Abstract
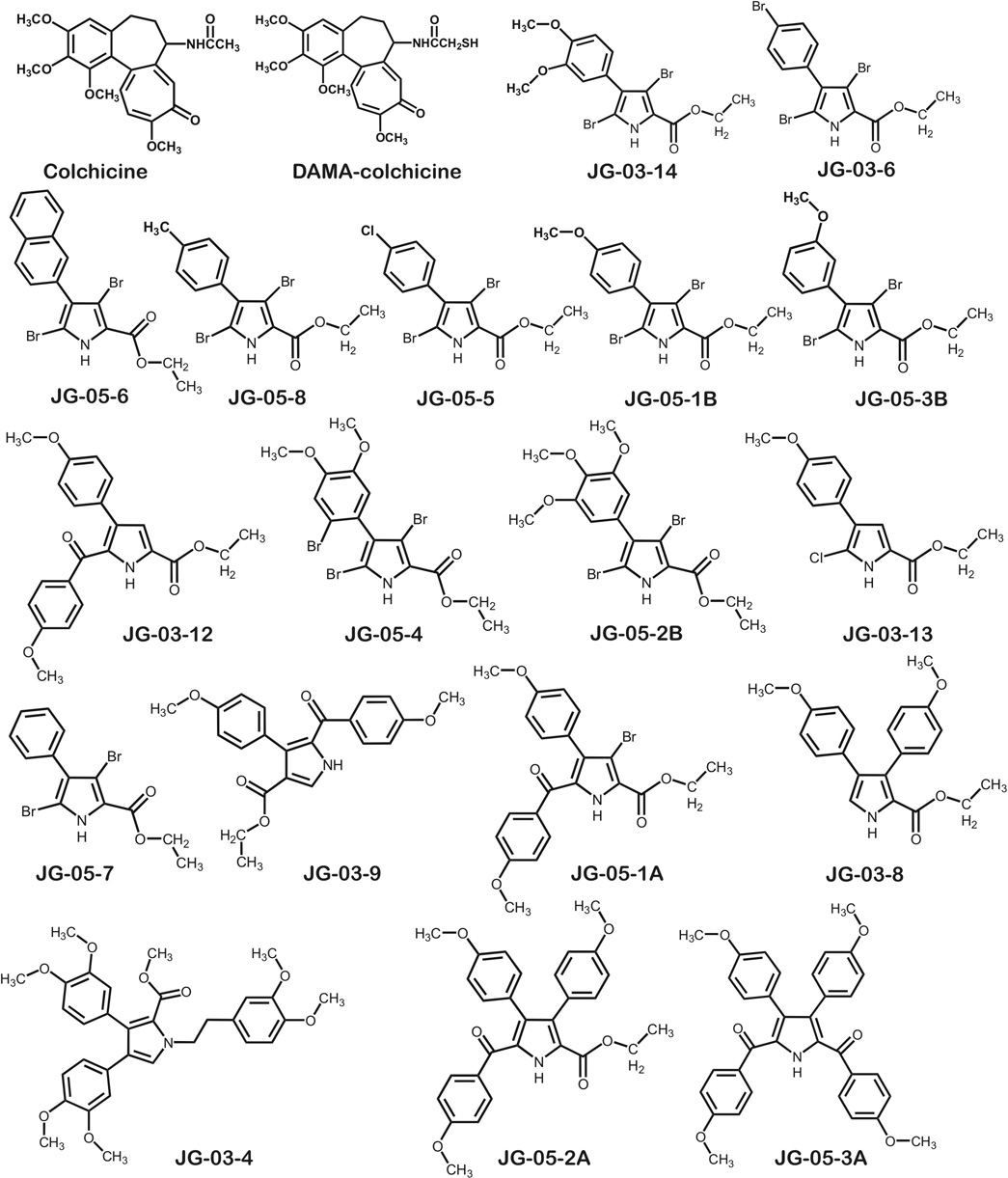
Compounds that bind at the colchicine site of tubulin have drawn considerable attention with studies indicating that these agents suppress microtubule dynamics and inhibit tubulin polymerization. Data for eighteen polysubstituted pyrrole compounds are reported, including antiproliferative activity against human MDA-MB-435 cells and calculated free energies of binding following docking the compounds into models of αβ-tubulin. These docking calculations coupled with HINT interaction analyses are able to represent the complex structures and the binding modes of inhibitors such that calculated and measured free energies of binding correlate with an r2 of 0.76. Structural analysis of the binding pocket identifies important intermolecular contacts that mediate binding. As seen experimentally, the complex with JG-03-14 (3,5-dibromo-4-(3,4-dimethoxyphenyl)-1H-pyrrole-2- carboxylic acid ethyl ester) is the most stable. These results illuminate the binding process and should be valuable in the design of new pyrrole-based colchicine site inhibitors as these compounds have very accessible syntheses.
