Abstract
RNA editing and cytoplasmic male sterility are two important phenomena in higher plant mitochondria. To determine whether correlations might exist between the two, RNA editing in different tissues of Sorghum bicolor was compared employing reverse transcription–PCR and subsequent sequence analysis. In etiolated shoots, RNA editing of transcripts of plant mitochondrial atp6, atp9, nad3, nad4, and rps12 genes was identical among fertile or cytoplasmic male sterile plants. We then established a protocol for mitochondrial RNA isolation from plant anthers and pollen to include in these studies. Whereas RNA editing of atp9, nad3, nad4, and rps12 transcripts in anthers was similar to etiolated shoots, mitochondrial atp6 RNA editing was strongly reduced in anthers of the A3Tx398 male sterile line of S. bicolor. atp6 transcripts of wheat and selected plastid transcripts in S. bicolor showed normal RNA editing, indicating that loss of atp6 RNA editing is specific for cytoplasmic male sterility S. bicolor mitochondria. Restoration of fertility in F1 and F2 lines correlated with an increase in RNA editing of atp6 transcripts. Our data suggest that loss of atp6 RNA editing contributes to or causes cytoplasmic male sterility in S. bicolor. Further analysis of the mechanism of cell type-specific loss of atp6 RNA editing activity may advance our understanding of the mechanism of RNA editing.
Mitochondria are found in almost all eukaryotes, with the exception of a few parasitic unicellular organisms. Complete sequences of mitochondrial (mt) genomes are now available from a large number of different eukaryotes, including the liverwort Marchantia polymorpha (1) and the higher plant Arabidopsis thaliana (2). The mt genome of liverwort consists of a single circular molecule (3, 4). In remarkable contrast, the mt genome of A. thaliana is split into several different subcircles (5). A multipartite mt structure due to recombination events was first observed in Brassica campestris (6), and appears to be a general feature of higher plants (7). One of the consequences of recombinational activity in higher plant mitochondria is the generation of chimeric reading frames (crf), e.g., the crf pcf found in petunia (8), orf107 of Sorghum bicolor line IS1112C (9), and T-urf13 found in the T cytoplasm of Zea mays (10–12), which is generated from nonprotein-coding sequences only.
Crfs are often associated with cytoplasmic male sterility (cms), a genetic trait that has an important economic relevance for the generation of hybrid seed. cms is characterized by aberrations in microsporogenesis or microgametogenesis that result in microspore abortion in higher plants, and are often inherited through the mitochondria of the maternal cytoplasm. Plants carrying the cms trait show a more or less normal phenotype during vegetative growth, but fail to shed viable pollen.
Crfs are usually transcribed and subjected to the mt RNA maturation process. The mechanisms of gene expression in plant mitochondria are extremely complex systems involving site-specific cleavage, cis- and trans-splicing, and most recently RNA editing. The latter process is characterized by C-to-U and rare U-to-C exchanges and influences transcripts of virtually every known gene (13, 14), except for some of the crfs, e.g., T-urf13 (15).
When RNA editing was established as a new step of RNA processing in higher plants several attempts were made to correlate RNA editing and cms. Only one report (16) has described an increased amount of unedited or partially edited atp9 transcripts in cms wheat; however, other investigators could not confirm this result (17). With a different approach, a correlation of transcript processing and reduced RNA editing with cms was reported in rice (18). A direct experimental approach used ATP9 protein that was synthesized from unedited atp9 transcripts and targeted into the mitochondria of tobacco. These transgenic tobacco plants exhibit a cms phenotype (19), implying a potential role of RNA editing in causing cms.
Because the cms trait occurs in microspores, we compared RNA editing of transcripts of selected mt genes in etiolated shoots and anthers. Loss of mt atp6 RNA editing was observed in a cell type-specific manner in anthers of a male sterile line of S. bicolor. Moreover, the restoration of fertility in F1 and F2 lines (see ref. 9) correlates with an increase of RNA editing of atp6 transcripts. Analysis of F2 plants indicates the involvement of dominant genes, which resemble cms restorer genes. This report links two important genetic phenomena in higher plant mitochondria, namely, RNA editing and cytoplasmic male sterility, and represents a starting point for dissection of the mechanism of RNA editing.
MATERIALS AND METHODS
Sorghum and Triticum Lines.
Five Sorghum lines were used in this study: Tx398, a normal cytoplasm line; IS1112C, a fertile line carrying a male-sterile cytoplasm; A3Tx398, a male-sterile line carrying the IS1112C cytoplasm; and F1 and F2 lines. A3Tx398 represents a near-isogenic line carrying the IS1112C cytoplasm in the Tx398 background; this line had been backcrossed at least 10 generations. The F1 was produced by pollinating line A3Tx398 with IS1112C (20). This F1 line was selfed to produce the F2 line. For wheat, a fertile Triticum durum line (D30) and a cms line of T. durum with Triticum timopheevi cytoplasm (cmsD30) were used. Seedlings were grown at 22°C or 28°C in the dark for 7 days before harvest. Anthers were collected from flowering plants.
mtRNA Isolation.
