Abstract
Heme oxygenase (HO) catalyzes the opening of the heme ring with the release of iron in both plants and animals. In cyanobacteria, red algae, and cryptophyceae, HO is a key enzyme in the synthesis of the chromophoric part of the photosynthetic antennae. In an attempt to study the regulation of this key metabolic step, we cloned and sequenced the pbsA gene encoding this enzyme from the red alga Rhodella violacea. The gene is located on the chloroplast genome, split into three distant exons, and is presumably expressed by a trans-splicing mechanism. The deduced polypeptide sequence is homologous to other reported HOs from organisms containing phycobilisomes (Porphyra purpurea and Synechocystis sp. strain PCC 6803) and, to a lesser extent, to vertebrate enzymes. The expression is transcriptionally activated under iron deprivation, a stress condition frequently encountered by algae, suggesting a second role for HO as an iron-mobilizing agent in photosynthetic organisms.
Keywords: rhodophyta, chloroplast, genome, intron, phycobilin
In the complex pathway of tetrapyrrole biosynthesis, heme oxygenase (HO) catalyzes the opening of the heme (iron protoporphyrin IX) ring at the alpha methene bridge to form biliverdin IX, Fe3+, and carbon monoxide. It is a highly conserved enzyme throughout the animal and plant kingdoms. In animals, HO is involved in a catabolic process: its primary role is to degrade hemoglobin in senescent red blood cells with the concomitant recycling of iron. HO from at least 12 animal tissues (reviewed in ref. 1) has been characterized. In higher plants and algae, HO is involved in a metabolic process, the synthesis of tetrapyrrole chromophores (2), phycobilins, and phytochromobilins. The phycobilins are the chromophores of the phycobiliproteins, light harvesting complexes of various photosynthetic organisms such as cyanobacteria and rhodophytes, where they are assembled in phycobilisomes (PBS), and cryptophytes. The phytochromobilins are the chromophores of the phytochromes, photoreceptors encountered in higher plants, chlorophytes, and presumably also in red algae. HO from one red alga, Cyanidium caldarium (3), has been extensively characterized. In higher plants, where the pathway of phytochrome synthesis (4) is proposed to be the same as for the related bilins in algae (2), no enzymatic characterization of this first metabolic step has been performed, although mutants deficient in phytochrome chromophore biosynthesis have been described in Arabidopsis (5, 6) and pea (7).
In mammals, HO is a microsomal enzyme; activity in vitro is associated with the electron donor NADPH-cytochrome P450 reductase (8, 9). Animal HO exists as two isoenzymes: HO-1 (32 kDa), inducible by a large number of environmental agents (including heme), and the constitutive HO-2 (34 kDa). In contrast to the animal HO anchored to the microsomal fraction, the algal enzyme is soluble and requires ferredoxin and ferredoxin-NADP reductase for activity (3). Algal heme oxygenase from C. caldarium has an apparent native molecular weight of 38 kDa (3) and is inducible by δ-aminolevulinic acid or another precursor of heme (10).
At the molecular level, several animal HO-1 and HO-2 genes or corresponding cDNA have been cloned and sequenced. The deduced amino acid sequences of HO-1 and HO-2 are about 40% similar. The 5′ untranslated region of HO-1 genes contains numerous regulatory sites presumably involved in its complex regulation (1). The HO gene has not yet been isolated from higher plants. In algae, an HO gene (called pbsA for phycobilin synthesis) has been identified by systematic mapping, cloning, and sequencing of the chloroplast genome of the rhodophyte Porphyra purpurea (11). The gene of C. caldarium has not been isolated, but the use of protein and RNA synthesis inhibitors has shown that the gene, in contrast to that of P. purpurea pbsA, is nuclear and that the protein is synthesized on cytoplasmic ribosomes (10). Two different HO genes, named ho, were identified in the cyanobacterium Synechocystis sp. strain PCC 6803 by homology with P. purpurea pbsA (12). A bacterial gene product (HmuO), homologous to heme oxygenase has been reported in Corynebacterium diphthriae (13).
