.
Aetiology: Some well-established reasons for human HCCs, such as chronic alcoholic liver disease, genetic haemochromatosis, and aflatoxin B1 exposure, are either ill defined and/or have not been evaluated in a sufficient number of cases studied by CGH. Therefore, we restricted our analysis to the potential associations with the HBV and HCV status. Analyses of the viral status carried some inconsistency by itself. Especially, in most studies HBV aetiology was evaluated by assaying for HBsAg in the serum. In contrast, HBV may exhibit its protumorigenic effects despite HBV seroconversion (Paterlini and Brechot, 1991). Significant correlations of chromosomal rearrangements with HBV aetiology have been unravelled, but in our analysis they may have been even more pronounced, if complete HBV serology would have been performed in all studies.
Lack of a uniform histological grading: Several grading schemes are in use for HCC (Edmondson and Steiner, 1954), and in many studies it remains unclear which grading scheme has been applied. Due to this fact and in order to generate groups large enough for statistical analysis, it was necessary to combine grades 1 and 2 (well differentiated) and 3 and 4 (poorly differentiated), which is in accordance which numerous studies in other carcinomas.
Despite these restrictions, a number of high-frequency genomic imbalances have been clearly identified (Figure 1). With some of these aberrations the expression of tumour relevant genes may be correlated, such as RB1 (13q), CDH1 (16q), SIAH1 (16q), and TP53 (17p). Interestingly, some of these genes have been described to participate in pivotal signalling pathways frequently dysregulated in HCCs (reviewed in Ozturk, 1999). While the p53 protein is involved in different cellular processes (e.g. apoptosis, cell cycle, and differentiation; Levine, 1997), dysregulation of retinoblastoma protein (RB1), p21WAF (CDKN1A), and CyclinA2 (CCNA) directly regulate initiation and progression of DNA synthesis (Zhang et al, 1994; Qin et al, 1998). Notably, several components of the WNT-signalling pathway were localised on aberrant genomic regions (WNT14, LEF1, FZD3, WISP1, SIAH-1, and AXIN2). This is of particular interest since upto 40% of all HCCs exhibit nuclear enrichment of β-catenin, the transcriptional activator of the WNT pathway (Cui et al, 2003; Prange et al, 2003). However, higher resolution techniques, such as matrix-CGH (Wessendorf et al, 2002) and expression analyses, will have to verify the expected expression changes for most genes.
Losses of 4q, 13q, 16q, and 8p were significantly correlated with HBV aetiology. Although the responsible genes are currently unknown, HBV-related chromosomal rearrangements have been mapped to 4q (Blanquet et al, 1988; Pasquinelli et al, 1988; Buetow et al, 1989). This suggests that in addition to well-established HBV-associated oncogenic mechanisms such as transcriptional transactivation by viral proteins (Wollersheim et al, 1988; Caselmann et al, 1990; Kekule et al, 1990) and nonhomologous chromosomal integration of viral DNA (Nagaya et al, 1987), specific corresponding host factors involving tumour suppressor genes positioned at the respective chromosomal loci may exist. Identification of these responsible genes may generate further insight into the mechanisms of HBV-induced oncogenesis.
Although the number of analysed premalignant lesions (DNs) is still low, several conclusions can already been drawn: Significant genomic imbalances can be detected in DNs and they partly resemble the changes present in HCCs, although at a lower frequency. This further supports the current hypothesis that DNs are indeed the immediate premalignant precursors of HCCs.
Another important question is the time point at which the different genomic imbalances occur during the process of hepatocarcinogenesis. Gains of 1q are the most frequent alteration in DNs and appear at equal frequencies in well and poorly differentiated HCCs; this suggests that amplifications of 1q represent an early protumorigenic change that mostly precedes malignant transformation. In contrast, deletions of 4q and 13q are found significantly more frequent in poorly differentiated HCCs. This suggests that both alterations are late progression events that typically occur after malignant transformation.
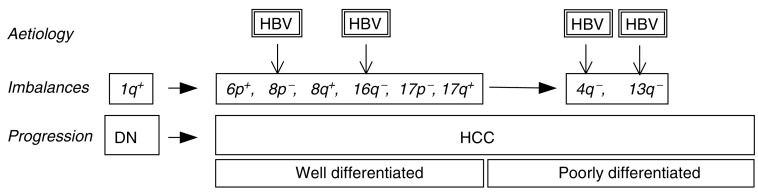
Statistical analyses demonstrate that 1q gains are positively correlated with all other high-frequency alterations, suggesting that they may predispose to chromosomal alterations. Thus, the status of 1q may distinguish between two different molecular pathways in hepatocarcinogenesis, of which cases with 1q gains are characterised by early-on acquired genomic imbalances (‘mutator phenotype’, Figure 4). This hypothesis is further supported by our finding that HCCs with 1q gains carry significantly more additional chromosomal alterations as compared to HCCs without 1q amplifications.