Keywords: Anti-tubulin, cytotoxicity, HINT, molecular docking, pyrroles
Introduction
A large number of targets are under exploration for chemotherapeutic treatments for cancer. In the past several years, based on the efficacy and commercial successes of paclitaxel and the vinca alkaloids, there have been major efforts to design inhibitors that bind and interfere with the function of microtubules. Microtubules are essential elements of the cytoskeleton and extremely important in mitosis and cell division. Colchicine, the first drug known to bind to the tubulin protein [1,2], inhibits microtubule formation and causes loss of cellular microtubules. In contrast, paclitaxel and its analogues actually promote microtubule polymer formation [3–5], albeit by acting at a different site on tubulin than colchicine. A variety of small molecules with diverse molecular scaffolds have been shown to bind tubulin at the colchicine site [6–9]. One class of these compounds receiving particular attention has been that based on the natural product combretastatin A-4 discovered by Pettit [10,11]. Despite some successes, the discovery of new, more efficacious inhibitors is becoming increasingly important because of multi-drug resistance to tubulin-binding antimitotic agents [12]. Furthermore, chemical synthesis of combretastatin analogues is multi-step and difficult. In any case, the true therapeutic potential of the colchicine site on tubulin has not been fully explored because of the lack of truly atomic level knowledge of the site.
In 2000, Hamel and colleagues mapped the binding site of colchicinoids on β-tubulin [13]. Using molecular modeling and computational docking of colchicinoids into the electron crystallographic model of β-tubulin in protofilaments [13], they found two potential binding sites. The first was entirely encompassed within β-tubulin with the colchicinoids forming adducts with Cys 356. The second potential site was located at the α/β interface and involved hydrogen bonding with Cys 241. More recently, Nguyen and colleagues [14] developed a comprehensive pharmacophore model for structurally diverse colchicine-like tubulin inhibitors using a structure-based approach on the newly available α/β-tubulin:DAMA-colchicine X-ray structure [15]. This crystal structure definitively identified a cleft at the α / β interface as the colchicine binding site, but has a resolution of only 3.58 Å and thus requires considerable computational effort before models derived from it can be considered “all-atom” [14].
While investigating the antiproliferative activity of compounds in a series of synthetic polysubstituted pyrroles (Scheme 1), our interest in the colchicine binding site of tubulin as a putative target for computational drug design studies was piqued after a COMPARE[16] analysis showed a correlation between one of the compounds (JG-03-14) and colchicine of 0.681 over the 45 cell lines that were assayed for both compounds. COMPARE evaluates similarities in activity profiles across the NCI cancer cell line panel, and has been used to elucidate modes of action for new anticancer agents [16]. In this work we report the results of docking this set of putative ligands into the colchicine site of tubulin to build stereochemically reasonable models. We evaluated these docking models with the HINT free energy force field [17] and found a good correlation between HINT scores and measured IC50s of cell proliferation by the compounds. While the measured IC50s represent a downstream biological effect and we are making the pragmatic assumption that the modes of action for all compounds in this series are the same, these results do allow us to appropriately characterize the colchicine binding site and will also serve in design and validation of new compounds similar to JG-03-14 in later stages of this research. This is particularly relevant since these and other polysubstituted pyrrole compounds are synthetically accessible.
Scheme 1.
Results and Discussion
While the character of the colchicine binding site was investigated by Nguyen et al. [14], their study was directed at deriving a generalized pharmacophore for the site and consequently the data set included only two polysubstituted pyrroles. These compounds represent an emerging class of agents with potential activity against a variety of human tumors with activity expressed at nM or µM concentrations in human tumor cell lines [18,19], but having advantages over natural products in terms of drug design and development. In particular, we have been exploring a series of brominated pyrroles whose structure suggests that they might interfere with tubulin function. One member of this series (JG-03-14, 3,5-dibromo-4-(3,4-dimethoxyphenyl)-1H-pyrrole-2-carboxylic acid ethyl ester), for which NCI tumor panel activity had been obtained [19], was suggested by COMPARE [16] to have an activity profile similar to colchicine. Cellular studies with JG-03-14 further support the contention that these compounds function as microtubule poisons [18]. In addition, JG-03-14 was found to have the capacity to promote both autophagic and apoptotic cell death, albeit in different cell lines, while retaining activity in tumor cells expressing the multidrug resistant pump P-glycoprotein [18, 24]. Because the development of additional synthetic or semi-synthetic pyrrole derivatives in this class is facilitated with their relatively facile syntheses, including modification of the molecule by adding and removing functional groups [19], we have both a rather extensive collection of molecules in-hand (Scheme 1) for building predictive molecular models and the potential for rational design and synthesis of many others.
Antiproliferative activity of polysubstituted pyrroles
Results from a number of assays have previously appeared regarding the antiproliferative and cytotoxic activities of the lead compound JG-03-14 [18, 19, 24]. However, most of the compounds in this series have not been examined in detail. An important component of structure-activity relationships and/or computational activity predictions is having reproducible and comprehensive data for a relatively large series of compounds, even those with comparatively poor activity, because understanding why particular compounds are inactive is potentially just as valuable as data on active compounds. Table 1 sets out the experimental antiproliferative assay results against human MDA-MB-435 cancer cells for the compounds in Scheme 1. While JG-03-14 remains the compound with the most potent (36 nM) activity, a few others (Table 1) have activities that are just 7–10 fold less potent, thus suggesting that design of additional new compounds with desirable properties is possible since we have only looked at a very small fraction of the possible analogs to date. Results of a second assay, microtubule depolymerizing activity EC50s for microtubule loss that serves as a partial check on mechanism of action, are also reported in Table 1.
Table 1.