We have studied the regulation of phycobilisome synthesis in cyanobacteria and red algae in our laboratory (14–17). Although the apoprotein part of the pigment has received much attention in the marine red alga Rhodella violacea, nothing is known about the genes involved in the synthesis of the chromophores of the phycobilisomes. We report here the isolation and characterization of the pbsA gene from this organism. We demonstrate that this gene is chloroplastic, split into three distant exons, and encodes a polypeptide of 27 kDa homologous to other previously described heme oxygenases. Due to its position at a key step in a heme- and ferredoxin- dependent pathway, it was reasonable to presume that the gene for heme oxygenase could be regulated by iron availability. We therefore studied the effect of iron deprivation on the pbsA mRNA level and show a transcriptional activation in response to iron deprivation. This finding suggests an additional role for heme oxygenase as an iron-mobilizing agent in photosynthetic organisms during iron-stress conditions.
MATERIALS AND METHODS
Materials.
Restriction enzymes were purchased from either Eurogentech or Amersham. [α-32P]dATP or dCTP (3,000 Ci/mmol; 1 Ci = 37 GBq), [α-33P]dATP (1,000–3,000 Ci/mmol), Hybond N membrane, and multiprime kits were from Amersham. The kilobase sequencing system was from BRL. Plasmid pTZ18 was from Pharmacia. Enzymes were used according to the manufacturer’s instructions.
Culture Conditions and Growth Media.
The unicellular marine red alga R. violacea (strain 115-79) was obtained from Sammlung von Algenkulturen, Pflanzenphysiologisches Institut der Universität (Göttingen Germany). Cells were grown photoautotrophically in sterile artificial seawater (18) with the addition of vitamin B12 at 25 μg/liter. Cultures were incubated at 21°C in glass culture flasks continuously flushed with sterile air, illuminated with fluorescent tubes (120 μM quanta/m−2/s−1) with a 16 hr light/8 hr dark photoperiod.
For iron regulation studies, growth was measure under the same conditions with different iron concentrations ranging from 10 μM (standard artificial seawater) to 0.05 μM FeCl3.
DNA Purification, Genomic Library Construction, and Hybridization.
Plastid DNA [chloroplast DNA (cpDNA)] from R. violacea was isolated by procedures described for chromophyte algae (19). Total or partial DNA libraries were constructed by ligation of HindIII- or EcoRI-digested plastid DNA into pTZ18R. Escherichia coli strain GT869 (20) was used as bacterial host. Standard methods (21) were used for subcloning, in situ colony hybridization and Southern blot analysis.
The heterologous probes were the total pbsA gene from P. purpurea (11) or a part of the gene encoding the last 175 amino acids, obtained by PCR. Homologous probes were either a 0.9-kb HindIII fragment from clone 1 (Fig. 1A) or a cDNA in the region covering the first exon (174 nt), the second exon (75-nt) and the first 42 nt of the third exon, obtained by reverse transcription–PCR (RT-PCR) (see below).
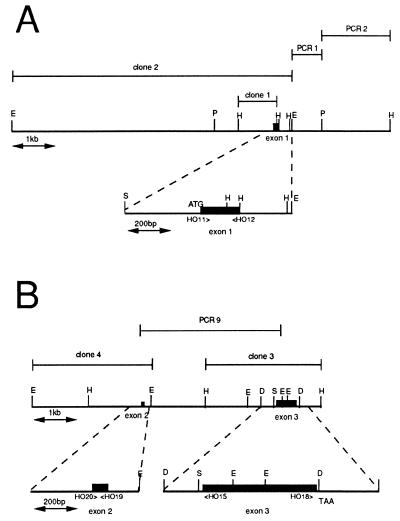
Figure 1.
Restriction maps of the two regions from R. violacea plastid DNA containing respectively exon 1 and exons 2 + 3 of the HO-encoding gene. Representative restriction sites: D, DraI, E, EcoRI, H, HindIII, P, PvuII, S, Sau3A. (A) The 8.8-kb region containing exon 1 obtained by cloning a 6.8-kb EcoRI fragment and its 2-kb flanking region by PCR walk on plastid DNA. (Lower) Sequenced region presented in Fig. 2A. (B) The 7-kb region containing exons 2 and 3. The whole was obtained by overlapping clones: a 2.7-kb HindIII clone (clone 3) containing exon 3, a 3.8-kb PCR product (PCR9) obtained between exon 2 and exon 3, and a 2.7-kb EcoRI clone (clone 4) isolated by screening a total EcoRI library with the 75-nt exon 2 obtained by PCR. (Lower) Sequenced regions presented in Fig. 2 B and C.
DNA Sequencing.
The DNA sequences were determined by the dideoxyribonucleotide chain termination method (22) on single- or double-stranded DNA with the use of reverse or “-40” primers from M13 or with individual oligonucleotides (Genset, Paris) designed from the R. violacea sequence. All sequences were determined on both strands.