RNA isolation from etiolated shoots was done as described (21). mtRNA isolation from anthers was done as follows: anthers and pollen were ground in buffer A (0.5 M sucrose/5 mM EDTA/0.1% BSA/0.05 M Tris·HCl, pH 7.5) in an Eppendorf tube. Nuclei and debris was removed by centrifugation at 4,000 rpm for 6 min at 4°C. The supernatant was transferred to a new tube and centrifugation was carried out for 17 min at 14,000 rpm at 4°C. The supernatant was removed and the crude mt pellet dissolved in 500 μl of 60°C hot lysis buffer (0.2 M boric acid/30 mM EDTA/1% SDS, pH 9.0). Phenol extraction and LiCl precipitation were performed to obtain the RNA pellet.
Oligonucleotides.
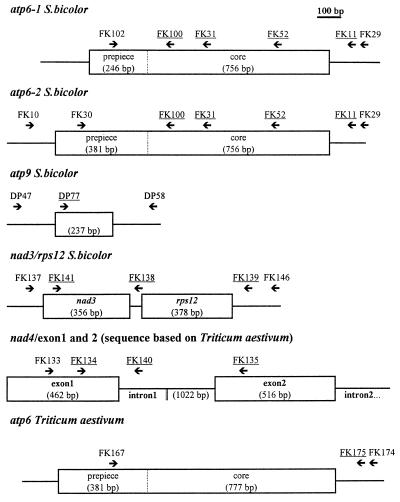
For localization of oligonucleotides see Fig. 1. atp6-specific oligos are as follows: FK10, 5′-AAGTCTCCCTTTCTTTCGG-3′; FK11, 5′-GTAGTTCTCTTCCAGTTCTCG-3′; FK29, 5′-TTCAAGGAAGTGAGTCTCAGA-3′; FK30, 5′-CGATGCCCCCGGCGCCGATACC-3′; FK52, 5′-TGACCGGCCATCATATTAGC-3′; FK100, 5′-GGACCAAACCGAGAGTGAG-3′; and FK102, 5′-TTTATCGCGCTCATCATAACC-3′.
Figure 1.
Schematic representations of genes and localization of oligonucleotides employed for specific PCR and RT-PCR experiments. Underlined oligonucleotides were used as primers for sequencing.
The following oligonucleotides were used for wheat atp6: FK167, 5′-CCCGATCTACATGATTTTTTCCC-3′; FK174, 5′-CCATGTTCAGTGCTGCAAAAGCGC-3′; FK175, and 5′-CCCTAGAATCTTCGGCTCCTCG-3′. atp9-specific oligos are as follows: DP47, 5′-CAACTAAGGAGAGGAGGAAGG-3′; DP58, 5′-GCGACCATGAAAGAAGCC-3′; DP77, 5′-GGCAGAATTCATGTCGCGACTCTACAT-3′. nad3- and rps12-specific oligos are: FK137, 5′-GAGAACGAAGTGGGCTTTGG-3′; FK138, 5′-CCTCTTTCCTATGTCCTTCCCCCC-3′; FK139, 5′-CAGTCCGGAATTCCCAGCAAATCC-3′; FK141, 5′-GGAATTTGCACCTATTTGTATC-3′; FK146, 5′-AAGTTCCAGAGGCATCTTCC-3′. nad4-specific oligos are: FK133, 5′-GAATGCTATTTCGATCTAAGTGG-3′; FK134, 5′-GGTCTGTGCGTTTCTCTTATTAC-3′; FK135, 5′-CCTTGATCTTTCTTTGTCTCG-3′; FK140, 5′-GCAGAGGAAAAGCTTGATCCCAGC-3′.
PCR and Reverse Transcription (RT)-PCR Amplification.
PCR amplification was done as described (21). For RT-PCR amplification we used a modified protocol; anther RNA was treated with RNase-free DNase (Boehringer Mannheim) for 1 hr. Reverse transcription was done for 1 hr at 37°C according to previously published procedures (21). The reaction was then split in aliquots of 2 μl for subsequent PCR amplifications. For sequencing, 0.2–5.0 μl of a successful PCR amplification was used for asymmetric PCR. Asymmetric PCR was done as described (21). Use of oligonucleotides for specific PCR amplifications is indicated in Figs. 1 and 2.
