Abstract
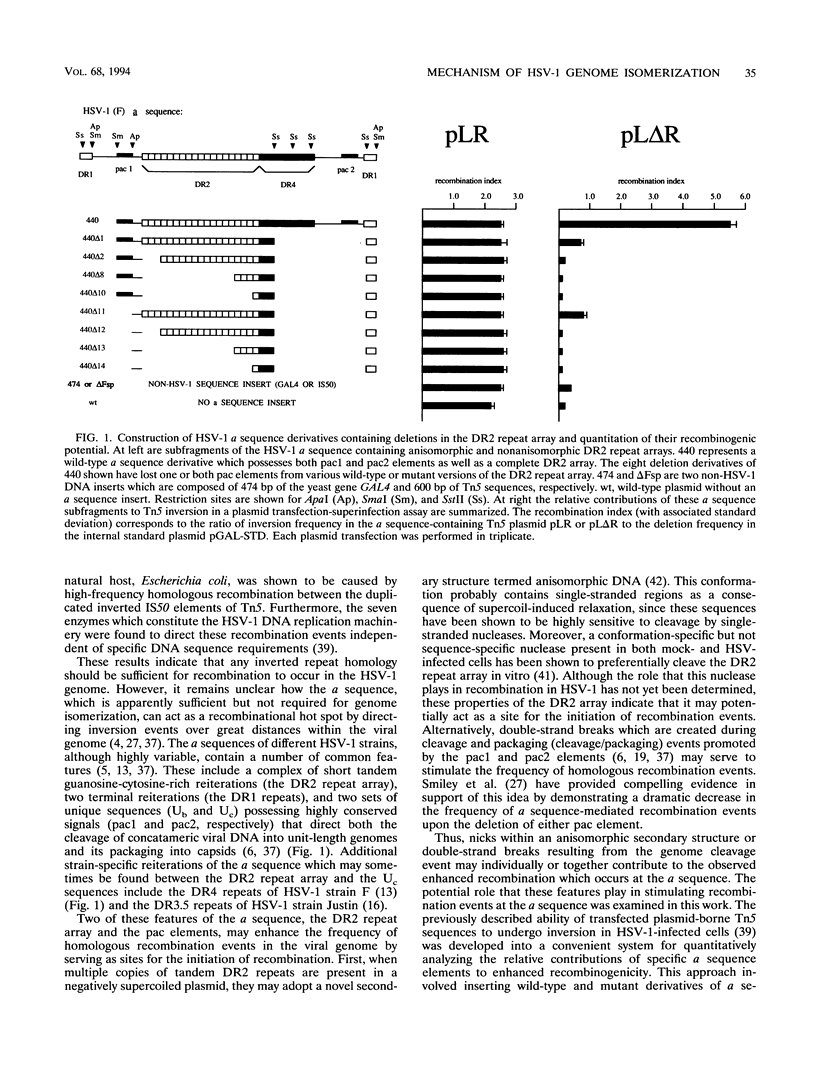
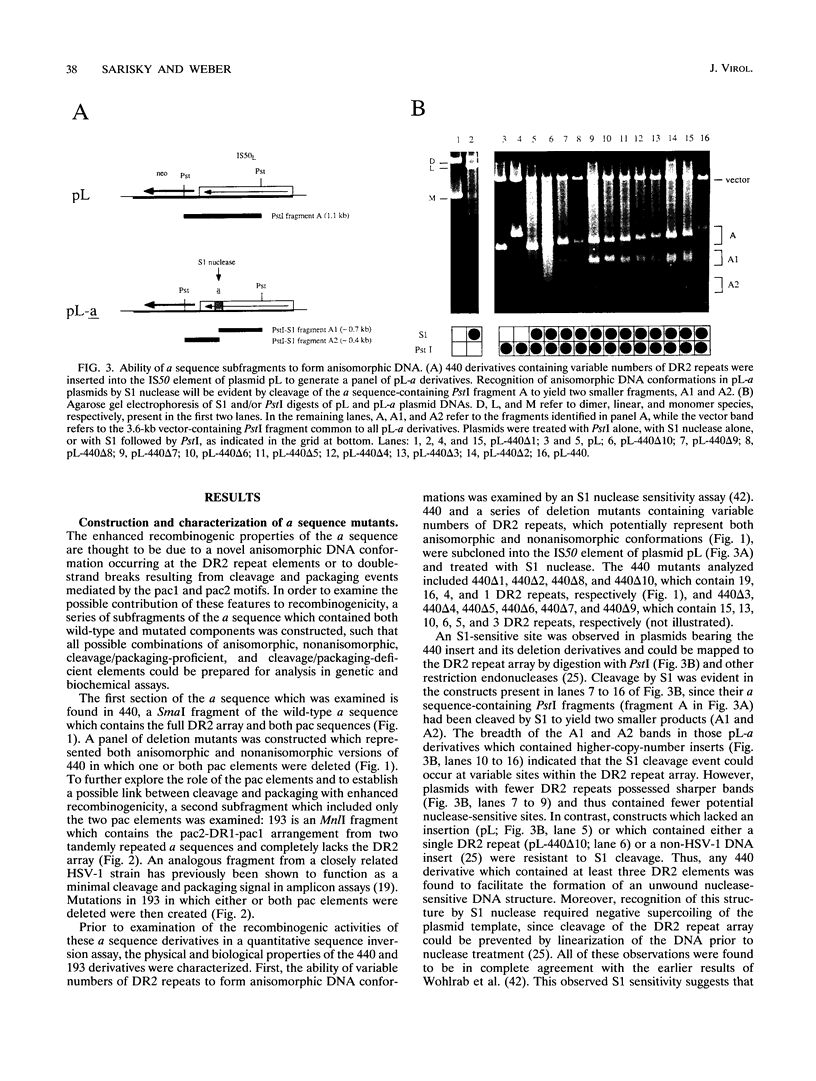
Herpes simplex virus type 1 (HSV-1) genome isomerization occurs as a result of DNA replication-mediated homologous recombination between several sets of inverted repeat sequences present in the viral DNA. The frequency with which this recombination occurs has been demonstrated to be dependent upon DNA homology length rather than specific sequences. However, the smallest of the viral inverted repeats, the alpha sequence, has been shown to function as a recombinational hot spot, leading to speculation that this sequence may represent a specific element through which genome isomerization is mediated. To investigate this apparent paradox, a quantitative transient recombination assay system was developed and used to examine the recombinogenic properties of a panel of alpha sequence mutants. This analysis revealed that the presence of both the pac1 and pac2 elements was both necessary and sufficient for the induction of high-frequency recombination events by the alpha sequence. However, it was the double-strand break promoted by pac1 and pac2 during cleavage and packaging at the alpha sequence, and not the DNA sequences of the elements themselves, which appeared to be critical for recombination. This was illustrated (i) by the inability of the same pac1 and pac2 sequences to mediate inversion events in cells infected with an HSV-1 mutant which was competent for DNA replication-dependent recombination but defective for the cleavage and packaging process and (ii) by the ability of double-strand breaks generated in non-HSV-1 DNA by an in vivo-expressed restriction endonuclease to significantly stimulate the initiation of recombination events in virus-infected cells. Thus, the alpha sequence appears to act as a hot spot for homologous recombination simply because it happens to coincide with the site of the double-strand break which is generated during the cleavage and packaging process, not because it contains discrete sequences which are required for this activity. However, it was found that this enhanced recombinogenicity disappeared when the element was flanked by regions of extensive sequence homology, particularly that of the large inverted repeats which flank the alpha sequence at its natural site in the HSV-1 genome. These findings are consistent with a model for HSV-1 genome isomerization in which recombination is initiated primarily by multiple random double-strand breaks which arise during DNA replication across the inverted repeats of the genome, rather than by a single specific break which occurs at the alpha sequence during the cleavage and packaging process.