Experimental IC50 , EC50 and docking results for polysubstituted pyrrole compounds.
| Compound | Activity Set | IC50 for anti-proliferationa | pIC50 | ΔGbinding (kcal mol−1) | HINT score | HINT LogP | EC50 for loss of microtubulesb |
|---|---|---|---|---|---|---|---|
| JG-03-14 | A | 36 nMc | 7.74 | −10.14 | 626 | 2.60 | 490nM |
| JG-03-6 | 312 nM | 6.51 | −8.87 | 609 | 3.17 | >50µMf | |
| JG-05-1B | 361 nM | 6.44 | −8.78 | 483 | 2.36 | 5.1 µM | |
| JG-05-3B | 278 nM | 6.55 | −8.94 | 410 | 2.36 | 2.4 µM | |
| JG-05-5 | 919 nM | 6.04 | −8.23 | 559 | 3.02 | >50 µMf | |
| JG-05-8 | B | 2.2 µM | 5.70 | −7.71 | 433 | 2.97 | >50 µMf |
| JG-03-12 | 2.6 µM | 5.58 | −7.61 | 221 | 4.88 | >50 µMf | |
| JG-05-4 | 1.1µM | 5.95 | −8.12 | 455 | 3.34 | 7.1 µM | |
| JG-05-6 | 1.4 µM | 5.85 | −7.98 | 351 | 3.59 | 7.5 µM | |
| JG-03-13 | 5 µM | 5.30 | −7.23 | 163 | 1.48 | >50 µMf | |
| JG-05-1A | 1.9 µM | 5.72 | −7.80 | 508 | 5.62 | >50 µMf | |
| JG-05-2A | 4.2 µM | 5.37 | −7.33 | 149 | 6.58 | >50 µM | |
| JG-05-7 | C | 10 µM | 5.00 | −6.82 | 152 | 2.43 | >50 µM |
| JG-05-2B | 13 µM | 4.89 | −6.66 | 136 | 2.90 | >50 µM | |
| JG-03-9 | 10 µM | 5.00 | −6.82 | 54 | 4.63 | >50 µM | |
| JG-03-4 | 10 µM | 5.00 | −6.82 | 27 | 6.66 | >50 µM | |
| JG-03-8d | >10 µM | 4.00 | −5.45 | −241 | 3.69 | >50 µM | |
| JG-05-3Ad | >20 µM | 3.70 | −5.04 | 296 | 9.02 | >50 µM | |
| Colchicinee | N/A | 2 µM | 5.70 | −7.77 | 563 | 3.24 | N/D |
| DAMA-colchicinee | 2 µM | 5.70 | −7.77 | 455 | 3.70 | N/D |
Antiproliferative activity against human MDA-MB-435 cells using the SRB assay.
Microtubule depolymerizing activity for microtubule loss.
From reference 18.
pIC50 and ΔGbinding calculated for 10×IC50.
Reported colchicine IC50 data was an average of values reported previously in the literature, reference 18, reference 31–reference 33. For calculation purposes DAMA-colchicine was assumed to have same binding as colchicine.
Mechanism of cytotoxicity appears to be unrelated to microtubule disrupting activity.
N/A – not applicable; N/D – not determined.
The colchicine binding site
Binding models for each pyrrole analogue were investigated to delineate steric, electrostatic and hydropathic features of the colchicine binding site. Because we have focused on a series of eighteen compounds with IC50s ranging over more than three orders of magnitude (see Table 1), we performed detailed docking studies with GOLD[21,22] followed by free energy scoring using the HINT protocol[17,27] to assess the binding modes. Without added constraints GOLD was found to reliably re-dock the crystallographic DAMA-colchicine ligand (RMSD = 0.76 Å) that was then used as the reference for all other docking experiments. The HINT score for co-crystallized DAMA-colchicine was 139; in the re-docked pose this score was 455. However, docking of the pyrrole analogues with GOLD produced a mixture of orientations that could not be rationalized with the GOLD docking score. Thus, as we have described in an earlier report,[28] docked poses were re-scored with HINT and we chose the highest HINT-scored pose for further analysis (see Table 1). Docking poses created using a variety of constraints (see Methods) did not yield higher scoring models and were less interpretable than the “freely” bound models we are using. These docked models of substituted pyrroles fit within the pharmacophoric model proposed by Nguyen et al. [14], and for the structural features in common between the substituted pyrroles and in the Nguyen et. al. study, the docking models are in generally good agreement. Key is that the hydrophobic methoxy substituted ring of the pyrrole analogues sits at the hydrophobic center where the TMP moiety of colchicine is found. Note that, although the pyrrole compounds have quite similar structures and are generally positioned in the binding pocket with essentially the same mode, the HINT scores are very sensitive and slight positional differences are detectable in the scores. This sensitivity combined with the number of compounds in the data set allowed us to analyze the site in considerable detail.