Walk-On Plastid DNA.
To sequence the DNA region situated in the vicinity of the 6.8-kb EcoRI clone (clone 2 in Fig. 1A), we progressed along the plastid DNA by random cloning of PvuII fragments (total digest of plastid DNA) in pTZ18R followed by amplification with a primer designed at the end of the EcoRI fragment and the reverse primer of pTZ18R. Using these two primers, we amplified a 0.8-kb fragment (PCR 1 in Fig. 1A); we then progressed by random cloning of HindIII fragments in pTZ18R followed by amplification with a primer designed at the end of the previous PCR 1 fragment and the reverse primer of pTZ18R. With these two primers we amplified a 1.2-kb fragment (PCR 2 in Fig. 1A).
Amplification of Large Fragments on Plastid DNA.
To determine the respective location of each of the three exons of R. violacea pbsA, we attempted to amplify by means of the Expand Long Template PCR System from Boehringer Mannheim (supposed to amplify up to 30 or 40 kb DNA) the plastid DNA between exons, using 6 oligonucleotides shown in Figs. 1 and 2 and associated in the 12 relevant combinations: HO11 (5′-CTATGATATATGACACTAATTTAG-3′), HO12 (5′-CATTAAGCTTACTTCATCTTCAATA-3′), HO20 (5′-ATGAAAAGACATCAAGAAAGT-3′), HO19 (5′-TTCTAAACTAGATTTTCTATT-3′), HO15 (5′-CTTGTCTTGCCATTCTAATCC-3′), and HO18 (5′-GCTTTTA AGTT- AAATATGAAA-3′).
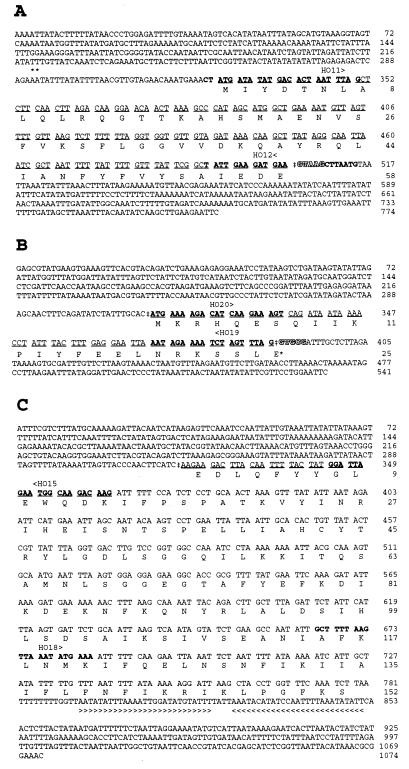
Figure 2.
DNA and amino acid sequences encoded by the exon 1 (A), exon 2 (B), and exon 3 (C) and flanking regions of the pbsA gene from R. violacea. DNA sequences underlined were confirmed by cDNA sequencing. The resulting RNA splice sites are marked by a †. Pentanucleotides characteristic of type II introns 5′ sequences are shaded. Nucleotides (or complementary sequences) in bold type correspond to oligonucleotides used as primers for PCR and RT-PCR. The 5′ end of mRNA is indicated by asterisks (∗) in A. Glutamic E*25 in B is encoded by GAA after the splicing event joining exons 2 and 3. Inverted repeat at the 3′ end of pbsA is indicated by the arrowed line in C.
RNA Isolation and Northern Hybridization.
Total RNA from R. violacea was isolated as described for cyanobacteria (23). RNA (5–10 μg) was electrophoresed on 1.2% denaturing agarose gels in HF buffer (0.5 M Hepes/10 mM EDTA/16% formaldehyde) and transferred to nylon membrane with 20× standard saline citrate (SSC); blots were prehybridized, hybridized (42°C), and washed according to Damerval et al. (24).
The probes used in Northern hybridizations were exon 1 (174 nt), exon 2 (75 nt) obtained by PCR amplification, and exon 3 (575 nt Sau3A–DraI in Fig. 1B).
RT-PCR.
To sequence the cDNA in the junction region between exons 1 and 3, we did coupled RT-PCR amplification. Total RNA (5 μg) from R. violacea was used with 0.02 pmol of oligonucleotide HO15 (Figs. 1B and 2C) as antisense primer, to synthesize the first strand with 400 units of reverse transcriptase (GIBCO) in the presence of 20 nanomol of each dNTP and 20 units of RNasin (Promega), an RNase inhibitor. After inhibition for 5 min at 95°C, the product of retrotranscription was amplified using HO11 (see Figs. 1A and 2A) as forward primer and HO15 as reverse primer, using Promega Taq polymerase in buffer and salt conditions recommended by the manufacturer. The program of the thermal cycler was the following: 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 50°C for 1 min, and extension at 72°C for 1 min.