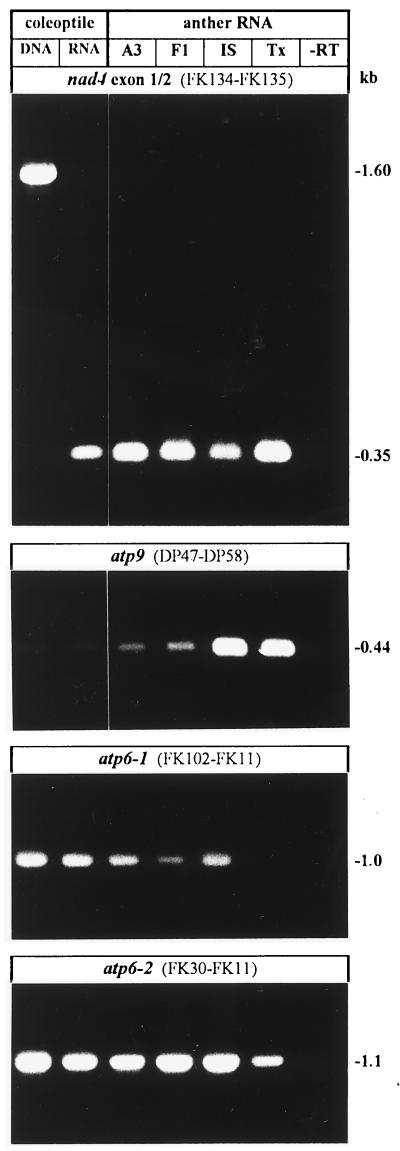
Figure 2.
PCR amplifications of specific transcripts from anther mRNA. Amplifications were done with FK102, FK11, or FK29 for atp6–1; FK10 and FK29 for atp6–2; DP47 or DP77 and DP58 for atp9; FK133 and FK135 for nad4 (some experiments not shown). For localization of oligonucleotides see Fig. 1. A3, line A3Tx398; IS, line IS1112C; Tx, line Tx398; −RT, control experiment without reverse transcription prior to PCR.
Quantitative RT-PCR.
RT-PCR reactions were as described above, but fewer cycles (15–35) were performed to compare the results in the logarithmic phase of chain reaction. Amplifications were done in one tube reaction from a single cDNA pool, which allowed direct comparison of data for atp6–1 (FK102-FK52), atp6–2 (FK10-FK52), and nad4 (FK134-FK135). One of the oligonucleotides for each amplification (FK52 and FK135) was radiolabeled with [[gamme]-32P]ATP by the use of T4 polynucleotide kinase (Boehringer Mannheim).
Standard Methods.
Sequencing, gel electrophoresis, and other standard methods were done according to Sambrook et al. (22). The degree of editing at the various sites was determined densitometrically.
RESULTS AND DISCUSSION
Anther-Specific Loss of atp6 RNA Editing in a cms Line.
Cytoplasmic male sterility in S. bicolor is used extensively in production of hybrid seed. Several cms groups are distinguished, based on the mode of fertility restoration (23, 24); among these, one member of the A3 group has several unusual characteristics (reviewed in ref. 23). Earlier, we demonstrated that RNA editing of atp6 transcripts is the same in fertile and A3 cms S. bicolor lines (21); however, in that study we used mtRNA isolated from etiolated shoots only. Therefore, tissue-specific differences in RNA processing events, such as RNA editing, could not be ruled out. We sought to include anther tissues in our analysis, because the cms phenotype is characterized by the inability to produce viable pollen. Whereas the amplification of DNA sequences has been described for single cells or single pollen grains (e.g., ref. 25), we attempted to analyze RNA processing events. We developed a protocol to isolate fractions enriched in mitochondria and mtRNA from anthers and pollen of S. bicolor. The average yield was 10 μg of enriched mtRNA from 16 mg of fresh-weight anther tissue.
cDNA pools were made with random primers, and specific oligonucleotides were employed for PCR amplifications (see Fig. 1). Specific amplifications were done for cDNAs of atp6, atp9, nad3, nad4, and rps12, which encode components of different mt enzyme complexes and a ribosomal protein (examples shown in Fig. 2). For nad4 the amplified region included an intron, thus providing an additional control for DNA contaminations. As shown in Fig. 2, no amplified product was obtained when RT was omitted. Therefore, the anther mtRNA preparations appear to be suitable for molecular analysis by RT-PCR and subsequent sequence analysis. Sequence data for Sorghum nad3-rps12 and nad4 are new and will be deposited in the appropriate sequence libraries.
