Abstract
In skeletal muscle myosin, the reactive thiols (SH1 and SH2) are close to a proposed fulcrum region that is thought to undergo a large conformational change. The reactive thiol region is thought to transmit the conformational changes induced by the actin–myosin–ATP interactions to the lever arm, which amplifies the power stroke. In skeletal muscle myosin, SH1 and SH2 can be chemically cross-linked in the presence of nucleotide, trapping the nucleotide in its pocket. Although the flexibility of the reactive thiol region has been well studied in skeletal muscle myosin, crystal structures of truncated nonmuscle myosin II from Dictyostelium in the presence of various ATP analogs do not show changes at the reactive thiol region that would be consistent with the SH1–SH2 cross-linking observed for muscle myosin. To examine the dynamics of the reactive thiol region in Dictyostelium myosin II, we have examined a modified myosin II that has cysteines at the muscle myosin SH1 and SH2 positions. This myosin is specifically cross-linked at SH1–SH2 by a chemical cross-linker in the presence of ADP, but not in its absence. Furthermore, the cross-linked species traps the nucleotide, as in the case of muscle myosin. Thus, the Dictyostelium myosin II shares the same dynamic behavior in the fulcrum region of the molecule as the skeletal muscle myosin. This result emphasizes the importance of nucleotide-dependent changes in this part of the molecule.
Myosin II is a hexamer consisting of two identical heavy chains and two pairs of essential and regulatory light chains. The head, or subfragment 1 (S1), consists of an N-terminal catalytic domain and a C-terminal α-helix around which two light chains wrap. S1 is conventionally divided into three domain segments, amino-terminal 25-kDa, central 50-kDa, and carboxyl-terminal 20-kDa segments. The S1 domain, which gives rise to the crossbridges that connect the thick and thin filaments in muscle, is the motor domain of myosin. It moves actin filaments and generates force in vitro in the presence of ATP (1).
The swinging lever arm hypothesis assumes that a swinging motion of the light chain binding domain (neck) relative to the catalytic domain of S1 generates movement. High-resolution x-ray crystal structures (2–4), as well as other biophysical studies (5, 6) of the head domain, provide a structural basis for this model. It is proposed that a series of conformational changes, induced by movement of the lower domain of the 50-kDa segment, are transmitted by means of the fulcrum to the neck to generate a swing. This pathway is suggested to be fundamental to the conversion of chemical energy into directed movement (3). Such multiconformational changes have been proposed to be initiated from either the movement between the two 50-kDa domains (3, 4) or between the lower 50-kDa and the reactive thiol region (see below) of the 20-kDa domain (7). The fulcrum has been suggested to be in the vicinity of (5) or right at (6) the so-called reactive thiol region, which is located at the 20-kDa segment and contains two reactive cysteines (SH1 and SH2) in skeletal muscle myosin. The reactive thiol region has been thought to be a critical part of this pathway because it appears to undergo a large conformational change upon binding of ATP. Based on chemical cross-linking studies on skeletal muscle myosin, SH1 and SH2 move from farther than 18 Å apart to as close as 3 Å when ATP or ADP is present. In the absence of nucleotides, SH1 and SH2 cannot be cross-linked (8, 9).
Although the flexibility of the reactive thiol region in the presence of nucleotides has been well studied in muscle myosin, little is known about the flexibility of this region in other myosins. Recently, Dictyostelium has become widely used for myosin structure–function studies. A series of crystal structures of truncated Dictyostelium myosin II in the presence of various ATP analogs have been solved, but they do not show changes at the reactive thiol region that can account for the SH1–SH2 cross-linking in muscle myosin (3, 4). Because crystallographic studies provide static and not dynamic information, we wished to test whether the reactive thiol region in Dictyostelium myosin II showed the same flexibility as its skeletal counterpart, using a chemical cross-linking approach. To reduce the complication due to nonspecific cross-linking from other cysteines existing in the myosin head domain, we used a plasmid containing a myosin II gene from Dictyostelium in which five cysteines in the head were replaced with other residues (10). Cysteines corresponding to the desired position were then engineered into this “cysteine-light” species. Protein expressed from these plasmids was subjected to chemical cross-linking reactions.