RT-PCR was also used to synthesize the cDNA corresponding to a putative pre-mRNA common to exon 2 and 3 separated by 3.8 kb. For cDNA synthesis HO15 was used as antisense primer, and PCR amplification was then performed with HO20 and HO15 as forward and reverse primers, respectively.
5′ mRNA Determination.
Primer extension was used to map the 5′ terminus of mRNAs. The 25- or 21-mer oligonucleotides were used: HO12, HO19, and HO15, respectively complementary to exon 1, exon 2, and exon 3 (see Fig. 2). A modification of the method described (21) was followed: 30 μg of total RNA from an exponential culture of R. violacea was annealed overnight at 30°C (after 10 min denaturation at 85°C) with 10 pmol of each primer in 40 mM Pipes (pH 6.4), 1 mM EDTA (pH 8), 0.4 M NaCl, and 80% deionized formamide. After ethanol precipitation, the hybrid was incubated in 10 μl with 400 units of superscript reverse transcriptase (GIBCO) in the appropriate buffer in the presence of 1 mM DTT, 0.15 μM dCTP, dGTP, and DTTP, and 10 μCi of [33P]dATP. After 10 min of labeling, extension was performed for 2 h by addition of 10 μM of each dNTP. Nonhybridized RNA was digested for 30 min by 0.5 μg of RNase. The samples were then ethanol-precipitated and loaded on a sequencing gel.
RESULTS
Gene Cloning and Determination of Nucleic Acid Sequences.
The expected phylogenetic relationships between the two rhodophytes R. violacea and P. purpurea led us to attempt isolation of the pbsA gene of R. violacea by means of heterologous DNA hybridization experiments. A probe containing the pbsA gene of P. purpurea (11) was hybridized to Southern blots of nuclear and plastid (cpDNA) DNA fractions of R. violacea digested with different restriction enzymes (data not shown). Only the cpDNA showed positive signals, and the most appropriate enzyme was HindIII, giving two positive fragments of 2.7 and 0.9 kb. The 0.9-kb fragment was first isolated from a total HindIII library constructed in pTZ18R (clone 1 in Fig. 1A). This HindIII fragment was then used as an homologous probe to screen a total EcoRI library in pTZ18R where a 6.8-kb insert clone (clone 2 in Fig. 1A) was selected, in agreement with the result obtained with the heterologous P. purpurea probe on Southern blotting.
The DNA sequence of 774 nt of this region (Figs. 1A and 2A) revealed the presence of an ORF that corresponded to only the first 58 amino acids of the R. violacea pbsA gene, as confirmed by similarity with the protein deduced from the P. purpurea gene (GenBank accession no. P51271). This sequence provided an indication of the presence of an intron within the heme oxygenase gene of R. violacea. Analysis of the region downstream from the EcoRI site (Fig. 1A) was performed by PCR walking on the cpDNA, and 2 kb were thus cloned (PCR1+ PCR2, Fig. 1A). The remaining part of the pbsA gene was not encoded within this 2-kb region, as shown by hybridization with the P. purpurea probe, and by partial sequencing.
A 2.7-kb HindIII fragment (clone 3, Fig. 1B) was further isolated from a partial pTZ18R library by heterologous hybridization with a probe containing the 3′ part of the P. purpurea pbsA gene. From this second region (Fig. 1B Right) a continuous DNA sequence of 1,074 bp was obtained (see Fig. 2C). By comparison with P. purpurea HO, we identified in this sequence the coding information for the C-terminal 152 residues. Between these two coding regions (Fig. 2 A and C), a gap of 25 amino acids was observed in the R. violacea polypeptide in comparison to other HOs. To verify the polypeptide sequence of R. violacea HO deduced from the sequencing of the two plastid DNAs already cloned, we synthesized and sequenced the cDNA corresponding to the junction of these regions (see Materials and Methods). A product of 300 bp was obtained and sequenced. The sequences of the entire exon 1 and the beginning of the distal exon were recovered, but separated by a new coding sequence of 75 nt that covered the gap of the 25 “missing” amino acids, thus confirming the presence of a third central exon.