Figure 4.
Schematic display of the CGH meta-analysis showing high-frequency chromosomal changes (imbalances) in association with aetiology and progression.
Taking these considerations into account, the following hypotheses can be formulated:
Dysplastic nodules are premalignant precursor lesions that already carry fixed genomic alterations.
Specific aetiologies, especially chronic HBV infection, may lead to characteristic host genomic alterations. Deletions on chromosome 4q, 13q, and 16q strongly correlate with HBV aetiology and tumour progression (4q and 13q), and may therefore contribute to the functional loss of tumour suppressors.
Gains of 1q are the predominant early genomic alterations. They are aetiology-independent and may further predispose to other chromosomal imbalances (‘mutator phenotype’).
These hypotheses will have to be tested experimentally by several means. Firstly, the data basis in regard to DNs is still restricted and it is worthwhile to increase the number of CGH analyses of DNs. Secondly, the resolution of conventional CGH analysis to identify regions of interest is limited (approximately 3–10 Mbp). This gap may in part be closed by high-resolution techniques such as matrix-CGH (down to approximately 100 kbp (Wessendorf et al, 2002; Pestova et al, 2004). Identification and functional analyses of potential target genes may finally unravel the mechanisms that predispose to secondary chromosomal changes (e.g. 1q) or aetiology-specific alterations (e.g. 4q).
Acknowledgments
The study was supported by grants from the Dr Mildred Scheel-Stiftung für Krebsforschung, and the Deutsche Forschungsgemeinschaft to PS.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc).
Supplementary Material
References
- Blanquet V, Garreau F, Chenivesse X, Brechot C, Turleau C (1988) Regional mapping to 4q32.1 by in situ hybridization of a DNA domain rearranged in human liver cancer. Hum Genet 80: 274–276 [DOI] [PubMed] [Google Scholar]
- Breuhahn K, Vreden S, Haddad R, Beckebaum S, Stippel D, Flemming P, Nussbaum T, Caselmann WH, Haab BB, Schirmacher P (2004) Molecular profiling of human hepatocellular carcinoma defines mutually exclusive interferon regulation and insulin-like growth factor II overexpression. Cancer Res 64: 6058–6064 [DOI] [PubMed] [Google Scholar]
- Buetow KH, Murray JC, Israel JL, London WT, Smith M, Kew M, Blanquet V, Brechot C, Redeker A, Govindarajah S (1989) Loss of heterozygosity suggests tumor suppressor gene responsible for primary hepatocellular carcinoma. Proc Natl Acad Sci USA 86: 8852–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselmann WH, Meyer M, Kekule AS, Lauer U, Hofschneider PH, Koshy R (1990) A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc Natl Acad Sci USA 87: 2970–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhou X, Liu Y, Tang Z, Romeih M (2003) Wnt signaling in hepatocellular carcinoma: analysis of mutation and expression of beta-catenin, T-cell factor-4 and glycogen synthase kinase 3-beta genes. J Gastroenterol Hepatol 18: 280–287 [DOI] [PubMed] [Google Scholar]
- Edmondson HA, Steiner PE (1954) Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 7: 462–503 [DOI] [PubMed] [Google Scholar]
- El-Serag HB (2002) Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol 35: S72–S78 [DOI] [PubMed] [Google Scholar]
- Feitelson MA (1998) Hepatitis B x antigen and p53 in the development of hepatocellular carcinoma. J Hepatobiliary Pancreat Surg 5: 367–374 [DOI] [PubMed] [Google Scholar]
- Kato N, Lan KH, Ono-Nita SK, Shiratori Y, Omata M (1997) Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol 71: 8856–8859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kekule AS, Lauer U, Meyer M, Caselmann WH, Hofschneider PH, Koshy R (1990) The preS2/S region of integrated hepatitis B virus DNA encodes a transcriptional transactivator. Nature 343: 457–461 [DOI] [PubMed] [Google Scholar]