Full text
PDF


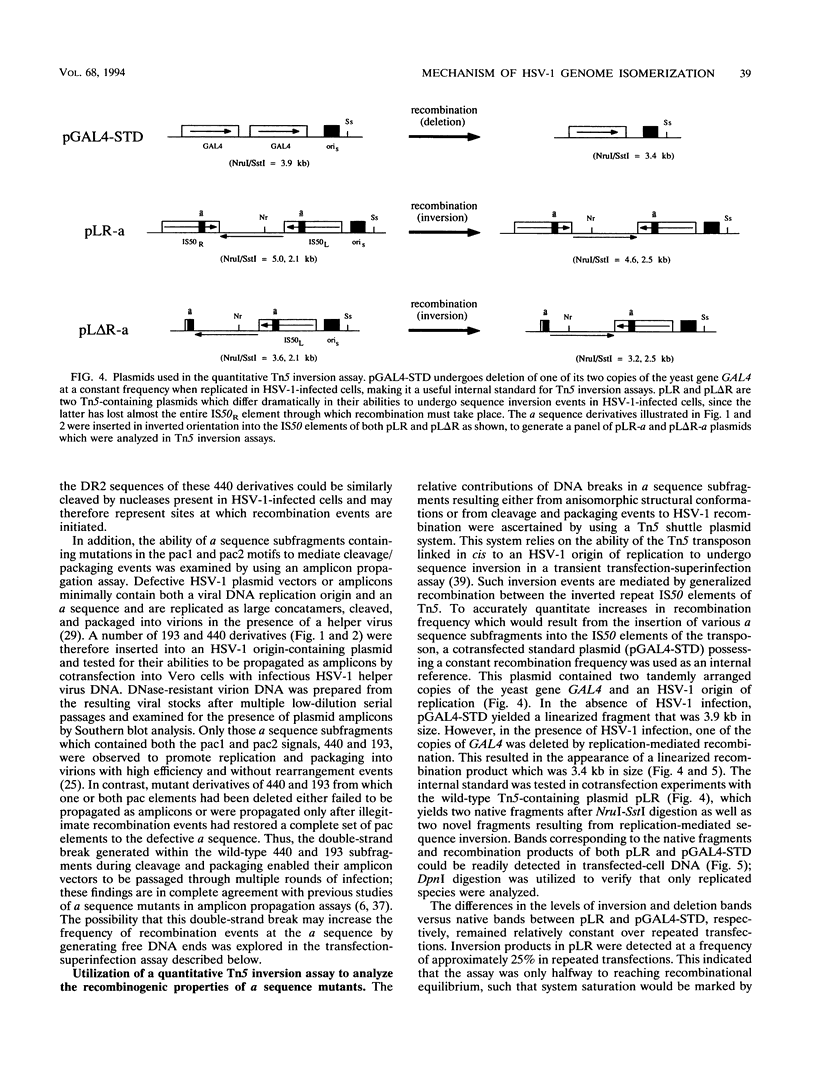




Images in this article
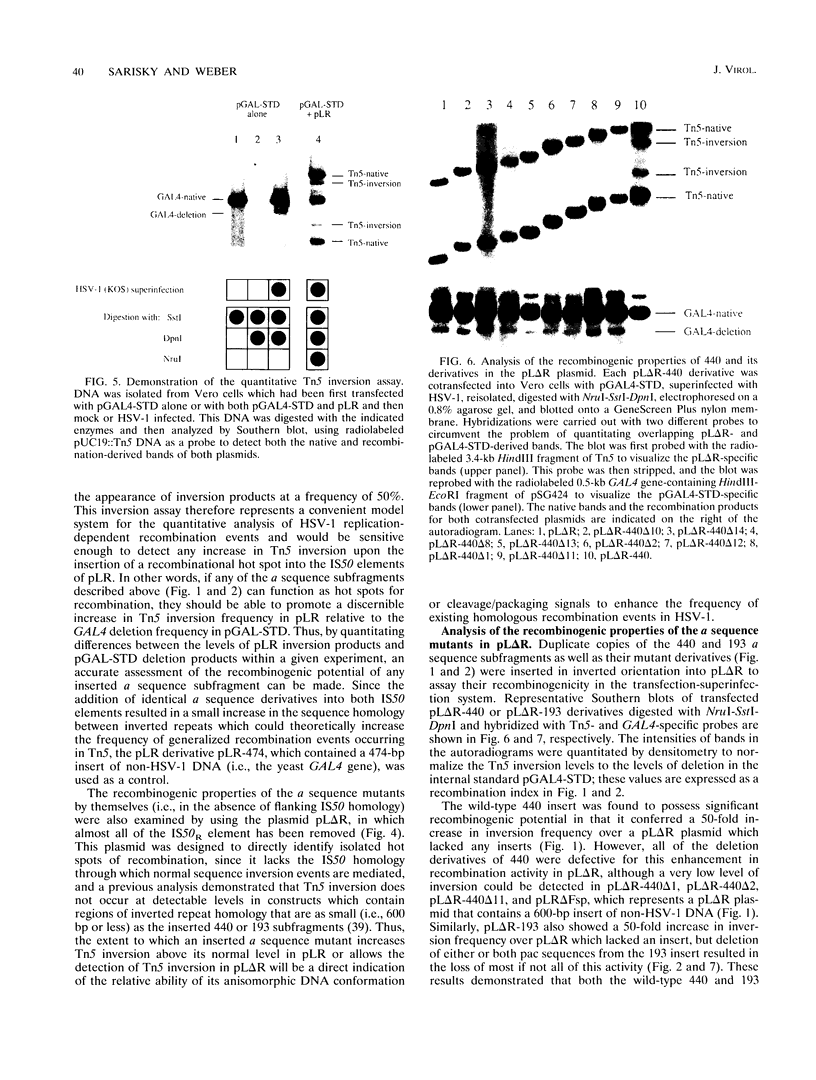
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
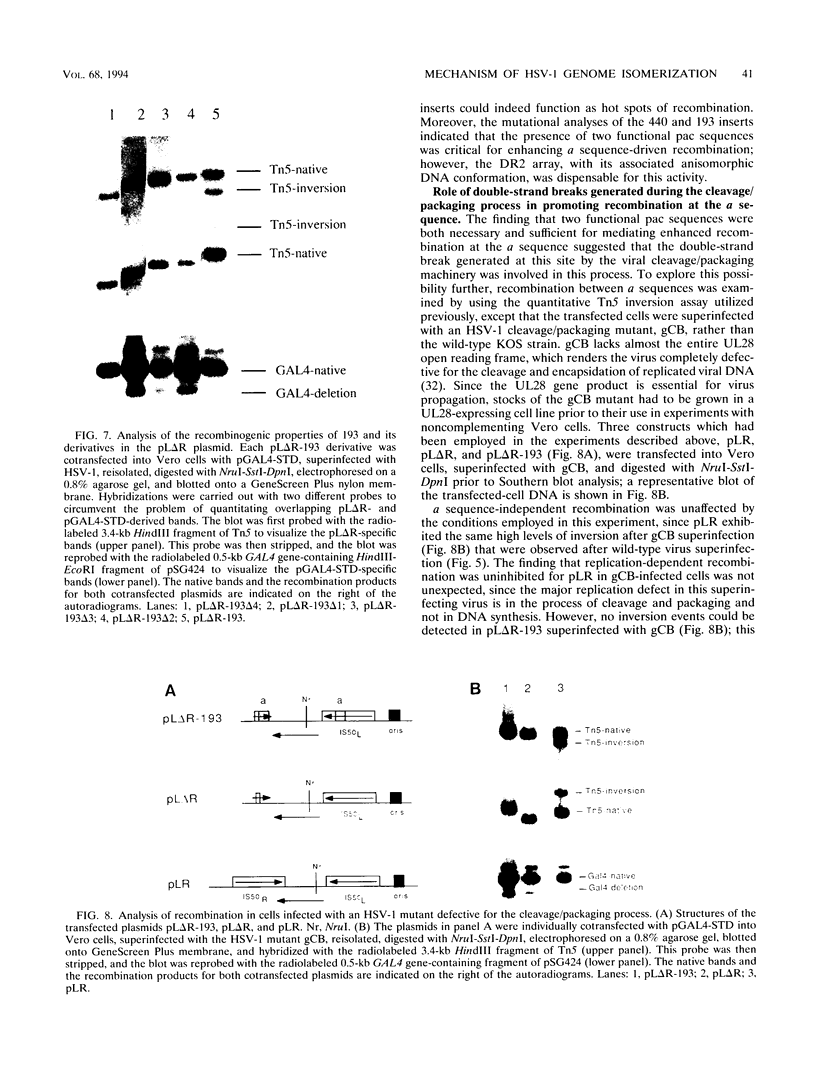
- Bortner C., Hernandez T. R., Lehman I. R., Griffith J. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J Mol Biol. 1993 May 20;231(2):241–250. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- Bruckner R. C., Dutch R. E., Zemelman B. V., Mocarski E. S., Lehman I. R. Recombination in vitro between herpes simplex virus type 1 a sequences. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10950–10954. doi: 10.1073/pnas.89.22.10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
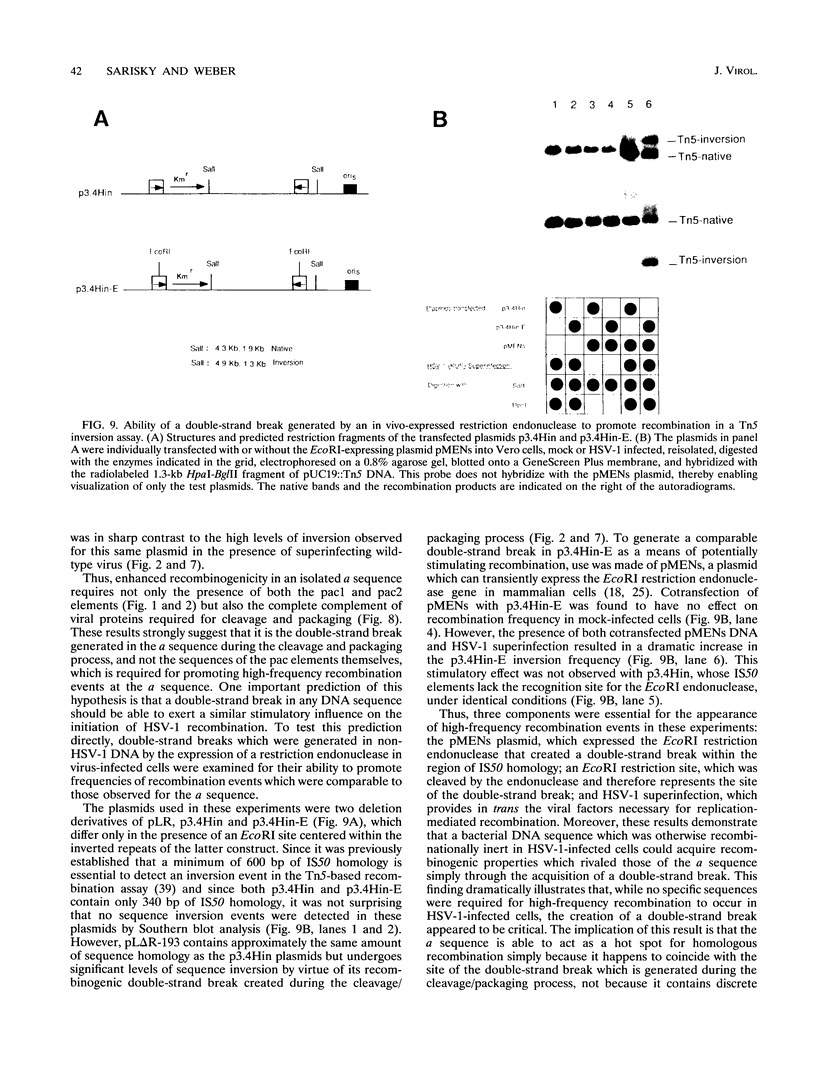
- Challberg M. D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985 Jul;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Deiss L. P., Chou J., Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986 Sep;59(3):605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Dutch R. E., Bruckner R. C., Mocarski E. S., Lehman I. R. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992 Jan;66(1):277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins F. J., Casadaban M. J., Roizman B. Application of the mini-Mu-phage for target-sequence-specific insertional mutagenesis of the herpes simplex virus genome. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4773–4777. doi: 10.1073/pnas.82.14.