Using oligonucleotides HO20 and HO19 (Figs. 1B and 2B) we synthesized the central 75-nt exon 2 and used it as a probe against a Southern blot of cpDNA. We thus detected a positive 2.7-kb EcoRI fragment and screened a total EcoRI library into which a clone (clone 4, Fig. 1B) containing this insert was found and partly sequenced. The sequence of exon 2 and flanking regions is presented in Fig. 2B.
Positioning the Three Different Exons Within the R. violacea Chloroplast Genome.
To determine the distance between the three exons of R. violacea pbsA, we attempted to amplify the plastid DNA present between the different coding regions using the 12 relevant combinations of oligonucleotides in the two orientations of the three exons by means of an Expand Long Template PCR System (Boehringer Mannheim). Only the pair of oligonucleotides HO15 and HO20 resulted in an amplification product of 3.8 kb (see PCR 9 in Fig. 1B) that was cloned and sequenced in the vicinity of exon 2. This result confirmed the distance of about 3.8 kb and orientation between exon 2 and exon 3. The fact that no amplification product was obtained when HO11 and HO12 were used allows us to suggest that the distance between exon1 and exons 2–3 is more than 20 kb.
Transcriptional Analysis.
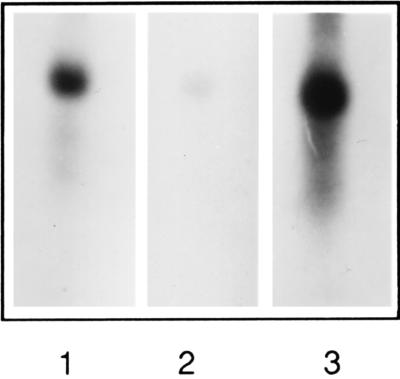
To identify the transcription product of the HO gene parts, we used specific DNA probes of exons 1, 2, and 3 of the pbsA gene in Northern blot analysis. The three probes hybridized with a common 0.85-kb mRNA transcript (Fig. 3) confirming the assumption of a functional gene represented by these three exons.
Figure 3.
Autoradiogram of Northern blots of total RNA (5 μg) from R. violacea hybridized with pbsA exon 1 (lane 1), exon 2 (lane 2), and exon 3 (lane 3) probes. Autoradiography was for 20 h for lanes 1 and 3, and 72 h for lane 2.
We attempted to characterize other RNA representing pre-mRNA species by hybridization with the three probes specific to exons 1, 2, and 3. Growing cells in the cold, conditions which should slow down splicing (25), and long exposure of the autoradiogram did not permit the detection of any other significant transcript. We also attempted to detect a common pre-mRNA between exons 2 and 3 by RT-PCR using oligonucleotides HO20 and HO15 (Fig. 1B); no such species was obtained.
5′ End of mRNA.
The 5′ end of the pbsA transcript was mapped by primer extension with oligonucleotides hybridizing with exon 1 (HO12), exon 2 (HO19), and exon 3 (HO15). The three oligonucleotides give a 5′ end for the mature mRNA occurring 37 or 38 bp before the initiating ATG (nucleotides 292 and 293; see the ∗ in Fig. 2A).
pbsA, a Gene Activated by Iron Stress.
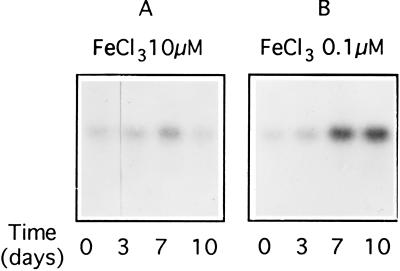
To determine the effect of iron on pbsA transcription, cells were grown for several generations in artificial sea water medium containing iron concentrations ranging from 10 to 0.05 μM FeCl3. Iron limitation did not affect growth rate; the doubling time was ≈26–30 h for both control and deficient cells. However, for iron concentrations below 0.2 μM, cell size and pigment content (chlorophyll a and phycobiliproteins) decreased. R. violacea cells responded to a reduction of Fe concentration from 10 μM (Fig. 4A) to 0.1 μM (Fig. 4B) by increasing the levels of the pbsA transcript. We assume that the level of HO activity is consequently increased.
Figure 4.
Autoradiogram of Northern blots of total RNA (5 μg) from R. violacea grown either in 10 μM (A) or in 0.1 μM FeCl3 (B) in artificial sea water medium. Hybridization was performed with pbsA exon 1 probe.