MATERIALS AND METHODS
Plasmid Construction.
A cysteine-light myosin II expression plasmid (10), pCL, was generated from pTIKL-Myo (a Bluescript vector containing a wild-type Dictyostelium mhcA gene driven by an actin-15 promoter and a neomycin gene for G418 selection; X. Liu, K. Ito, S. Morimoto, A. Hikkoshi-Iwane, T. Yanagida & T. Q. P. Uyeda, personal communication) by overlapping-extension PCR mutagenesis (11). The wild-type myosin does not have a cysteine corresponding to the SH1 reactive thiol in the chicken counterpart. Myosin expressed from pCL contains the following five mutations: C312Y, C442S, C470I, C599L, and C678Y. Plasmids pCLSH1/SH2 and pCLSH1 were constructed as follows. One-kilobase mhcA gene fragments (corresponding to residues 476–830) containing mutations corresponding to Y678C+T688C (for pCLSH1/SH2) or T688C alone (for pCLSH1) were obtained by overlapping-extension PCR mutagenesis using pCL as a template. Purified 0.65-kb BglII/NcoI fragments from the 1-kb PCR product were then ligated back to pCL. The presence of the mutations was verified by DNA sequencing.
Growth of Dictyostelium Cells.
Dictyostelium discoideum cells were grown in HL-5 medium as described (12). Plasmids were electroporated into HS1, a myosin II null strain derived from JH10 (12). Transformants were selected and grown in the presence of 5 μg/ml G418.
Protein Purification and ATPase Assay.
Myosin II proteins were purified from Dictyostelium cells as described (13). The purified myosin was treated with myosin light-chain kinase A (14). Usually a Bio-Gel A15 (Bio-Rad) gel filtration column was used to further purify the myosin (15). The concentration of the purified myosin II was determined by the Bradford method with rabbit skeletal muscle myosin as the standard (16). Actin was purified from chicken skeletal muscle according to the method of Pardee and Spudich (17). The concentration of actin was determined by absorbance at 290 nm by using the extinction coefficient of 0.62 (mg/ml)−1⋅cm−1 (13). The molecular masses of actin and myosin II were 42 and 500 kDa, respectively. ATPase assays were performed as described (12). Typical protein concentrations were 0.1 mg/ml (0.2 μM) myosin with 0.8 mg/ml (19 μM) actin in a volume of 50 μl.
Chemical Cross-Linking.
Cross-linking of myosins was performed as described (9). Myosin was allowed to react with N,N′-p-phenylenedimaleimide (pPDM), the cross-linking reagent, for 1 hr at 0°C in the presence of ADP (1 mM) or ATP-coated agarose beads [Sigma; ATP molecules were attached at ribose hydroxyls by way of an 11-atom spacer (adipic acid dihydrazide) to the agarose beads]. Typically, 3–5 μl of the bead solution was used in each reaction. The reaction was quenched by adding DTT to a final concentration of 1 mM. Typical myosin concentrations were 4–8 μM in a volume of 50–100 μl. The typical molar ratio of pPDM to myosin was 2.3.
RESULTS
Creation of pCLSH1/SH2 and pCLSH1.
The plasmids used in this paper are as follows. pCL is the plasmid expressing cysteine-light myosin II (see Materials and Methods). Derived from pCL, pCLSH1 expresses myosin II with a cysteine at the position corresponding to SH1 in skeletal muscle myosin, and pCLSH1/SH2 expresses myosin II with cysteines at both positions corresponding to SH1 and SH2. pTIKL-Myo, serving as a control, contains a wild-type myosin II heavy chain gene. Note that wild-type Dictyostelium myosin II does not have a cysteine at the SH1 position, but does at the SH2 position. Myosin II expressed from pCLSH1/SH2 was denoted as CLSH1/SH2, and the same denotation was applied to the rest of the proteins.