To avoid time-consuming steps, such as cloning, aliquots from RT-PCR amplifications were directly sequenced. This was achieved by producing excess single-stranded DNA through asymmetric PCR followed by sequence analysis. By sequencing the entire cDNA pool for each transcript we were able to circumvent the problem of PCR artifacts and can determine RNA editing efficiency at every site densitometrically, which is advantageous compared with sequencing a large number of cloned cDNAs and has been used in other studies as well (26). However, as an additional control, some experiments were successfully reconfirmed by sequencing individually cloned cDNA clones. These data are summarized in Table 1 and typical results for nad4 and atp6 transcripts are shown in Fig. 3. The extent of RNA editing of transcripts from anthers was similar to that of etiolated shoots for all analyzed transcripts with the exception of atp6. The atp6 gene exists in two copies in the IS1112C cytoplasm, designated atp6–1 and atp6–2, which share the same conserved core region, but differ in their 5′ sequences (20). In both cases, RNA editing was drastically reduced in cms anthers of line A3Tx398 compared with the previously published data from etiolated shoots (21, 27) for atp6–1 and atp6–2 (Figs. 3B, 4, and 5). We sampled the anthers at anthesis, when the diameter of the sterile pollen is 75% of the fertile and the pollen has not collapsed. However, our samples reflect a mixture of mostly haploid pollen and diploid cells. To demonstrate whether the effect is restricted to pollen or a general feature of the anther tissues, haploid pollen and diploid anther tissues were separated, RNA was isolated, and RT-PCR and sequence analysis were performed. Reduced atp6 RNA editing was observed in both diploid and haploid cells. The loss of RNA editing in diploid tissue might be correlated to the observation that anthers of A3Tx398 do not normally dehisce and do not shed pollen. This may be due to (i) a specific defect in the A3Tx398 anther tissue, (ii) because the volume of the infertile pollen does not place enough stress on the anther to cause it to dehisce, or (iii) because there is no signal from the cms pollen.
Table 1.
Tissue-specific RNA editing of transcripts in different S. bicolor lines
| Transcript | Coleoptiles, % | Anthers/pollen, % |
|---|---|---|
| atp6-1 | 100 | A3Tx398 (20), F1 (36), Tx398 and IS1112C (100) |
| atp6-2 | 100 | Same as for atp6-1 |
| atp9 | 50 | 50 |
| nad3* | 25 | 25 |
| nad4 | 100 | 100 |
| rps12* | 25 | 25 |
Editing identical for all lines unless otherwise stated.
nad3–rps12 cotranscript was analyzed, which may explain partial editing.
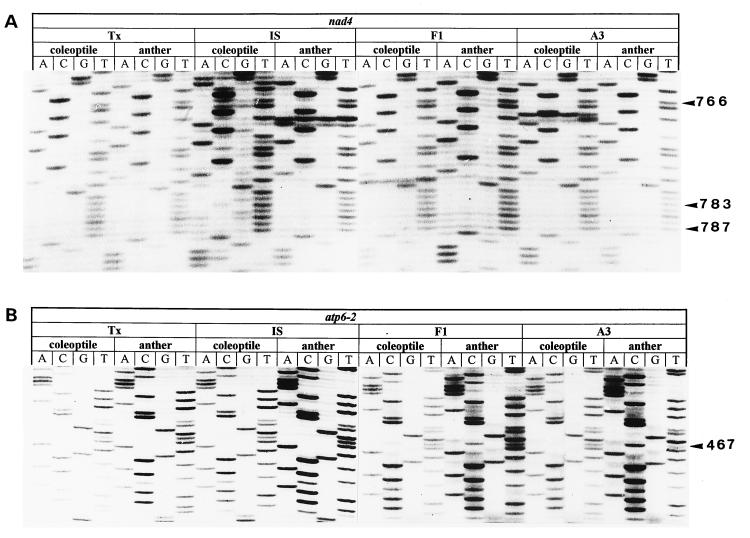
Figure 3.
Sequence data from cDNA pools of RT-PCRs of coleoptile and anther mRNAs. Arrowheads indicate RNA editing sites at the indicated positions. Sequence numbering is according to ref. 20. (A) cDNAs from nad4 transcripts; for line designation see Fig. 2. (B) cDNAs from atp6 transcripts; same lines and tissues as in A.
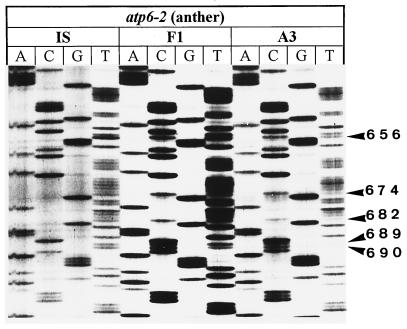
Figure 4.
Additional examples of the loss of atp6 RNA editing in line A3Tx398. Only anther tissue data is shown.
Figure 5.