DISCUSSION
In this paper we present results concerning the isolation, cloning and sequencing of the HO gene, pbsA, from the unicellular red alga R. violacea.
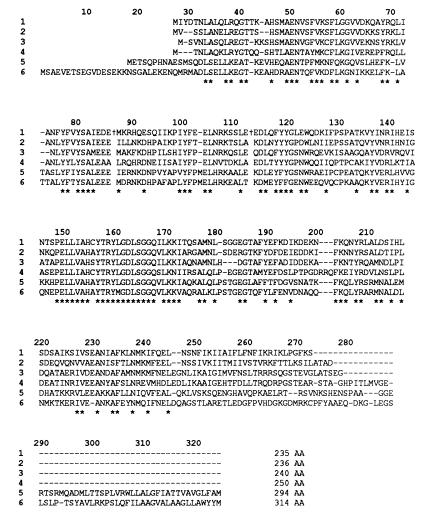
The coding region of the R. violacea pbsA gene is highly similar to the corresponding genes from a variety of organisms (see Fig. 5). The R. violacea HO deduced from the nucleotide sequence shows identity of 59%, 52% ,and 43% with P. purpurea, Synechocystis sp. strain PCC 6803 (product of gene 1, sll1184), and Synechocystis sp. strain PCC 6803 (product of gene 2, sll1875), respectively; taking into account conservative substitutions of amino acids, similarities reach 76%, 73%, and 65% respectively. The HO from these four organisms are very close in size (27–29 kDa), the shortest being the R. violacea enzyme, and significantly shorter than the heme-binding component (38 kDa) of C. caldarium estimated by gel filtration on a Sephadex G75 column (3). Similarity is greatest around the active site that contains highly conserved hydrophobic residues 148–173 (Fig. 5) and includes the essential His-154 supposed to bind heme (9). Animal HO are longer than algal polypeptides; they contain a C-terminal tail that is involved in the anchoring of the protein to the microsomal fraction, a tryptic digest of the C terminus resulting in a catalytically active soluble enzyme. Furthermore, HO-2 has a N-terminal extension of 20 or so residues compared with the HO-1. In the region where HO from R. violacea and animals are aligned, a significant (around 33%) similarity is observed. Similarity in the same range is found between R. violacea pbsA and C. diphteriae hmuO (13) gene products.
Figure 5.
Comparison of amino acid sequences derived from the homologous heme oxygenase genes from different sources: line 1, R. violacea, this work; line 2, Porphyra purpurea (26); line 3, Synechocystis sp. strain PCC 6803, gene sll1184 (12); line 4, Synechocystis sp. strain PCC 6803, gene sll1875 (12); line 5, chicken HO-1 (27); and line 6, human kidney HO-2 (28). Stars indicate identical or conservative amino acid residues in the six different sequences. The † in R. violacea HO indicate the break points of exons 1–2 and exons 2–3.
Interestingly, we have demonstrated that in R. violacea, pbsA is split into three exons whereas in P. purpurea (11), the other rhodophyte from which the gene has been isolated, pbsA is continuous. Furthermore, R. violacea pbsA is spread over the chloroplast genome, exon 2 and exon 3 being separated by 3.8 kb as determined by PCR. The location of the first exon with respect to the other two still remains unknown, though PCR experiments indicate that it is at least 20 kb away.
These results suggest that a complex mechanism produces the mature transcript. We could propose a trans-splicing mechanism between the first exon and exons 2 and 3, taking into account the possible distance (presumably >20 kb) between them. Such a splicing mechanism has already been described in chloroplast genes such as psaA1 of Chlamydomonas reinhardtii (29) or rpsl12 of tobacco (30) and in plant mitochondrial genes (reviewed in ref. 31). On the contrary, for exons 2 and 3, separated by 3.8 kb and in the same orientation, joining of the two exons could be possible from a single pre-mRNA subject to cis-splicing, though a common transcript of ≈4 kb was not detected by either Northern hybridization or RT-PCR. The probable occurrence of such a complex splicing mechanism (involving cis- and trans-splicing) has been previously reported (reviewed in ref. 32).
The pbsA gene of R. violacea is, to our knowledge, only the second example together with the R. violacea rpeB gene (14) of a split gene in a rhodophyte plastid genome. The detection of chloroplast introns only in R. violacea and not in the other rhodophytes studied so far suggests that either (according to the “introns late” theory), they entered the R. violacea plastid after the separation from other rhodophytes or that (according to the “intron early” theory) they were lost in the other red algae studied previously (see ref. 33 for review).