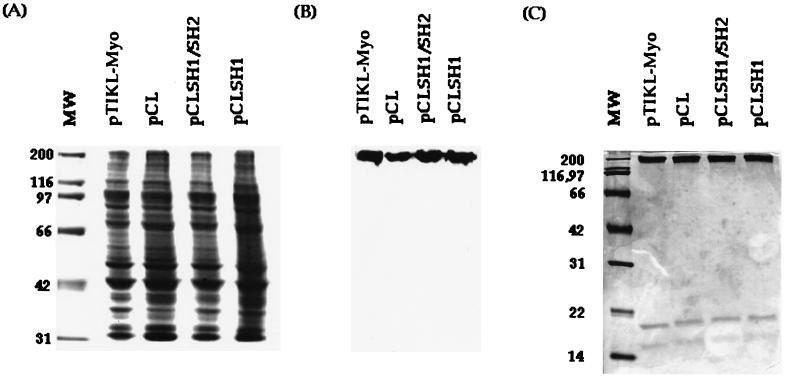
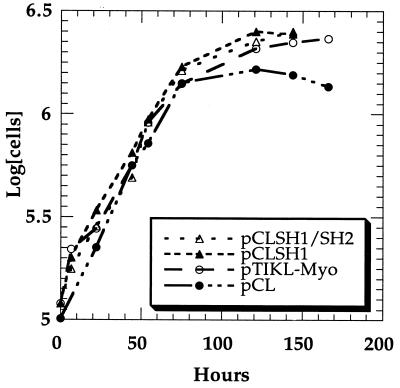
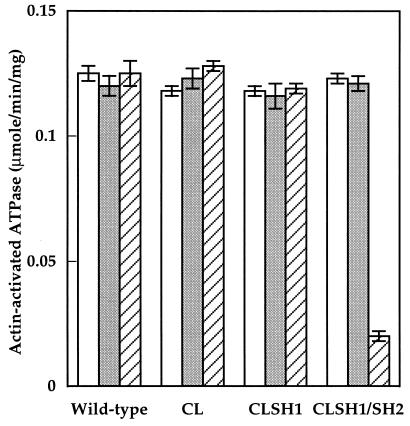
To test whether the pCL-derived myosins have defects in vivo, the pCL-derived myosin II constructs as well as pTIKL-Myo (wild-type construct) were transformed into myosin-null cells and the myosin expression levels were estimated by immunoblot (Fig. 1). The levels of expression and the yields after purification were similar in all cases. All the transformants grew in suspension with a doubling time of 16–20 hr (Fig. 2). Cells complemented with pTIKL-Myo and with pCL-derived constructs developed normally and formed sorocarps within 3–4 days at 22°C (Fig. 3). The morphologies of the fruiting bodies from pTIKL-Myo, pCLSH1, and pCLSH1/SH2 cells were basically indistinguishable. pCL cells had a higher population of smaller, slightly deformed sorocarps than the other transformants. All of the pCL-derived myosins were stable through the course of purification. No aggregation was observed, and the purified proteins had ATPase activities that were similar to the normal cysteine-containing myosin (Fig. 4).
Figure 1.
Expression of wild-type (pTIKL-Myo) and pCL-derived myosins in Dictyostelium cells. (A) Coomassie-stained gel from whole-cell lysates. (B) Immunoblot. (C) Coomassie-stained gel for purified proteins. CLSH1/SH2 and CLSH1 were expressed at approximately 95–100% of that of wild-type myosin. CL was expressed at about 85% of the wild-type level.
Figure 2.
Growth curves for pCLSH1/SH2, pCLSH1, pCL, and pTIKL-Myo (wild-type) cells. At time zero, parallel aliquots of cells growing on plates were placed into suspension culture or prepared for the Western blots shown in Fig. 1. All transformants grew in suspension with a doubling time of 16–20 hr.
Figure 3.
Development of fruiting bodies. pTIKL-Myo (wild-type) (A), pCL (B), pSH1 (C), and pSH1/SH2 (D) cells were grown and developed on a lawn of Klebsiella aerogenes for 5 days at 22°C.
Figure 4.