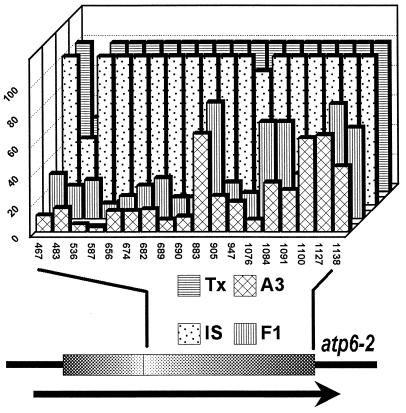
Comparison of atp6 RNA editing in lines Tx398, IS1112C, F1, and A3Tx398. The degree of editing at each of the 18 atp6–2 RNA editing sites present in the core region (atp6–2 gene and transcript shown at bottom of figure) was measured densitometrically and is given as a percentage (vertical axis). Data are the same for atp6–1 transcripts (not shown). Whereas editing activity is very low in cms A3Tx398 (hatched boxes), it somewhat increases in partially fertile F1 plants (vertically striped boxes), compared with full editing in IS1112C (dotted boxes) and Tx398 (horizontally striped boxes). Editing site 483 is a silent site that exhibits only 50% editing in fertile lines and much less in sterile lines. For unknown reasons, loss of editing activity at some sites is not as drastic as at other sites. Numbers at the horizontal axis correspond to editing sites of atp6–2 (28). For abbreviations see Fig. 2. Resulting amino acid residue changes due to RNA editing at the following positions: 467, S ⇒ F; 483, silent; 536, S ⇒ L; 587, P ⇒ L; 656, S ⇒ L; 674, S ⇒ L; 682, R ⇒ C; 689+690, P ⇒ L; 883, H ⇒ Y; 905, S ⇒ L; 947, S ⇒ L; 1076,S ⇒ L; 1084, H ⇒ Y; 1091, S ⇒ F; 1100, S ⇒ L; 1127, T ⇒ I; 1138, T ⇒ stop codon.
RNA editing of atp6 in anthers and etiolated shoots of Tx398 and IS1112C was similar and both lines are fertile. Interestingly, the F1 line, which results from backcross A3Tx398 × IS1112C, exhibits both partial male fertility and partial RNA editing (see Figs. 4 and 5). Because PCR amplifications were performed from one cDNA pool we can exclude DNA contamination as a cause for this result, since the effect was limited to atp6 transcripts, e.g., full nad4 RNA editing was observed in all lines (Fig. 3A). We have verified these observations with RNA from several growth seasons.
Loss of Transcript-Specific RNA Editing Occurs Exclusively in Sorghum Mitochondria.
Transcript-specific loss of RNA editing activity has not yet been reported for mitochondria, although there are reports of tissue-specific RNA editing in mitochondria and in plastids (28, 29). Plastid RNA editing of transcripts of ndhB and rpl2 was analyzed in anthers of fertile and male sterile S. bicolor lines; however, no differences were observed (R. M. Maier and F. Albertazzi, personal communication). In addition, we analyzed RNA editing of atp6 transcripts in T. timopheevi mitochondria. In Triticum, eight editing sites at the 3′ end of the transcript were analyzed in cms and fertile lines, including the conserved CAA codon, which is edited to the new stop codon ′TAA′. Thus, the polypeptide deduced from wheat atp6 cDNAs is seven amino acids shorter compared with the genomic version, as proposed in an earlier study of higher plant atp6 RNA editing (30). No differences were detected between etiolated shoots and anther tissues (data not shown) of fertile and male sterile lines. Therefore we conclude that atp6 transcript-specific loss of RNA editing is not a general phenomenon, but apparently occurs in mitochondria from anthers of S. bicolor exclusively.
Correlation of RNA Editing and cms in Sorghum.
The findings reported here are unique in several ways. (i) This is the first case of cell type-specific loss of RNA editing of a particular transcript in plant mitochondria, although evidence for developmental and tissue specificity of single nad3 RNA editing sites was observed in maize (29). (ii) The phenomenon also appears to be determined by the nuclear background, since all lines except Tx398 share the same cytoplasm. (iii) The effect is transcript specific (atp6) and occurs only in mitochondria of cms S. bicolor. (iv) An increase in RNA editing in a partially male fertile F1 line is observed.
From the above data we conclude that a correlation exists between cms and RNA editing in S. bicolor.
Our interpretation includes the concept of cell type-specific effects involved in restoration of cms or RNA editing. Most investigations on cms in higher plants are conducted with etiolated shoots or tissue cultures, with the exception of a few reports (31, 32) that showed a cell type-specific effect of fertility restoration of a cms-related gene. Based on our data, it appears that cell- and tissue-specific effects in higher plants may be more common than previously believed.
Involvement of Nuclear Genes.