Flanking regions of exons 1, 2 and 3 on the cpDNA (see Fig. 2 A, B, and C) reveal some features of group II introns (33): exon 1 is followed by GTAAG and exon 2 is followed by GTGCG, similar to the 5-nt GTGYG encountered at the 5′ end of group II introns. At the 3′ end of the introns a C is found immediately preceding exon 2 and exon 3, and the A is found 8 nt upstream from exon 2 and exon 3, is presumably involved in the lariat formation, also specific to group II introns (34).
The 5′ end of mature mRNA determined by primer extension with three different primers is situated at nucleotides A 292 and 293 (Fig. 2A). Sequences weakly similar to −35 (nt 251–256) and −10 (nt 273–278) promoter elements of bacterial and plastid genes are found upstream from these 5′ end nucleotides. However, we cannot assert that this is the transcription initiation site rather than a processing or splicing site. Downstream from exon 3 (Fig. 2C), an inverted repeat of 27 nt able to form a stable stem configuration could be a barrier against exonucleolytic attack of the mRNA and/or correspond to a transcription terminator. The length of ≈850 nt of the mature RNA obtained by Northern hybridization is in agreement with the size determined between the 5′ end of the mature mRNA and this putative 3′ end.
We have shown that the pbsA gene (shown at the mRNA level and presumed at the enzyme activity level) from R. violacea is more highly expressed under iron limitation. Iron is of special importance for photosynthetic organisms as it is present as a cofactor in many protein complexes of the photosynthetic apparatus such as reaction centers and cytochromes. Iron is also necessary for the synthesis (via heme) of bilin pigments. The biological availability of iron is often limiting due to the low solubility of Fe3+ above neutral pH in marine and fresh waters (35). Algae, as cyanobacteria, must increase their iron content to compensate for growth and cell division. As a necessity, they have evolved a number of responses to deal with frequently occurring iron deficiencies. HO transcriptional activation could represent one of these responses. This physiological response, never reported before in photosynthetic organisms, suggests a new and interesting role for the product of pbsA as a mobilizing agent during iron limitation conditions. We suggest that the pbsA gene product, besides its anabolic function in bilin synthesis, plays a role in a catabolic process by removal of iron from heme.
The transcriptional regulation of pbsA by iron limitation remains to be studied at the molecular level. The current model involving a Fur-repressor binding to “Fur boxes” for turning off the expression of iron regulated genes in microorganisms (reviewed in ref. 36) may be considered, although no such sequence is present in the vicinity of the pbsA putative promoter region. Alternatively, the mechanism involved can be totally different for pbsA where a basal level of expression is needed in iron-replete conditions in contrast to Fur-regulated genes in bacteria, whose transcription is either on (low iron) or off (iron excess).
The unambiguous chloroplastic origin of P. purpurea and R. violacea pbsA genes has to be compared with the very likely nuclear location of the corresponding gene in another red alga, C. caldarium (10). This result might give useful information about evolution: in R. violacea and P. purpurea the export of genes from the chloroplast to the nucleus is less extensive than in Cyanidiophyceae. In addition, in higher plants and presumably also in the green alga C. reinhardtii, the HO encoding gene has not been found on plastid DNA so far sequenced and is therefore probably also nuclear.
Thus the role of HO appears to be multiple: in pathogenic bacteria, its enzymatic activity is essential in the processes of host infection where iron is needed (13); in animals, HO is not only used for degrading hemoglobin but also serves in modulation of oxidative stress and in the regulation of cell growth and differentiation (1); in plants and other photosynthetic organisms such as algae and cyanobacteria, it could have a dual role: (i) biosynthesis of bilin chromophores for photosynthetic antennae or photoreceptor synthesis and (ii) release of iron from available cell components under deprivation conditions.
Acknowledgments
We thank Michael Reith for providing the P. purpurea pbsA probe, Annette Joder for cultures of R. violacea, and Iain Old and Michael Herdman for helpful reading of the manuscript. We also are specially indebted to Jean-Claude Thomas, Miguel Alfonso, and our colleagues of the laboratory headed by Anne-Lise Etienne for fruitful discussions and comments throughout this work.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HO, heme oxygenase; cpDNA, chloroplast DNA; RT-PCR, reverse transcription–PCR.