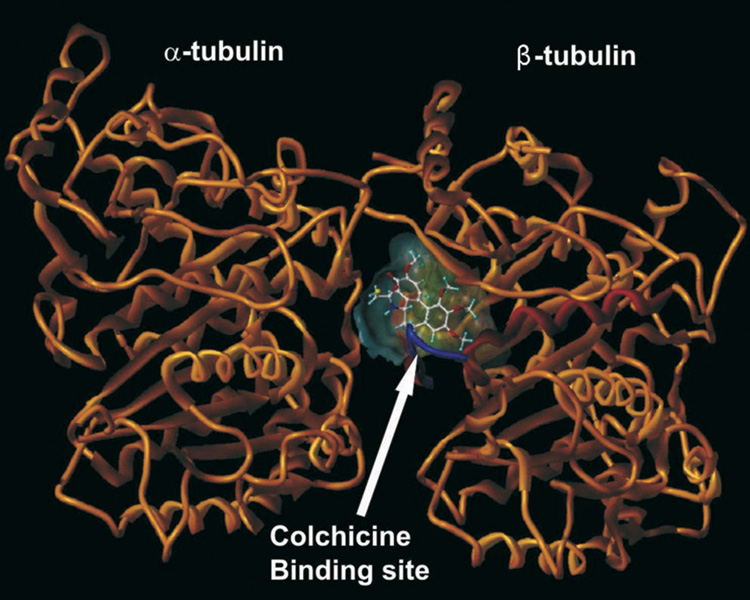
The focus of these computational investigations was on structural aspects of the interactions. The colchicine binding site lies at the interface between the α and β subunits of tubulin, mostly in the β subunit lined by helices 7 and 8 (see Figure 1). The funnel-shaped binding cavity has a volume of about 600 Å3. Residues Tyr202β, Val238β, Thr239β, Cys241β, Leu242β, Leu248β, Leu252β, Leu255β, Ile378β, and Val318β form the narrow funnel end-like part and confer a strong hydrophobic character to this part of the cavity. At the wider portion, the cavity is surrounded by Ala250β, Asp251β, Lys254β, Asn258β, Met259β, Ala316β, Ala317β, Thr353β and Ala354β making it moderately polar/moderately hydrophobic. The open mouth end is surrounded by Asn101α, Thr179α, Ala180α, Val181α and Thr314β, Asn349β, Asn350β, Lys352β. The crystal structure for the complex indicates that DAMA-colchicine (and presumably colchicine) is positioned in the pocket such that its tri-methoxyphenyl (TMP) moiety sits snugly in the narrow hydrophobic pocket. Colchicine also forms hydrogen bonds with the backbone amides of Ala180α and Val181α.
Figure 1.
Colchicine binding site at the interface between the α and β subunits of tubulin.
Structure-activity-binding relationships
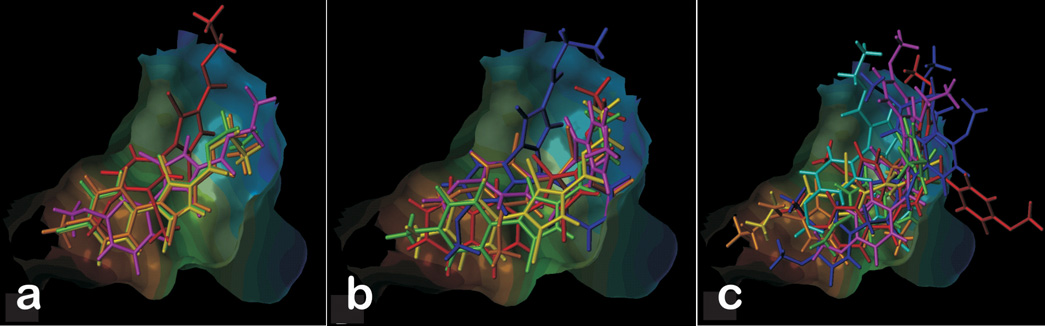
The pyrrole analogues were clustered into three activity sets in order to study them in detail (see Table 1). The first set (A) was comprised of substituted pyrroles that showed antiproliferative activity with sub-µM IC50s. The second set (B) consisted of ligands with IC50 values ranging from 1 µM to 5 µM. The remaining ligands, with IC50 values above 5 µM, comprised the third set (C). The analogs from subset A have noticeable similarity in their structures and are relatively simpler molecules than those in sets B and C. For all of these (set A) compounds the pyrrole ring is substituted by bromines at the 3 and 5 positions and an ethyl ester group at position 2. The differences among this group are substitutions to the phenyl ring at the 4 position of pyrrole. In these, the more potent compounds, most substituents to the phenyl ring, i.e., Cl, Br and methoxy, serve to make this portion of the ligand hydrophobic. Figure 2A illustrates the final docked orientations of the high-affinity pyrroles in the colchicine site of tubulin. The hydrophobic substituted phenyl ring fits snugly in the hydrophobic (narrow funnel) region of the binding pocket. The docked model for JG-03-14 is qualitatively similar to one reported earlier [18].
Figure 2.
Pyrrole analogues docked at colchicine binding site. (A) Substituted pyrroles with activity in sub-µM IC50. (B) Ligands with IC50 ranging from 1 µM to 5 µM. (C) Ligands with IC50 value above 5 µM.