Actin-activation of Mg2+ ATPase activity of myosin. Rates of ATP hydrolysis were measured at 30°C and basal ATPase rates (0.02–0.04 μmol/min per mg in all cases, except for CLSH1/SH2 after cross-linking with pPDM in the presence of ADP, which consistently was >50% inhibited) were subtracted, as described previously (12). Symbols represent nontreated (open), pPDM-treated without adding ADP (shaded), and pPDM-treated in the presence of ADP (hatched) myosin. Error bars show standard deviations. Data shown were compiled from 4–10 measurements from 2–7 independent preparations.
Chemical Cross-Linking.
When skeletal muscle myosin SH1 and SH2 are chemically cross-linked with pPDM in the presence of ADP, there is a complete loss of ATPase activity, and the nucleotide is trapped in the active site (8, 9). We used the same technique to verify the extent of cross-linking using our Dictyostelium proteins (Fig. 4).
All myosins hydrolyzed ATP at a similar rate without chemical modification. The actin-activated ATPase activities for the nontreated wild-type myosins, CL, CLSH1, and CLSH1/SH2 were all about 0.12 μmol/min per mg (0.5 s−1 per head). Incubation of these proteins with pPDM in the presence of ADP at 0°C for 1 hr resulted in little change of the ATPase activity for the wild-type myosins CL and CLSH1. However, for CLSH1/SH2, the ATPase activity was inhibited by 85%.
To test whether the activity reduction of CLSH1/SH2 was simply due to the presence of cross-linking reagents, reactions were performed under the same conditions in the absence of ADP. The ATPase activity of CLSH1/SH2 was not inhibited under these conditions (Fig. 2). For the wild-type myosin, we also performed the cross-linking reactions in the presence of ADP and a 10- to 15-fold excess of pPDM to ensure modification at the SH2 position. The hydrolysis activity remained normal, with a value of 0.12 ± 0.01 μmol/min per mg (not shown).
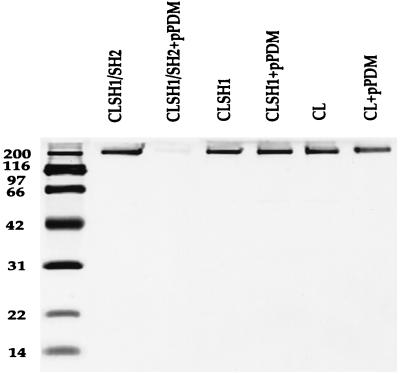
For some experiments, cross-linking reactions were performed in the presence of ATP-coated beads. The reactions were then quenched, the beads were sedimented by centrifugation, and the supernatant was subjected to gel electrophoresis. For CL and CLSH1, most of the myosin remained in the supernatant after treatment with pPDM (Fig. 5). However, the majority of the CLSH1/SH2 sedimented with the beads. Trapped myosin on the beads was detected by gel electrophoresis after denaturing the myosin-bead pellets (data not shown).
Figure 5.
Cross-linking reactions in the presence of ATP-coated beads. Beads were sedimented by centrifugation for 15 min at 100,000 × g and aliquots of the supernatants were loaded onto a gel. Only CLSH1/SH2 in the presence of cross-linking reagent pPDM was spun down with the ATP-coated beads.
DISCUSSION
With 1.3-fold molar excess of pPDM over chicken skeletal S1 protein, approximately 3–5% of the wild-type ATPase activity was reported after chemical cross-linking (8, 9). For pPDM inhibition of Dictyostelium myosin, typical residual ATPase activity was around 15%. This difference in residual activity could be explained by differences in protein structure, given that there is only 45% conservation in amino acid sequence between skeletal and Dictyostelium myosin (18). On the other hand, this difference may also arise from 15% of the Dictyostelium myosin molecules failing to cross-link because of the formation of undesired higher oxidation products of cysteine (19).