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins F. J., Roizman B. Herpes simplex virus 1 recombinants with noninverting genomes frozen in different isomeric arrangements are capable of independent replication. J Virol. 1986 Aug;59(2):494–499. doi: 10.1128/jvi.59.2.494-499.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J Virol. 1986 May;58(2):583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Deiss L. P., Frenkel N. Nucleotide sequence and structural features of a novel US-a junction present in a defective herpes simplex virus genome. J Virol. 1985 Jul;55(1):140–146. doi: 10.1128/jvi.55.1.140-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Morgan W. F., Fero M. L., Land M. C., Winegar R. A. Inducible expression and cytogenetic effects of the EcoRI restriction endonuclease in Chinese hamster ovary cells. Mol Cell Biol. 1988 Oct;8(10):4204–4211. doi: 10.1128/mcb.8.10.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger K. L., Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985 Feb;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger K. L., Tabares E., Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci U S A. 1983 May;80(9):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Lee G. T., Spear P. G. Novel rearrangements of herpes simplex virus DNA sequences resulting from duplication of a sequence within the unique region of the L component. J Virol. 1985 Feb;53(2):456–461. doi: 10.1128/jvi.53.2.456-461.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Spear P. G. Enhanced rate of conversion or recombination of markers within a region of unique sequence in the herpes simplex virus genome. J Virol. 1986 May;58(2):704–708. doi: 10.1128/jvi.58.2.704-708.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989 Sep 25;17(18):7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Duncan J., Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990 Oct;64(10):5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J. R., Lavery C., Howes M. The herpes simplex virus type 1 (HSV-1) a sequence serves as a cleavage/packaging signal but does not drive recombinational genome isomerization when it is inserted into the HSV-2 genome. J Virol. 1992 Dec;66(12):7505–7510. doi: 10.1128/jvi.66.12.7505-7510.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Kobayashi I., Stahl M. M. In phage lambda, cos is a recombinator in the red pathway. J Mol Biol. 1985 Jan 20;181(2):199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tengelsen L. A., Pederson N. E., Shaver P. R., Wathen M. W., Homa F. L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993 Jun;67(6):3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler D. S., Stahl M. M., Stahl F. W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage lambda. J Mol Biol. 1987 May 5;195(1):75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl M. M., Stahl F. W. Evidence that the normal route of replication-allowed Red-mediated recombination involves double-chain ends. EMBO J. 1987 Oct;6(10):3171–3176. doi: 10.1002/j.1460-2075.1987.tb02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umene K. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J Gen Virol. 1985 Dec;66(Pt 12):2659–2670. doi: 10.1099/0022-1317-66-12-2659. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Unstable heterozygosity in a diploid region of herpes simplex virus DNA. J Virol. 1984 Feb;49(2):356–362. doi: 10.1128/jvi.49.2.356-362.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Challberg M. D., Nelson N. J., Levine M., Glorioso J. C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988 Jul 29;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Levine M., Glorioso J. C. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J Virol. 1990 Jan;64(1):300–306. doi: 10.1128/jvi.64.1.300-306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlrab F., Chatterjee S., Wells R. D. The herpes simplex virus 1 segment inversion site is specifically cleaved by a virus-induced nuclear endonuclease. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6432–6436. doi: 10.1073/pnas.88.15.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlrab F., McLean M. J., Wells R. D. The segment inversion site of herpes simplex virus type 1 adopts a novel DNA structure. J Biol Chem. 1987 May 5;262(13):6407–6416. [PubMed] [Google Scholar]