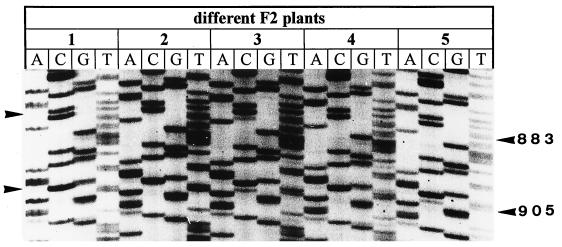
To study the correlation of anther-specific RNA editing and cms, individual plants of an F2 line were analyzed for anther RNA editing. None of the F2 plants were male sterile, and range from partial fertile to fully restored (D. R. Pring, personal communication). RNA from individual plants exhibiting different degrees of fertility restoration was analyzed as described above. Typical examples of sequencing runs are shown in Fig. 6. All F2 plants exhibit normal atp6 RNA editing activity. This result can be explained by the gametophytic mode of fertility restoration in S. bicolor (9), wherein only gametes carrying requisite fertility restoration genes are viable. Thus, no sterile plants are expected in the F2 generation, and these fertile plants should be able to edit atp6 transcripts, if one assumes that pollen is not viable without atp6 RNA editing. For several reasons it appears that pollen unable to edit atp6 transcripts is not viable. (i) The atp6 core sequence is strongly conserved in animals and fungi. This conserved sequence is retained by only RNA editing. ATP6 protein from unedited RNA would exhibit a different hydropathy profile and different deduced secondary structures. Therefore, unedited RNA-derived ATP6 would not be functional or not even translated (see below). (ii) The same conclusion may be drawn from an experiment done in tobacco: Transgenic tobacco plants expressing unedited ATP9 protein that was targeted to the mitochondria were male sterile, possibly because the introduced unedited ATP9 protein competes with endogenous ATP9 for assembly into the ATP-synthase complex (19). (iii) Moreover, in petunia, it was shown that unedited atp6 transcript is not translated, or if translated, proteins do not accumulate (33). Sometimes, protein polymorphism occurs for differentially edited transcripts (e.g., rps12) (34); therefore, it is not clear whether unedited atp6 transcripts are translated. Because suitable antibodies are not available we cannot confirm the absence of “edited” ATP6 protein experimentally. Nevertheless, we believe it is likely that pollen is not viable and cannot pollinate if atp6 transcripts remain unedited for reasons outlined in (i) and (ii).
Figure 6.
Sequence data from RT-PCRs of anther atp6–2 transcripts from five fully or partial fertile F2 plants. All plants exhibit normal RNA editing (data from one growing season).
If all F2 seed is produced from editing-competent pollen, high or normal RNA editing of atp6 in F2 plants may be explained by the activity of dominant restorer genes. To date, no experimental data are available to determine the actual number of genes involved. These potential genes may provide editing factors that simply interfere with the editing machinery; e.g., from petunia it is known that a single nuclear gene influences both transcript abundance and RNA editing (35).
Toward the Mechanism of atp6-Specific Loss of RNA Editing.
In the above model the dominant genes would either suppress an inhibitor of RNA editing or provide an essential cofactor. Loss of atp6 RNA editing occurs only in a combination of IS1112C cytoplasm and Tx398 nucleus. Differences in IS1112C and Tx398 mitochondria may provide answers to this problem. One example is the existence of crf orf107 in the IS1112C cytoplasm, which correlates with cms in the A3 group (9). The above model for loss of atp6 RNA editing is similar to that for the processing of crf orf107 in cms S. bicolor (Table 2). To date, we cannot confirm a functional relationship between RNA processing of orf107 and atp6 RNA editing. However, the absence of orf107 RNA processing correlates with the failure of atp6 RNA editing in anthers, as does the restoration of processing activity in F1 and F2 (see table 2 orf107 data from ref. 9). The putative polypeptide encoded by the unprocessed mRNA of orf107 may interfere with atp6 RNA editing in A3Tx398, although this hypothesis does not easily explain the cell type specificity of atp6 RNA editing.
Table 2.
Comparison of fertility status, orf107 processing, and atp6 RNA editing activity
| Line | Fertility status | orf107 mRNA processing (9) | Anther atp6 RNA editing |
|---|---|---|---|
| Tx398 | Full | Not known | Yes |
| IS1112C | Full | Yes | Yes |
| A3Tx398 | Sterile | Trace | Trace |
| F1 | Partial | Yes | Yes (partial) |
| F2 | Partial/full | Yes | Yes |
A second difference concerns the atp6 genes themselves. Tx398 carries only the atp6–2 gene copy (20). The presence of a second (atp6–1) copy in the IS1112C cytoplasm may contribute to the effect seen in the A3Tx398 line, because the Tx398 nuclear background may lack genes to balance the expression of both atp6 gene copies in a cell type-specific manner, leading to reduced atp6 transcription. In petunia, certain nuclear backgrounds cause a drastic decrease in the abundance of nad3 transcript, which correlates with the loss of RNA editing (35). A similar effect could be responsible for the results found in the mitochondria of cms S. bicolor anthers. The small amount of anther RNA available did not allow us to perform Northern hybridization to check for transcript abundances in anthers. Therefore, quantitative PCR was employed using specific oligonucleotides for amplification of atp6–1, atp6–2, and nad4 transcripts. This experimental setup allowed us to compare the abundance of the atp6–1 and atp6–2 transcripts, using nad4 as an internal control. For this approach, RNA from fertile IS1112C, the sterile A3Tx398, and the partial-fertile F1 lines was used. However, copy numbers for atp6–1 and atp6–2 transcripts were the same in anthers (data not shown) and in etiolated shoots (27) in all lines. Consequently, we can exclude differences in transcript abundance as a cause for this phenomenon.