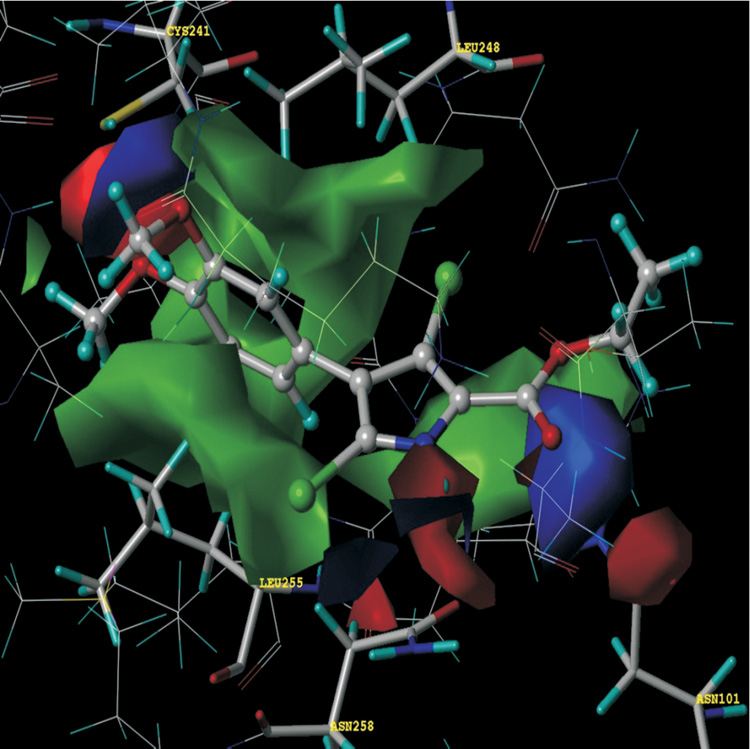
HINT hydropathic analysis reveals more detail concerning the forces orienting these ligands in the binding site. First, hydrophobic interactions are the dominating force contributing towards the stability of the complexes, with additional hydrogen-bonding interactions anchoring the ligands in the cavity. As listed in Table 1, the most potent-binding ligand has the highest HINT score (vide infra), i.e., JG-03-14 interacts with the binding site residues forming the most stable complex. The methoxy-substituted phenyls are positioned deep in the hydrophobic cavity surrounded by Cys241β, Leu242β, Leu248β, Ala250β, Leu255β, Ala354β and Ile378β, all of which contribute to favorable hydrophobic-hydrophobic binding. Figure 3 illustrates these interactions in a HINT map, where the relative sizes of the displayed contours represent the strength, and the colors represent the character, of the interactions between JG-03-14 and the tubulin colchicine binding site. The phenyl ring of JG-03-14 fits in a hydrophobic glove formed by the Leu248 and Leu255.Favorable polar interaction with Asn101, Cys241 and Asn258 also contribute in binding. The ethyl ester tail of the ligand faces towards the polar opening and is stabilized with a strong hydrogen-bond to the amide oxygen of Asp258β with a length of 2.41 Å. Another set of strong hydrogen-bonds are formed between the amine of Asn101α and the carbonyl oxygen (1.48 Å) and alcoholic oxygen (2.72 Å) of the ligand’s ethyl ester substituent. This latter feature, anchoring of the flexible ethyl ester tail, is somewhat different in our models as compared to those of Nguyen et al. [14], probably due to the lack of steric constraints at the open polar end of the cavity.
Figure 3.
HINT interaction maps for JG-03-14 (ball and stick rendering) at colchicine binding site. Blue contours represent regions of favorable polar interactions, e.g., hydrogen bonds, red contours represent unfavorable polar interactions and green contours represent favorable hydrophobic interactions.
On analyzing subset B, docked ligands in the low µM range, it can be seen that these ligands are somewhat similar to the subset A ligands, but with slightly bulkier groups overall as in JG-03-12, JG-05-6 and JG-05-1A, more highly substituted at the pyrrole ring as in JG- 03-12, and/or with less hydrophobic substituents as in JG-03-13, JG-05-4, and JG-05-8. For example, in JG-03-13 the single chlorine substitution is less hydrophobic than the two bromines of JG-03-14 and having only one methoxy also reduces this compound’s hydrophobicity. In the case of JG-05-8 only a single methyl group substitutes the phenyl ring at the para position. JG-03-12 and JG-05-1A have bulkier substitutions at the 5 position of the pyrrole, likely leading to their higher (less potent) IC50 values. The docked models (Figure 2B) and detailed HINT analysis confirm this SAR by showing a relatively poor fit in the active site for the bulkier ligands, and poorer hydrophobic HINT scores for the less optimally-substituted ligands. A single exception, the napthyl-substituted JG-05-6 produces a high HINT score inconsistent with its relatively low potency, but this may be due to this compound being too hydrophobic (Table 1) for solubility or transport to the site (vide infra).
Lastly, many of the subset C (inactive) ligands (Figure 2C) do not fit well in the site, while others are inappropriately decorated to make the required contacts with the site residues. Many of them have one or more bulkier substituents on the pyrrole ring, and only fit in the binding pocket with their side chains protruding out of the pocket.