References
- 1.Abraham N G, Drummond G S, Lutton J D, Kappas A. Cell Physiol Biochem. 1996;6:129–168. [Google Scholar]
- 2.Beale S I, Cornejo J. Arch Biochem Biophys. 1983;22:279– 286. doi: 10.1016/0003-9861(83)90372-7. [DOI] [PubMed] [Google Scholar]
- 3.Cornejo J, Beale S I. J Biol Chem. 1988;263:11915–11921. [PubMed] [Google Scholar]
- 4.Terry M J, Wahleithner J A, Lagarias J C. Arch Biochem Biophys. 1993;306:1–15. doi: 10.1006/abbi.1993.1473. [DOI] [PubMed] [Google Scholar]
- 5.Parks B M, Quail P H. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagatani A, Reed J W, Chory J. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weller J L, Terry M J, Rameau C, Reid J B, Kendrick R E. Plant Cell. 1996;8:55–67. doi: 10.1105/tpc.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida T, Kikuchi G. J Biol Chem. 1978;253:4225–4229. [PubMed] [Google Scholar]
- 9.Yoshinaga T, Sassa S, Kappas A. J Biol Chem. 1982;257:7778–7785. [PubMed] [Google Scholar]
- 10.Rhie G, Beale S I. J Biol Chem. 1994;269:9620–9626. [PubMed] [Google Scholar]
- 11.Reith M, Munholland J. Plant Cell. 1993;5:465–475. doi: 10.1105/tpc.5.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt M P. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard C, Thomas J C, Mazel D, Mousseau A, Castets A M, Tandeau de Marsac N, Dubacq J P. Proc Natl Acad Sci USA. 1992;89:9564–9568. doi: 10.1073/pnas.89.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnier F, Dubacq J P, Thomas J C. Plant Physiol. 1994;106:747–754. doi: 10.1104/pp.106.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernard C, Etienne A L, Thomas J C. J Phycol. 1996;32:265–271. [Google Scholar]
- 17.Lichtlé C, Garnier F, Bernard C, Zabulon G, Spilar A, Thomas J C, Etienne A L. Plant Physiol. 1996;112:1045–1054. doi: 10.1104/pp.112.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones R F, Speer H L, Kury W. Physiol Plant. 1963;16:636–643. [Google Scholar]
- 19.Douglas S E. Curr Genet. 1988;14:591–598. [Google Scholar]
- 20.Parsot C. EMBO J. 1986;5:3013–3019. doi: 10.1002/j.1460-2075.1986.tb04600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1979;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazel D, Gugliemi G, Houmard J, Sidler W, Bryant D A, Tandeau de Marsac N. Nucleic Acids Res. 1986;14:8279–8290. doi: 10.1093/nar/14.21.8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damerval T, Castets A M, Gugliemi G, Houmard J, Tandeau de Marsac N. J Bacteriol. 1989;171:1445–1452. doi: 10.1128/jb.171.3.1445-1452.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjellm L, Loiseaux S, Rozier C, Dalmon J. Plant Sci Lett. 1980;18:381–388. [Google Scholar]
- 26.Reith M. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:549–575. [Google Scholar]
- 27.Evans C O, Healey J F, Greene Y, Bonkovsky H L. Biochem J. 1991;273:659–666. doi: 10.1042/bj2730659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoubrey W K, Jr, Ewing J F, Maines M D. Arch Biochem Biophys. 1992;295:13–20. doi: 10.1016/0003-9861(92)90481-b. [DOI] [PubMed] [Google Scholar]
- 29.Kuck U, Choquet Y, Schneider M, Dron M, Bennoun P. EMBO J. 1987;6:2185–2195. doi: 10.1002/j.1460-2075.1987.tb02489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torazawa K, Hayashida N, Obokata J, Shinozaki K, Sugiura M. Nucleic Acids Res. 1986;14:3143. doi: 10.1093/nar/14.7.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonen L. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 32.Rochaix J–. Annu Rev Cell Biol. 1992;8:1–28. doi: 10.1146/annurev.cb.08.110192.000245. [DOI] [PubMed] [Google Scholar]
- 33.Cerff R. In: Tracing Biological Evolution in Protein and Gene Structures. Go M, Schimmel P, editors; Go M, Schimmel P, editors. New York: Elsevier; 1995. pp. 205–227. [Google Scholar]
- 34.Michel F, Umesono K, Ozeki H. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 35.Behrenfeld M J, Bale A J, Kolber Z S, Aiken J, Falkowski P G. Nature (London) 1996;383:508–511. [Google Scholar]
- 36.Straus N A. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor; Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 731–750. [Google Scholar]