Several papers suggest the presence of trans-acting factors that could provide sequence specificity (27, 36, 37); recently, direct evidence for the presence of trans-acting site-specific factors that mediate chloroplast editing site recognition was presented. Interspecific protoplast fusion experiments suggest that these transfactors are of extraplastidic origin (38). A similar mechanism may exist for plant mitochondria. If transfactors exist, the Tx398 nuclear background may be unable to generate sufficient amounts of site-specific editing factors for two atp6 transcripts in a tissue-specific manner. This may be due to an interacting factor that is expressed in anthers. An anther-specific compound was suggested for maize T cytoplasm (39) and, indeed, our data suggest loss of RNA editing in both haploid and diploid cells.
Lack of sufficient amounts of trans-acting factors, as suggested above, may lead to reduced or complete loss of RNA editing. This hypothesis is supported by experiments in transgenic plastids. In that approach the introduction of sequences, including a known editing site into the plastid genome, led to a significant reduction of RNA editing at the endogenous editing site (37). Unfortunately, this hypothesis cannot be tested experimentally in Sorghum, because of the inability to transform plant mitochondria.
Differential display analysis is currently being used to further analyze the mechanism responsible for anther-specific loss of atp6 RNA editing in a cms line. Hopefully, cDNAs will be identified that encode proteins specifically expressed in cms or fertile lines. It is likely that the factors involved therein may also be part of or interact with the RNA editing machinery.
Acknowledgments
We are especially grateful to Dr. Daryl R. Pring (Gainesville) for kindly providing Sorghum seed stocks, atp9-specific oligonucleotides, critical discussion, and reading of the manuscript. We thank the staff of the botanical garden of the Ruhr University (Bochum), particularly Mr. K. Slowak, for cultivating Sorghum. We wish to acknowledge a collaboration to analyze plastid RNA editing in S. bicolor anthers with the late Dr. Hans Kössel, which was continued by his former coworker Dr. Rainer Maier (Freiburg), who kindly provided plastid RNA editing data. We also thank Dr. Ulrich Kück for basic support and Triticum seed stocks and Mrs. Alexandra Wakenhut for help typing the manuscript. This work was funded by Grant Ke4093/ from the Deutsche Forschungsgemeinschaft to F.K.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: atp, subunit of the F0F1-ATP synthase; crf, chimeric reading frame; mt, mitochondrial; cms, cytoplasmic male sterility; RT, reverse transcription.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Z85978).
References
- 1.Oda K, Kohchi T, Ohyama K. Biosci Biotechnol Biochem. 1992;56:132–135. doi: 10.1271/bbb.56.132. [DOI] [PubMed] [Google Scholar]
- 2.Brennicke A, Klein M, Binder S, Knoop V, Grohmann L, Malek O, Marchfelder A, Marienfeld J, Unseld M. Naturwissenshaften. 1996;83:339–346. [Google Scholar]
- 3.Bendich A J. Curr Genet. 1993;24:279–290. doi: 10.1007/BF00336777. [DOI] [PubMed] [Google Scholar]
- 4.Gray M W. Curr Opin Genet Dev. 1993;3:884–890. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- 5.Klein M, Eckert-Ossenkopp U, Schmiedeberg I, Brandt P, Unseld M, Brennicke A, Schuster W. Plant J. 1994;6:447–455. doi: 10.1046/j.1365-313x.1994.06030447.x. [DOI] [PubMed] [Google Scholar]
- 6.Palmer J D, Shields C R. Nature (London) 1984;307:437–440. [Google Scholar]
- 7.Fauron C M R, Casper M. Genetics. 1994;137:875–882. doi: 10.1093/genetics/137.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson M R. Annu Rev Genet. 1991;25:461–486. doi: 10.1146/annurev.ge.25.120191.002333. [DOI] [PubMed] [Google Scholar]
- 9.Tang H V, Pring D R, Shaw L C, Salazar R A, Muza F R, Yan B, Schertz K F. Plant J. 1996;10:123–133. doi: 10.1046/j.1365-313x.1996.10010123.x. [DOI] [PubMed] [Google Scholar]