Predictive models for ligand binding
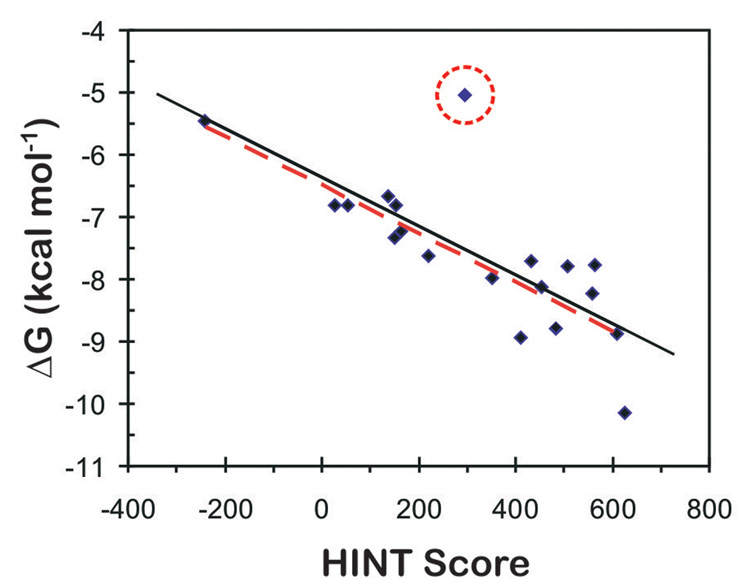
Figure 4 presents the correlation between the experimental binding (Δ Gbinding as calculated from IC50, see Methods) in kcal mol−1 and HINT scores for the eighteen synthetic pyrroles in this study. The IC50s, antiproliferative activities of the compounds, are being taken in this work as approximations of binding affinity, with the implicit assumption that the antiproliferative activity is wholly due to tubulin binding. The consequences of this assumption will be discussed below. The trend represented by the plot of Figure 4 indicates that higher scoring complexes are generally among those with more favorable free energies of binding, while lower scoring complexes are generally those with unfavorable binding. The correlation equation:
| (1) |
has an r2 = 0.5818 and a standard error of ± 0.52 kcal mol−1. A better correlation is observed after omitting the outlier JG-05-3A from the correlation. Although this compound shows a high HINT score suggesting optimal intermolecular interactions within the tubulin colchicine site, it has a very high logPo/w value of 9.02 (Table 1) suggesting that this compound would likely not be transported to the binding site and may even be insoluble. The scoring function does not take into account cell permeability and completely ignores whether or not the compound could in vivo or in vitro be accessible to the binding site. Thus, the unfavorable physiochemical properties of JG-05-3A, and not statistical evaluation, warrant excluding it from the model and justify treating it as an outlier. Ignoring this outlier gives an r2 = 0.76 and a standard error of ± 0.41 kcal mol−1, with a very similar correlation equation:
| (2) |
We believe that this model is predictive such that we can identify the active (subset A) ligands from the inactive (subset C) ligands with reasonable confidence and that further refinement of the model with additional data will improve its usefulness. However, it must be noted that the EC50 for tubulin depolymerization data (Table 1) suggest that several of the compounds (two in set A) that dock in the colchicine binding site with high HINT scores do not appear to cause perturbations of cellular microtubules, i.e., their interactions within the colchicine binding site may not be the mechanism of cytotoxicity. Thus, while we cannot state that all antiproliferative activity is due to tubulin binding in the pyrrole compounds, there is enough experimental information for several of the more active compounds, and a compelling case for JG-03-14, to believe that designing ligands to bind with optimimum interactions in the tubulin colchicine binding site will produce compounds that will likely disrupt cellular microtubules and cause antimitotic actions.
Figure 4.
Dependence of the experimental ΔG on HINT score units for Tubulin-pyrrole complexes. The solid black line represents the regression for ΔG vs. HINT score for all protein-ligand complexes. The red line represents the regression for ΔG vs. HINT score excluding the circled outlier (JG-05-3A).
Conclusions
The present communication demonstrates that the state-of-the-art molecular modeling calculations along with HINT interaction calculations are able to complement experimental studies of binding in many aspects, including accurate representation of the structure of the complex and the binding mode of putative drugs. The structural analysis of the binding pocket has identified important intermolecular contacts that mediate binding. The complex with JG-03-14 has the highest binding score corroborating the experimental data. In conclusion, the present series of pyrrole analogues have yielded representative compounds that are potent tubulin polymerization inhibitors and others that bind with less efficacy, but that still provide useful information for designing compounds with improved performance and selectivity.
Methods
Synthesis of pyrrole compounds
The synthetic methods used to prepare the highly functionalized pyrroles and related derivatives depicted in Scheme I can be found in previously reported work [19–23].
Antiproliferative activity of substituted pyrroles against human tumor cell lines
The antiproliferative effects of the compounds were evaluated in MDA-MB-435 cells using the SRB assay as previously described [29]. A 48 hr exposure time was used. The IC50 values, i.e., the concentration that causes 50% inhibition of proliferation, was calculated from the log dose-response curves and represents the mean of three independent experiments. The effects of the compounds on cellular microtubules were evaluated using indirect immunofluorescence techniques. Briefly, A-10 cells were exposed to the compounds for 18 hr and then the cells were fixed and microtubules visualized using a β-tubulin antibody and the DNA was visualized using DAPI. The SRB assay was also used to measure cytotoxicity by including a time zero point such that the loss of cells from the time of drug addition was monitored. All compounds except JG-05-3A were found to be cytotoxic. The EC50s for microtubule depolymerization (Table 1) were determined using visual observation as previously described [30]. A range of concentrations were tested for each compound and the percent microtubule loss determined for each concentration. The data from 3 independent experiments were averaged and plotted as percent microtubule loss vs. concentration and EC50 values calculated.
Model Building
The X-ray crystal structure (3.58 Å) of αβ-tubulin complexed with DAMA-colchicine [15] PDB code: 1SA0) was used in this study. The stathmin-like domain and the C and D subunits were removed from the model. After hydrogen atoms were added to the model, their positions were optimized to an energy gradient of 0.005 kcal-Å/mol with the Tripos force field (in Sybyl 7.1) while keeping heavy atom positions fixed. The models for pyrrole analogues were constructed using the Sybyl 7.3 (www.tripos.com) and optimized similarly.
