Abstract
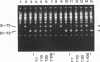
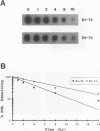
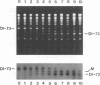
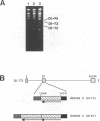
We used a protoplast system to study the mechanisms involved in the generation and evolution of defective interfering (DI) RNAs of tomato bushy stunt tombusvirus (TBSV). Synthetic transcripts corresponding to different naturally occurring TBSV DI RNAs, or to various artificially constructed TBSV defective RNAs, were analyzed. The relative levels of competitiveness of different DI RNAs were determined by coinoculating their corresponding transcripts into protoplasts along with helper genomic RNA transcripts and monitoring the level of DI RNA accumulation. Further studies were performed to assess the contribution of naked DI RNA stability and DI RNA encapsidation efficiency to the observed levels of competitiveness. In addition, the ability of various defective RNAs to evolve to alternative forms was tested by serially passaging protoplast infections initiated with transcripts corresponding to helper genomic RNA and a single type of defective RNA. These studies, and the analysis of the sequences of observed recombinants, indicate that (i) replication competence is a major factor dictating DI RNA competitiveness and is likely a primary determinant in DI RNA evolution, (ii) DI RNAs are capable of evolving to both smaller and larger forms, and the rates at which various transitions occur differ, (iii) DI RNA-DI RNA recombination and/or rearrangement is responsible for the formation of the evolved RNA molecules which were examined, and (iv) sequence complementarities between positive- and negative-sense strands in the regions of the junctions suggest that, in some cases, base pairing between an incomplete replicase-associated nascent strand and acceptor template may mediate selection of recombination sites. On the basis of our data, we propose a stepwise deletion model to describe the temporal order of events leading to the formation of tombusvirus DI RNAs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biebricher C. K., Luce R. In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J. 1992 Dec;11(13):5129–5135. doi: 10.1002/j.1460-2075.1992.tb05620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski J. J., Dzianott A. M. Generation and analysis of nonhomologous RNA-RNA recombinants in brome mosaic virus: sequence complementarities at crossover sites. J Virol. 1991 Aug;65(8):4153–4159. doi: 10.1128/jvi.65.8.4153-4159.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgyan J., Rubino L., Russo M. De novo generation of cymbidium ringspot virus defective interfering RNA. J Gen Virol. 1991 Mar;72(Pt 3):505–509. doi: 10.1099/0022-1317-72-3-505. [DOI] [PubMed] [Google Scholar]
- Cascone P. J., Carpenter C. D., Li X. H., Simon A. E. Recombination between satellite RNAs of turnip crinkle virus. EMBO J. 1990 Jun;9(6):1709–1715. doi: 10.1002/j.1460-2075.1990.tb08294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascone P. J., Haydar T. F., Simon A. E. Sequences and structures required for recombination between virus-associated RNAs. Science. 1993 May 7;260(5109):801–805. doi: 10.1126/science.8484119. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T., Macnaughton T. B., La Monica N., Lai M. M. Natural evolution of coronavirus defective-interfering RNA involves RNA recombination. Virology. 1993 May;194(1):408–413. doi: 10.1006/viro.1993.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearne P. Q., Knorr D. A., Hillman B. I., Morris T. J. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology. 1990 Jul;177(1):141–151. doi: 10.1016/0042-6822(90)90468-7. [DOI] [PubMed] [Google Scholar]
- Hillman B. I., Carrington J. C., Morris T. J. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell. 1987 Nov 6;51(3):427–433. doi: 10.1016/0092-8674(87)90638-6. [DOI] [PubMed] [Google Scholar]
- Jarvis T. C., Kirkegaard K. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 1992 Aug;11(8):3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. W., Jackson A. O., Morris T. J. Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology. 1990 Jun;176(2):539–545. doi: 10.1016/0042-6822(90)90024-l. [DOI] [PubMed] [Google Scholar]
- Kim Y. N., Lai M. M., Makino S. Generation and selection of coronavirus defective interfering RNA with large open reading frame by RNA recombination and possible editing. Virology. 1993 May;194(1):244–253. doi: 10.1006/viro.1993.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M. Preferred sites of recombination in poliovirus RNA: an analysis of 40 intertypic cross-over sequences. Nucleic Acids Res. 1988 Dec 23;16(24):11705–11723. doi: 10.1093/nar/16.24.11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr D. A., Mullin R. H., Hearne P. Q., Morris T. J. De novo generation of defective interfering RNAs of tomato bushy stunt virus by high multiplicity passage. Virology. 1991 Mar;181(1):193–202. doi: 10.1016/0042-6822(91)90484-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992 Mar;56(1):61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarini R. A., Keene J. D., Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981 Oct;26(2 Pt 2):145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- Li X. H., Heaton L. A., Morris T. J., Simon A. E. Turnip crinkle virus defective interfering RNAs intensify viral symptoms and are generated de novo. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9173–9177. doi: 10.1073/pnas.86.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ball L. A. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J Virol. 1993 Jul;67(7):3854–3860. doi: 10.1128/jvi.67.7.3854-3860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Fujioka N., Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985 May;54(2):329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Soe L. H., Baker S. C., Lai M. M. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988 Oct;166(2):550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. D., Bujarski J. J. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J Virol. 1992 Nov;66(11):6824–6828. doi: 10.1128/jvi.66.11.6824-6828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault J. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- Resende R. de O., de Haan P., van de Vossen E., de Avila A. C., Goldbach R., Peters D. Defective interfering L RNA segments of tomato spotted wilt virus retain both virus genome termini and have extensive internal deletions. J Gen Virol. 1992 Oct;73(Pt 10):2509–2516. doi: 10.1099/0022-1317-73-10-2509. [DOI] [PubMed] [Google Scholar]
- Rochon D. M., Johnston J. C. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology. 1991 Apr;181(2):656–665. doi: 10.1016/0042-6822(91)90899-m. [DOI] [PubMed] [Google Scholar]
- Rochon D. M. Rapid de novo generation of defective interfering RNA by cucumber necrosis virus mutants that do not express the 20-kDa nonstructural protein. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11153–11157. doi: 10.1073/pnas.88.24.11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon D. M., Tremaine J. H. Complete nucleotide sequence of the cucumber necrosis virus genome. Virology. 1989 Apr;169(2):251–259. doi: 10.1016/0042-6822(89)90150-5. [DOI] [PubMed] [Google Scholar]
- Romero J., Huang Q., Pogany J., Bujarski J. J. Characterization of defective interfering RNA components that increase symptom severity of broad bean mottle virus infections. Virology. 1993 Jun;194(2):576–584. doi: 10.1006/viro.1993.1297. [DOI] [PubMed] [Google Scholar]
- Roux L., Simon A. E., Holland J. J. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. A., Bancroft J. B., Mackie G. A. Coding capacity determines in vivo accumulation of a defective RNA of clover yellow mosaic virus. J Virol. 1992 May;66(5):3069–3076. doi: 10.1128/jvi.66.5.3069-3076.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. A., Bancroft J. B., Mackie G. A. Defective RNAs of clover yellow mosaic virus encode nonstructural/coat protein fusion products. Virology. 1991 Aug;183(2):479–486. doi: 10.1016/0042-6822(91)90977-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R. J., van der Most R. G., Spaan W. J. The fitness of defective interfering murine coronavirus DI-a and its derivatives is decreased by nonsense and frameshift mutations. J Virol. 1992 Oct;66(10):5898–5905. doi: 10.1128/jvi.66.10.5898-5905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]