- Kern MA, Breuhahn K, Schirmacher P (2002a) Molecular pathogenesis of human hepatocellular carcinoma. Adv Cancer Res 86: 67–112 [DOI] [PubMed] [Google Scholar]
- Kern MA, Schubert D, Sahi D, Schoneweiss MM, Moll I, Haugg AM, Dienes HP, Breuhahn K, Schirmacher P (2002b) Proapoptotic and antiproliferative potential of selective cyclooxygenase-2 inhibitors in human liver tumor cells. Hepatology 36: 885–894 [DOI] [PubMed] [Google Scholar]
- Koike K (1995) Hepatitis B virus HBx gene and hepatocarcinogenesis. Intervirology 38: 134–142 [DOI] [PubMed] [Google Scholar]
- Levine AJ (1997) p53, the cellular gatekeeper for growth and division. Cell 88: 323–331 [DOI] [PubMed] [Google Scholar]
- Luber B, Arnold N, Sturzl M, Hohne M, Schirmacher P, Lauer U, Wienberg J, Hofschneider PH, Kekule AS (1996) Hepatoma-derived integrated HBV DNA causes multi-stage transformation in vitro. Oncogene 12: 1597–1608 [PubMed] [Google Scholar]
- Matsumura T, Makino R, Mitamura K (2001) Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin Cancer Res 7: 594–599 [PubMed] [Google Scholar]
- Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K (1998) The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med 4: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Nagaya T, Nakamura T, Tokino T, Tsurimoto T, Imai M, Mayumi T, Kamino K, Yamamura K, Matsubara K (1987) The mode of hepatitis B virus DNA integration in chromosomes of human hepatocellular carcinoma. Genes Dev 1: 773–782 [DOI] [PubMed] [Google Scholar]
- Ozturk M (1999) Genetic aspects of hepatocellular carcinogenesis. Semin Liver Dis 19: 235–242 [DOI] [PubMed] [Google Scholar]
- Pasquinelli C, Garreau F, Bougueleret L, Cariani E, Grzeschik KH, Thiers V, Croissant O, Hadchouel M, Tiollais P, Brechot C (1988) Rearrangement of a common cellular DNA domain on chromosome 4 in human primary liver tumors. J Virol 62: 629–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini P, Brechot C (1991) The detection of hepatitis B virus (HBV) in HBsAG negative individuals with primary liver cancer. Dig Dis Sci 36: 1122–1129 [DOI] [PubMed] [Google Scholar]
- Pestova E, Wilber K, King W (2004) Microarray-based CGH in cancer. Methods Mol Med 97: 355–375 [DOI] [PubMed] [Google Scholar]
- Prange W, Breuhahn K, Fischer F, Zilkens C, Pietsch T, Petmecky K, Eilers R, Dienes HP, Schirmacher P (2003) Beta-catenin accumulation in the progression of human hepatocarcinogenesis correlates with loss of E-cadherin and accumulation of p53, but not with expression of conventional WNT-1 target genes. J Pathol 201: 250–259 [DOI] [PubMed] [Google Scholar]
- Qin LF, Ng IO, Fan ST, Ng M (1998) p21/WAF1, p53 and PCNA expression and p53 mutation status in hepatocellular carcinoma. Int J Cancer 79: 424–428 [DOI] [PubMed] [Google Scholar]
- Weiss MM, Hermsen MA, Meijer GA, van Diest PJ (2002) Comparative genomic hybridization in cancer investigations. Methods Mol Biol 204: 369–378 [DOI] [PubMed] [Google Scholar]
- Wessendorf S, Fritz B, Wrobel G, Nessling M, Lampel S, Goettel D, Kuepper M, Joos S, Hopman T, Kokocinski F, Dohner H, Bentz M, Schwaenen C, Lichter P (2002) Automated screening for genomic imbalances using matrix-based comparative genomic hybridization. Lab Invest 82: 47–60 [DOI] [PubMed] [Google Scholar]
- Wollersheim M, Debelka U, Hofschneider PH (1988) A transactivating function encoded in the hepatitis B virus X gene is conserved in the integrated state. Oncogene 3: 545–552 [PubMed] [Google Scholar]
- Zhang X, Xu HJ, Murakami Y, Sachse R, Yashima K, Hirohashi S, Hu SX, Benedict WF, Sekiya T (1994) Deletions of chromosome 13q, mutations in Retinoblastoma 1, and retinoblastoma protein state in human hepatocellular carcinoma. Cancer Res 54: 4177–4182 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.