Docking
Computational docking was carried out using the genetic algorithm-based ligand docking program GOLD 3.0 [25]. GOLD explores ligand conformations fairly exhaustively and also provides limited flexibility to protein side chains with hydroxyl groups by reorienting the hydrogen bond donor and acceptor groups. The GOLD scoring function is based on favorable conformations found in Cambridge Structural Database and on empirical results of weak chemical interactions [26]. The active site was defined by a single solvent accessible point near the center of the protein active site, an approximate radius of 10 Å, and the GOLD cavity detection algorithm. GOLD docking was carried out without constraints to get an unbiased result and to explore all possible binding modes of the ligands. The trimethoxy phenyl fragment of colchicine was used as the template for biasing the pose of all ligands. In this study we performed 100 GOLD genetic algorithm runs, as opposed to the default of 10 and early termination of ligand docking was switched off. All other parameters were as the defaults. To evaluate and validate GOLD performance the co-crystallized ligand DAMA-colchicine [15] was extracted and docked. GOLD accurately reproduced the experimentally observed binding mode of DAMA-colchicine in αβ -tubulin. All remaining ligands were docked using the same parameters.
Dockings with different/optional constraints such as enforced hydrogen bonds, hydrophobic regions and scaffold match were also explored. For hydrogen bond constraints, docking was biased so that the ligands make hydrogen bonds with Asn258, Ser178, Asn101, and the backbone amides of Ala180 and Val181. For region hydrophobic constraints the ligand positions were constrained by defining a hydrophobic sphere where the tri-methoxy phenyl moiety of colchicine was positioned. Then specific ligand atoms to be docked in the hydrophobic region of the active site were defined. Alternatively, scaffold match constraints were used to place the ligand at a specific position within the active site.
Hydropathic scoring
The HINT (Hydropathic INTeractions) scoring function [17] (version 3.11S β) was used to investigate the structural aspects of the interactions by analyzing and ranking the GOLD docking solutions. HINT evaluates and scores each atom-atom interaction in a biomolecular complex using a parameter set derived from solvation partition coefficients for 1-octanol/water. LogPo/w is a thermodynamic parameter that can be directly correlated with free energy [27]. The HINT model describes specific interactions between two molecules as,
| (3) |
where a is the hydrophobic atom constant derived from Logo/w, S is the solvent accessible surface area, T is a function that differentiates polar-polar interactions (acid-acid, acid-base or base-base), and R, r are functions of the distance between atoms i and j as previously described [28]. The binding score, bij, describes the specific atom-atom interaction between atoms i and j, whereas B describes the total interaction. For selection of the optimum docked conformation and to further differentiate the relative binding efficacy of the pyrrole ligands, interaction scores were calculated for each pose found by docking. The protein and ligands were partitioned as distinct molecules. “Essential” hydrogen atoms, i.e., only those attached to polar atoms (N, O, S, P) were explicitly considered in the model and assigned HINT constants. The inferred solvent model, where each residue is partitioned based on its hydrogen count, was applied. The solvent accessible surface area for the amide nitrogens of the protein backbone were corrected with the “+20” option. Finally, HINT scores were plotted against experimental binding free energies that were calculated using the standard Gibbs free energy equation:
| (4) |
where R is Boltzmann’s constant (1.9872 cal K−1 mol−1) and T is 298 K; Keq is an equilibrium binding constant, ideally KD. In this work measured IC50 values are being used as approximations for equilibrium constants.
Acknowledgements
Financial support from the National Institutes of Health with GM071894 to GEK and AREA grant CA067236 to JTG and from the William Randolph Hearst Foundation to SLM are gratefully acknowledged. The help of Dr. Alexander Bayden (VCU) and Ms. Patrice M. Hill (SFBR) are appreciated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Owellen RJ, Owens AH, Jr, Donigian DW. Biochem. Biophys. Res. Comm. 1972;47:685–691. doi: 10.1016/0006-291x(72)90546-3. [DOI] [PubMed] [Google Scholar]
- 2.Jordan A, Hadfield JA, Lawrence NJ, McGown AT. Med. Res. Rev. 1998;18:259–269. doi: 10.1002/(sici)1098-1128(199807)18:4<259::aid-med3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 3.Kumar N. J. Biol. Chem. 1981;256:10435–10441. [PubMed] [Google Scholar]
- 4.Sengupta S, Boge TC, Liu Y, Hepperle M, George GI, Himes RH. Biochemistry. 1997;36:5179–5184. doi: 10.1021/bi962891m. [DOI] [PubMed] [Google Scholar]