The results from the ATPase activities from the four myosins treated with pPDM and ADP show that both SH1 and SH2 are required for successful inhibition of ATPase activity, and hence confirm that SH1 and SH2 are indeed cross-linked after the reaction is performed in the presence of ADP but not in its absence. The results from the ATP-coated beads further confirm that nucleotides are trapped in the active site because of thiol cross-linking. The two reactive thiols are thought to be located in a flexible fulcrum point in the catalytic domain because they can be cross-linked in the presence of nucleotide. However, the three-dimensional structures of chicken S1 crystallized without nucleotide show that these two cysteines are separated by an α-helix and are approximately 18 Å apart. Surprisingly, the helix separating the two reactive thiols is still present in the structure of Dictyostelium myosin that contains nucleotide analogs. The presence of an intact helix between these two reactive thiols does not appear to allow close proximity for successful cross-linking. It has been suggested that the light chain binding domain is required to see the expected conformational change at the fulcrum point (3, 4). This is not likely because Dictyostelium myosin completely lacking the binding domain for both light chains is still a functional motor, i.e., able to move actin filaments and hydrolyze ATP in vitro (7). We think it is more likely that the static crystal structure represents one conformation that is preferably trapped by the crystallization process. Because the expected conformational change at the thiol fulcrum point has not been observed by x-ray crystallography, we have used dynamic chemical cross-linking as an alternative means to probe this domain movement in Dictyostelium myosin. Our results show that SH1 and SH2 in full-length Dictyostelium myosin can be indeed cross-linked in a nucleotide-dependent manner, suggesting an important role for dynamic behavior of the reactive thiol region during the ATPase cycle.
Acknowledgments
We thank Dr. Roger Cooke and Dr. Hans Warrick for comments on the manuscript. We are very grateful to William M. Shih for providing pCL and helpful discussions on the manuscript. This work was supported by National Institutes of Health Program Project Grant AR42895. W.L. was supported by a tumor training grant from the National Institutes of Health and a postdoctoral fellowship from the American Cancer Society.
ABBREVIATIONS
- S1
subfragment 1
- pPDM
N,N′-p-phenylenedimaleimide
References
- 1. Toyoshima Y, Kron J J, McNally E M, Niebling K, Toyoshima C, Spudich J A. Nature (London) 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 2.Rayment I, Holden H M, Whittaker M, Yohn C B, Lorenz M, Holmes K C, Milligan R A. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 3.Fisher A J, Smith C A, Thoden J, Smith R, Sutoh K, Holden H M, Rayment I. Biophys J. 1995;68:19s–28s. [PMC free article] [PubMed] [Google Scholar]
- 4.Smith C A, Rayment I. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 5.Holmes K C. Curr Opin Struct Biol. 1996;6:781–789. doi: 10.1016/s0959-440x(96)80008-x. [DOI] [PubMed] [Google Scholar]
- 6.Uyeda T Q P, Abramson P D, Spudich J A. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S, Ajtai K, Burghardt T P. Biochim Biophys Acta. 1996;1296:1–4. doi: 10.1016/0167-4838(96)00086-6. [DOI] [PubMed] [Google Scholar]
- 8.Wells J A, Sheldon M, Yount R G. J Biol Chem. 1980;255:1598–1602. [PubMed] [Google Scholar]
- 9.Wells J A, Yount R G. Proc Natl Acad Sci USA. 1979;76:4966–4970. doi: 10.1073/pnas.76.10.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih W M, Liang W, Spudich J A. Mol Biol Cell. 1996;7:196a. (abstr.). [Google Scholar]
- 11.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Giese K C, Spudich J A. Biochemistry. 1997;36:8465–8473. doi: 10.1021/bi963141f. [DOI] [PubMed] [Google Scholar]
- 13.Ruppel K M, Uyeda T Q-P, Spudich J A. J Biol Chem. 1994;269:18773–18780. [PubMed] [Google Scholar]
- 14.Smith J L, Silveira L A, Spudich J A. EMBO J. 1996;15:6075–6083. [PMC free article] [PubMed] [Google Scholar]
- 15.Moores S, Spudich J A. Mol Cell. 1998;1:1043–1050. doi: 10.1016/s1097-2765(00)80104-5. [DOI] [PubMed] [Google Scholar]
- 16.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Pardee J D, Spudich J A. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 18.Warrick H M, Spudich J A. Annu Rev Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- 19.Careaga C L, Sutherland J, Sabeti J, Falke J J. Biochemistry. 1995;34:3048–3055. doi: 10.1021/bi00009a036. [DOI] [PMC free article] [PubMed] [Google Scholar]