- 10.Dewey R E, Levings C S, III, Timothy D H. Cell. 1986;44:439–449. doi: 10.1016/0092-8674(86)90465-4. [DOI] [PubMed] [Google Scholar]
- 11.Dewey R E, Timothy D H, Levings C S., III Proc Natl Acad Sci USA. 1987;84:5374–5378. doi: 10.1073/pnas.84.15.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levings C S., III Science. 1990;250:942–947. doi: 10.1126/science.250.4983.942. [DOI] [PubMed] [Google Scholar]
- 13.Gray M W. Proc Natl Acad Sci USA. 1996;93:8157–8159. doi: 10.1073/pnas.93.16.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster W, Brennicke A. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:61–78. [Google Scholar]
- 15.Ward G C, Levings C S., III Plant Mol Biol. 1991;17:1083–1088. doi: 10.1007/BF00037148. [DOI] [PubMed] [Google Scholar]
- 16.Araya A, Begu D, Graves P V, Hernould M, Litvak S, Mouras A, Suharsono S. In: Plant Mitochondria. Brennicke A, Kück U, editors. New York: VCH; 1993. pp. 171–180. [Google Scholar]
- 17.Laser B, Oettler G, Kück U. Plant Physiol. 1995;107:663–664. doi: 10.1104/pp.107.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwabuchi M, Kyozuka J, Shimamoto K. EMBO J. 1993;12:1437–1446. doi: 10.1002/j.1460-2075.1993.tb05787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernould M, Suharsono S, Litvak S, Araya A, Mouras A. Proc Natl Acad Sci USA. 1993;90:2370–2374. doi: 10.1073/pnas.90.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullen J A, Pring D R, Kempken F, Ferguson J, Chase C D. Plant Mol Biol. 1992;20:71–79. doi: 10.1007/BF00029150. [DOI] [PubMed] [Google Scholar]
- 21.Kempken F, Höfken G, Pring D R. Curr Genet. 1995;27:555–558. doi: 10.1007/BF00314447. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Pring D R, Tang H V, Schertz K F. In: Molecular Biology of the Mitochondria. Levings C S III, Vasil K, editors. Dordrecht, The Netherlands: Kluwer; 1995. pp. 461–495. [Google Scholar]
- 24.Xu G-W, Cui Y-X, Schertz K F, Hart G E. Theor Appl Genet. 1996;90:1180–1187. doi: 10.1007/BF00222941. [DOI] [PubMed] [Google Scholar]
- 25.Petersen G, Johanson B, Seberg O. Plant Mol Biol. 1996;31:189–192. doi: 10.1007/BF00020620. [DOI] [PubMed] [Google Scholar]
- 26.Bock R, Hermann M, Kössel H. EMBO J. 1996;15:5052–5059. [PMC free article] [PubMed] [Google Scholar]
- 27.Kempken F, Howad W. Curr Genet. 1996;30:186–189. doi: 10.1007/s002940050119. [DOI] [PubMed] [Google Scholar]
- 28.Bock R, Hagemann R, Kössel H, Kudla J. Mol Gen Genet. 1993;240:238–244. doi: 10.1007/BF00277062. [DOI] [PubMed] [Google Scholar]
- 29.Grosskopf D, Mulligan R M. Curr Genet. 1996;29:556–563. doi: 10.1007/BF02426960. [DOI] [PubMed] [Google Scholar]
- 30.Kempken F, Mullen J A, Pring D R, Tang H V. Curr Genet. 1991;20:417–422. doi: 10.1007/BF00317071. [DOI] [PubMed] [Google Scholar]
- 31.Smart C J, Monéger F, Leaver C J. Plant Cell. 1994;6:811–825. doi: 10.1105/tpc.6.6.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnasamy S, Makaroff C A. Plant Mol Biol. 1994;26:935–946. doi: 10.1007/BF00028860. [DOI] [PubMed] [Google Scholar]
- 33.Lu B, Hanson M R. Plant Cell. 1994;6:1955–1968. doi: 10.1105/tpc.6.12.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu B, Wilson R K, Phreaner C G, Mulligan R M, Hanson M R. Mol Cell Biol. 1996;16:1543–1549. doi: 10.1128/mcb.16.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu B, Hanson M R. Nucleic Acids Res. 1992;20:5699–5703. doi: 10.1093/nar/20.21.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson R K, Hanson M R. Curr Genet. 1996;30:502–508. doi: 10.1007/s002940050162. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhuri S, Carrer H, Maliga P. EMBO J. 1995;14:2951–2957. doi: 10.1002/j.1460-2075.1995.tb07295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bock R, Koop H U. EMBO J. 1997;16:3282–3288. doi: 10.1093/emboj/16.11.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levings C S., III Science. 1996;272:1279–1280. doi: 10.1126/science.272.5266.1279. [DOI] [PubMed] [Google Scholar]