- 5.Altmann KH, Gertsch J. Nat. Prod. Rep. 2007;24:327–357. doi: 10.1039/b515619j. [DOI] [PubMed] [Google Scholar]
- 6.Sackett DL. Pharmacol. Therapeut. 1993;59(2):163–228. doi: 10.1016/0163-7258(93)90044-e. [DOI] [PubMed] [Google Scholar]
- 7.Verdier-Pinard P, Lai JY, Yoo HD, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Mol. Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 8.Kruse LI, Ladd DL, Harrsch PB, McCabe FL, Mong SM, Faucette L, Johnson R. J. Med. Chem. 1989;32:409–417. doi: 10.1021/jm00122a020. [DOI] [PubMed] [Google Scholar]
- 9.Mejillano MR, Shivanna BD, Himes RH. Arch. Biochem. Biophys. 1996;336:130–138. doi: 10.1006/abbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 10.Pettit GR, Cragg GM, Herald Delbert L, Schmidt JM, Lohavanijaya P. Can. J. Chem. 1982;60:1374. [Google Scholar]
- 11.Simoni D, Romagnoli R, Baruchello R, Rondanin R, Rizzi M, Pavani MG, Alloatti D, Giannini G, Marcellini M, Riccioni T, Castorina M, Guglielmi MB, Bucci F, Carminati P, Pisano C. J. Med. Chem. 2006;49:3143–3152. doi: 10.1021/jm0510732. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson RE, Jackson SM, Stanley AJ, Joyce AD, Harnden P, Morrison EE, Patel PM, Phillips RM, Selby PJ, Banks RE. Int. J. Cancer. 2005;115:155–163. doi: 10.1002/ijc.20816. [DOI] [PubMed] [Google Scholar]
- 13.Bai R, Covell DG, Pei XF, Ewell JB, Nguyeni NY, Brossi A, Hamel E. J. Biol. Chem. 2000;275(22):40443–40452. doi: 10.1074/jbc.M005299200. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen TL, McGrath C, Hermone AR, Burnett JC, Zaharevitz DW, Day BW, Wipf P, Hamel E, Gussio R. J. Med. Chem. 2005;48:6107–6116. doi: 10.1021/jm050502t. [DOI] [PubMed] [Google Scholar]
- 15.Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M, et al. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 16.Cleaveland ES, Monks A, Vaigro-Wolff A, Zaharevitz DW, Paull K, Ardalan K, Cooney DA, Ford H., Jr Biochem. Pharmacol. 1995;49:947–954. doi: 10.1016/0006-2952(95)00009-o. [DOI] [PubMed] [Google Scholar]
- 17.Kellogg GE, Abraham DJ. Eur. J. Med. Chem. 2000;35:651–661. doi: 10.1016/s0223-5234(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 18.Mooberry S, Wiederhold K, Dakshanamurthy S, Hamel E, Banner E, Kharlamova A, Hempel J, Gupton J, Brown M. Mol. Pharmacol. 2007;72:132–140. doi: 10.1124/mol.107.034876. [DOI] [PubMed] [Google Scholar]
- 19.Gupton JT, Burham BS, Krumpe K, Du K, Sikorski JA, Warren AE, Barnes CR, Hall IH. Archiv der Pharmazie (Weinheim, Germany) 2000;333:3–9. doi: 10.1002/(sici)1521-4184(200001)333:1<3::aid-ardp3>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Gupton JT, Krumpe KE, Burnham BS, Webb TM, Shuford JS, Sikorski J. Tetrahedron. 1999;55:14515–14522. [Google Scholar]
- 21.Gupton JT, Clough SC, Miller RB, Lukens JR, Henry CA, Kanters RPF, Sikorski JA. Tetrahedron. 2003;59:207–215. [Google Scholar]
- 22.Gupton JT, Miller RB, Clough SC, Krumpe KE, Banner EJ, Kanters RPF, Du KX, Keertikar KM, Lauerman NE, Solano JM, Adams BR, Callahan DW, Little BA, Scharf AB, Sikorski JA. Tetrahedron. 2005;61:1845–1854. [Google Scholar]
- 23.Gupton JT, Banner EJ, Scharf AB, Norwood BK, Kanters RPF, Dominey RN, Hempel JE, Kharlamova A, Bluhn-Chertudi I, Hickenboth CR, Little BA, Sartin MD, Coppock MB, Krumpe KE, Burnham BS, Holt H, Du KX, Keertikar KM, Diebes A, Ghassemi S, Sikorski JA. Tetrahedron. 2006;62:8243–8255. [Google Scholar]
- 24.Arthur CR, Gupton JT, Kellogg GE, Yeudall WAMC, Cabot MC, Newsham IF, Gewirtz DA. Biochem. Pharmacol. 2007;74:981–991. doi: 10.1016/j.bcp.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones G, Willett P, Glen RC, Leach AR, Taylor R. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 26.Bissantz C, Folkers G, Rognan D. J. Med. Chem. 2000;43:4759–4767. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]
- 27.Cozzini P, Fornabaio M, Marabotti A, Abraham DJ, Kellogg GE, Mozzarelli A. J. Med. Chem. 2002;45:2469–2483. doi: 10.1021/jm0200299. [DOI] [PubMed] [Google Scholar]
- 28.Spyrakis F, Amadasi A, Fornabaio M, Abraham DJ, Mozzarelli A, Kellogg GE, Cozzini P. Eur. J. Med. Chem. 2007;42:921–933. doi: 10.1016/j.ejmech.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 29.Clark EA, Hills PM, Davidson BS, Wender PA, Mooberry SL. Mol. Pharm. 2006;3:457–467. doi: 10.1021/mp060016h. [DOI] [PubMed] [Google Scholar]
- 30.Rao PN, Cessac JW, Tinley TL, Mooberry SL. Steroids. 2002;67:1079–1089. doi: 10.1016/s0039-128x(02)00066-1. [DOI] [PubMed] [Google Scholar]
- 31.Brown ML, Rieger JM, Macdonald TL. Bioorg. & Med. Chem. 2000;8:1433–1441. doi: 10.1016/s0968-0896(00)00068-7. [DOI] [PubMed] [Google Scholar]
- 32.Chang JY, Yang MF, Chang CY, Chen CM, Kuo CC, Liou JP. J. Med. Chem. 2006;49:6412–6415. doi: 10.1021/jm060616k. [DOI] [PubMed] [Google Scholar]
- 33. http://www.cytoskeleton.com/products/tubulins/h001.html.